Autophagy and Polyphenols in Osteoarthritis: A Focus on Epigenetic Regulation
Abstract
:1. Autophagy
2. Autophagy and Aging
3. Autophagy and Osteoarthritis
4. Autophagy as a Therapeutic Target in OA
5. Polyphenols and Autophagy
6. Polyphenols used to Regulate Autophagy in OA
7. Epigenetics and OA
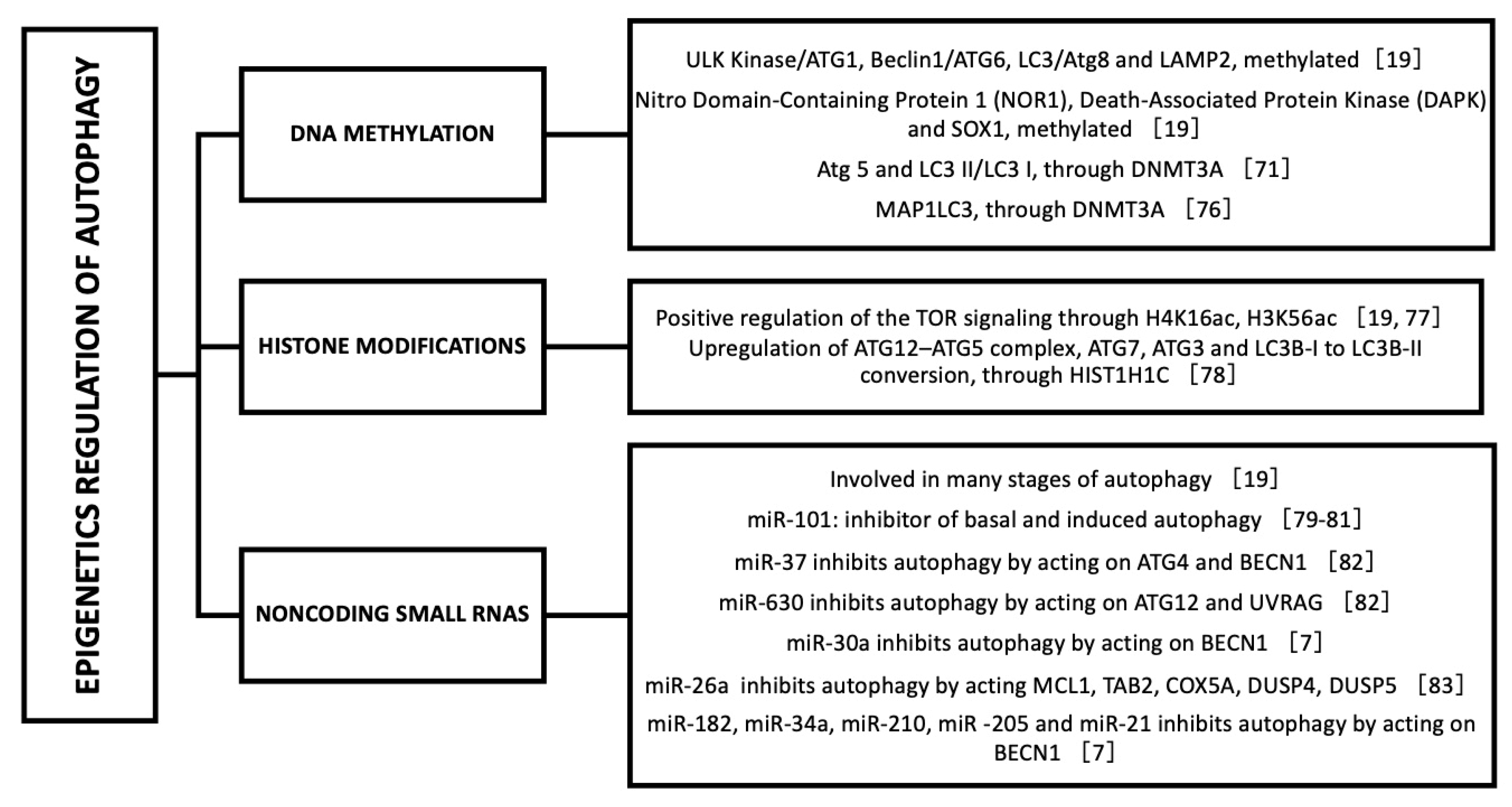
8. Epigenetic Regulation of Autophagy
9. Epigenetic Regulation of Autophagy in OA
10. Epigenetics, Polyphenols, Autophagy and OA
11. Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lotz, M.K.; Carames, B. Autophagy and cartilage homeostasis mechanisms in joint health, aging and OA. Nat. Rev. Rheumatol. 2011, 7, 579–587. [Google Scholar] [CrossRef] [Green Version]
- Zachari, M.; Ganley, I.G. The mammalian ULK1 complex and autophagy initiation. Essays Biochem. 2017, 61, 585–596. [Google Scholar] [CrossRef] [Green Version]
- Deter, R.L.; Baudhuin, P.; De Duve, C. Participation of lysosomes in cellular autophagy induced in rat liver by glucagon. J. Cell Biol. 1967, 35, C11–C16. [Google Scholar] [CrossRef] [Green Version]
- Jing, K.; Lim, K. Why is autophagy important in human diseases? Exp. Mol. Med. 2012, 44, 69–72. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.; Galluzzi, L.; Zitvogel, L.; Kroemer, G. Autophagy and Cellular Immune Responses. Immunity 2013, 39, 211–227. [Google Scholar] [CrossRef] [Green Version]
- Füllgrabe, J.; Klionsky, D.J.; Joseph, B. The return of the nucleus: Transcriptional and epigenetic control of autophagy. Nat. Rev. Mol. Cell Biol. 2013, 15, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Jing, Z.; Han, W.; Sui, X.; Xie, J.; Pan, H. Interaction of autophagy with microRNAs and their potential therapeutic implications in human cancers. Cancer Lett. 2015, 356, 332–338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nabavi, S.F.; Sureda, A.; Dehpour, A.R.; Shirooie, S.; Silva, A.S.; Devi, K.P.; Ahmed, T.; Ishaq, N.; Hashim, R.; Sobarzo-Sánchez, E.; et al. Regulation of autophagy by polyphenols: Paving the road for treatment of neurodegeneration. Biotechnol. Adv. 2018, 36, 1768–1778. [Google Scholar] [CrossRef] [PubMed]
- Carames, B.; Kiosses, W.B.; Akasaki, Y.; Brinson, D.C.; Eap, W.; Koziol, J.; Lotz, M.K. Glucosamine Activates Autophagy In Vitro and In Vivo. Arthritis Rheum. 2013, 65, 1843–1852. [Google Scholar] [CrossRef]
- Xu, H.-D.; Wu, D.; Gu, J.-H.; Ge, J.-B.; Wu, J.-C.; Han, R.; Liang, Z.-Q.; Qin, Z.-H. The Pro-Survival Role of Autophagy Depends on Bcl-2 Under Nutrition Stress Conditions. PLoS ONE 2013, 8, e63232. [Google Scholar] [CrossRef] [Green Version]
- Shintani, T.; Klionsky, D.J. Autophagy in Health and Disease: A Double-Edged Sword. Science 2004, 306, 990–995. [Google Scholar] [CrossRef] [Green Version]
- Dong, Y.; Chen, H.; Gao, J.; Liu, Y.; Li, J.; Wang, J. Molecular machinery and interplay of apoptosis and autophagy in coronary heart disease. J. Mol. Cell. Cardiol. 2019, 136, 27–41. [Google Scholar] [CrossRef] [Green Version]
- D’Arcy, M.S. Cell death: A review of the major forms of apoptosis, necrosis and autophagy. Cell Biol. Int. 2019, 43, 582–592. [Google Scholar] [CrossRef]
- Musumeci, G.; Castrogiovanni, P.; Trovato, F.M.; Weinberg, A.M.; Al-Wasiyah, M.K.; Alqahtani, M.H.; Mobasheri, A. Biomarkers of Chondrocyte Apoptosis and Autophagy in Osteoarthritis. Int. J. Mol. Sci. 2015, 16, 20560–20575. [Google Scholar] [CrossRef] [Green Version]
- Galluzzi, L.; Green, D.R. Autophagy-Independent Functions of the Autophagy Machinery. Cell 2019, 177, 1682–1699. [Google Scholar] [CrossRef]
- Yu, L.; Chen, Y.; Tooze, S.A. Autophagy pathway: Cellular and molecular mechanisms. Autophagy 2018, 14, 207–215. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.; Luo, Y.; Zou, J.; Ouyang, J.; Cai, Z.; Zeng, X.; Ling, H.; Zeng, T. Autophagy and its role in gastric cancer. Clin. Chim. Acta 2019, 489, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Legendre, F.; Ollitrault, D.; Hervieu, M.; Baugé, C.; Maneix, L.; Goux, D.; Chajra, H.; Mallein-Gerin, F.; Boumediene, K.; Galera, P.; et al. Enhanced Hyaline Cartilage Matrix Synthesis in Collagen Sponge Scaffolds by Using siRNA to Stabilize Chondrocytes Phenotype Cultured with Bone Morphogenetic Protein-2 Under Hypoxia. Tissue Eng. Part C Methods 2013, 19, 550–567. [Google Scholar] [CrossRef] [PubMed]
- Zheng-Hong, Q. Autophagy: Biology and Diseases; Springer: Berlin/Heidelberg, Germany, 2019; Volume 1206. [Google Scholar]
- Catherine, B. Regulation and Role of TGFβ Signaling Pathway in Aging and Osteoarthritis Joints. Aging Dis. 2014, 5, 394–405. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zhi, T.; Chao, H.; Jiang, K.; Liu, Y.; Bao, Z.; Fan, L.; Wang, D.; Li, Z.; Liu, N.; et al. ERK1/2/mTOR/Stat3 pathway-mediated autophagy alleviates traumatic brain injury-induced acute lung injury. Biochim. et Biophys. Acta (BBA) Mol. Basis Dis. 2018, 1864, 1663–1674. [Google Scholar] [CrossRef]
- Mobasheri, A.; Matta, C.; Zákány, R.; Musumeci, G. Chondrosenescence: Definition, hallmarks and potential role in the pathogenesis of osteoarthritis. Maturitas 2015, 80, 237–244. [Google Scholar] [CrossRef] [Green Version]
- Nikniaz, Z.; Ostadrahimi, A.; Mahdavi, R.; Ebrahimi, A.A.; Nikniaz, L. Effects of Elaeagnus angustifolia L. supplementation on serum levels of inflammatory cytokines and matrix metalloproteinases in females with knee osteoarthritis. Complement. Ther. Med. 2014, 22, 864–869. [Google Scholar] [CrossRef]
- Wang, J.; Li, J.; Cao, N.; Li, Z.; Han, J.; Li, L. Resveratrol, an activator of SIRT1, induces protective autophagy in non-small-cell lung cancer via inhibiting Akt/mTOR and activating p38-MAPK. OncoTargets Ther. 2018, 11, 7777–7786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loeser, R.F.; Collins, J.A.; Diekman, B.O. Ageing and the pathogenesis of osteoarthritis. Nat. Rev. Rheumatol. 2016, 12, 412–420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yi, W.; Wen, Y.; Tan, F.; Liu, X.; Lan, H.; Ye, H.; Liu, B. Impact of NF-κB pathway on the apoptosis-inflammation-autophagy crosstalk in human degenerative nucleus pulposus cells. Aging 2019, 11, 7294–7306. [Google Scholar] [CrossRef] [PubMed]
- Maiese, K. Moving to the Rhythm with Clock (Circadian) Genes, Autophagy, mTOR, and SIRT1 in Degenerative Disease and Cancer. Curr. Neurovascular Res. 2017, 14, 299–304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pallauf, K.; Rimbach, G. Autophagy, polyphenols and healthy ageing. Ageing Res. Rev. 2013, 12, 237–252. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Yang, Y.-H.; Jiang, S.-D.; Jiang, L.-S. Decreased activity of osteocyte autophagy with aging may contribute to the bone loss in senile population. Histochem. Cell Biol. 2014, 142, 285–295. [Google Scholar] [CrossRef]
- Zhao, G.; Zhang, W.; Li, L.; Wu, S.; Du, G. Pinocembrin Protects the Brain against Ischemia-Reperfusion Injury and Reverses the Autophagy Dysfunction in the Penumbra Area. Molecules 2014, 19, 15786–15798. [Google Scholar] [CrossRef] [Green Version]
- Matsuzaki, T.; Alvarez-Garcia, O.; Mokuda, S.; Nagira, K.; Olmer, M.; Gamini, R.; Miyata, K.; Akasaki, Y.; Su, A.I.; Asahara, H.; et al. FoxO transcription factors modulate autophagy and proteoglycan 4 in cartilage homeostasis and osteoarthritis. Sci. Transl. Med. 2018, 10. [Google Scholar] [CrossRef] [Green Version]
- Lapierre, L.; Kumsta, C.; Sandri, M.; Ballabio, A.; Hansen, M. Transcriptional and epigenetic regulation of autophagy in aging. Autophagy 2015, 11, 867–880. [Google Scholar] [CrossRef] [Green Version]
- Carames, B.; Olmer, M.; Kiosses, W.B.; Lotz, M.K. The Relationship of Autophagy Defects to Cartilage Damage During Joint Aging in a Mouse Model. Arthritis Rheumatol. 2015, 67, 1568–1576. [Google Scholar] [CrossRef]
- Weng, T.; Xie, Y.; Yi, L.; Huang, J.; Luo, F.; Du, X.; Chen, L.; Liu, C.; Chen, D. Loss of Vhl in cartilage accelerated the progression of age-associated and surgically induced murine osteoarthritis. Osteoarthr. Cartil. 2014, 22, 1197–1205. [Google Scholar] [CrossRef] [Green Version]
- Van der Kraan, P. Osteoarthritis year 2012 in review: Biology. Osteoarthr. Cartil. 2012, 20, 1447–1450. [Google Scholar] [CrossRef] [Green Version]
- Mobasheri, A.; Kalamegam, G.; Musumeci, G.; Batt, M.E. Chondrocyte and mesenchymal stem cell-based therapies for cartilage repair in osteoarthritis and related orthopaedic conditions. Maturitas 2014, 78, 188–198. [Google Scholar] [CrossRef] [PubMed]
- Hunter, D.J.; Bierma-Zeinstra, S. Osteoarthritis. Lancet 2019, 393, 1745–1759. [Google Scholar] [CrossRef]
- Loeser, R.F. Aging processes and the development of osteoarthritis. Curr. Opin. Rheumatol. 2013, 25, 108–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Musumeci, G.; Castrogiovanni, P.; Trovato, F.M.; Imbesi, R.; Giunta, S.; Szychlinska, M.A.; Loreto, C.; Castorina, S.; Mobasheri, A. Physical activity ameliorates cartilage degeneration in a rat model of aging: A study on lubricin expression. Scand. J. Med. Sci. Sports 2015, 25, e222–e230. [Google Scholar] [CrossRef] [PubMed]
- De Figueroa, P.L.; Lotz, M.K.; Blanco, F.J.; Caramés, B. Autophagy Activation and Protection From Mitochondrial Dysfunction in Human Chondrocytes. Arthritis Rheumatol. 2015, 67, 966–976. [Google Scholar] [CrossRef]
- Sasaki, H.; Takayama, K.; Matsushita, T.; Ishida, K.; Kubo, S.; Matsumoto, T.; Fujita, N.; Oka, S.; Kurosaka, M.; Kuroda, R. Autophagy modulates osteoarthritis-related gene expression in human chondrocytes. Arthritis Rheum. 2011, 64, 1920–1928. [Google Scholar] [CrossRef]
- Duan, R.; Xie, H.; Liu, Z.-Z. The Role of Autophagy in Osteoarthritis. Front. Cell Dev. Biol. 2020, 8, 8. [Google Scholar] [CrossRef]
- Li, Y.; Wu, Y.; Jiang, K.; Han, W.; Zhang, J.; Xie, L.; Liu, Y.; Xiao, J.; Wang, X. Mangiferin Prevents TBHP-Induced Apoptosis and ECM Degradation in Mouse Osteoarthritic Chondrocytes via Restoring Autophagy and Ameliorates Murine Osteoarthritis. Oxidative Med. Cell. Longev. 2019, 2019, 8783197-17. [Google Scholar] [CrossRef]
- Arias, C.; Saavedra, N.; Saavedra, K.; Alvear, M.; Cuevas, A.; Maria-Engler, S.S.; Abdalla, D.S.P.; Salazar, L.A. Propolis Reduces the Expression of Autophagy-Related Proteins in Chondrocytes under Interleukin-1β Stimulus. Int. J. Mol. Sci. 2019, 20, 3768. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Vasheghani, F.; Li, Y.-H.; Blati, M.; Simeone, K.; Fahmi, H.; Lussier, B.; Roughley, P.; Lagares, D.; Pelletier, J.-P.; et al. Cartilage-specific deletion of mTOR upregulates autophagy and protects mice from osteoarthritis. Ann. Rheum. Dis. 2015, 74, 1432–1440. [Google Scholar] [CrossRef]
- Abramoff, B.; Caldera, F.E. Osteoarthritis: Pathology, diagnosis, and treatment options. Med. Clin. N. Am. 2020, 104, 293–311. [Google Scholar] [CrossRef]
- Skou, S.T.; Roos, E.M. Physical therapy for patients with knee and hip osteoarthritis: Supervised, active treatment is current best practice. Clin. Exp. Rheumatol. 2019, 37, 112–117. [Google Scholar] [PubMed]
- Wu, T.-T.; Li, W.-M.; Yao, Y.-M. Interactions between Autophagy and Inhibitory Cytokines. Int. J. Biol. Sci. 2016, 12, 884–897. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Akhtar, N.; Haqqi, T.M. Green tea polyphenol epigallocatechi3-gallate: Inflammation and arthritis. Life Sci. 2010, 86, 907–918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopez, H.L. Nutritional Interventions to Prevent and Treat Osteoarthritis. Part II: Focus on Micronutrients and Supportive Nutraceuticals. PM&R 2012, 4, S155–S168. [Google Scholar] [CrossRef]
- Jia, G.; Cheng, G.; Gangahar, D.M.; Agrawal, D.K. Insulin-like growth factor-1 and TNF-α regulate autophagy through c- jun N-terminal kinase and Akt pathways in human atherosclerotic vascular smooth cells. Immunol. Cell Biol. 2006, 84, 448–454. [Google Scholar] [CrossRef]
- Shukla, S.; Meeran, S.M.; Katiyar, S.K. Epigenetic regulation by selected dietary phytochemicals in cancer chemoprevention. Cancer Lett. 2014, 355, 9–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imagawa, K.; De Andrés, M.C.; Hashimoto, K.; Pitt, D.; Itoi, E.; Goldring, M.B.; Roach, H.I.; Oreffo, R.O. The epigenetic effect of glucosamine and a nuclear factor-kappa B (NF-kB) inhibitor on primary human chondrocytes–implications for osteoarthritis. Biochem. Biophys. Res. Commun. 2011, 405, 362–367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, G.; Long, H.; Ma, S.; Shunhan, Y.; Li, J.; Yao, J. miRNA-335-5p relieves chondrocyte inflammation by activating autophagy in osteoarthritis. Life Sci. 2019, 226, 164–172. [Google Scholar] [CrossRef]
- Lian, W.-S.; Ko, J.-Y.; Wu, R.-W.; Sun, Y.-C.; Chen, Y.-S.; Wu, S.-L.; Weng, L.-H.; Jahr, H.; Wang, F.-S. MicroRNA-128a represses chondrocyte autophagy and exacerbates knee osteoarthritis by disrupting Atg12. Cell Death Dis. 2018, 9, 919. [Google Scholar] [CrossRef]
- Bannuru, R.R.; Osani, M.C.; Vaysbrot, E.E.; Arden, N.K.; Bennell, K.; Bierma-Zeinstra, S.M.A.; Kraus, V.B.; Lohmander, L.S.; Abbott, J.H.; Bhandari, M.; et al. OARSI guidelines for the non-surgical management of knee, hip, and polyarticular osteoarthritis. Osteoarthr. Cartil. 2019, 27, 1578–1589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, M.-H.; Lai, C.-S.; Wu, J.-C.; Ho, C.-T. Epigenetic and Disease Targets by Polyphenols. Curr. Pharm. Des. 2013, 19, 6156–6185. [Google Scholar] [CrossRef] [PubMed]
- Joven, J.; Micol, V.; Segura-Carretero, A.; Alonso-Villaverde, C.; Menéndez, J.A.; Aragonès, G.; Barrajón-Catalán, E.; Beltrán-Debón, R.; Camps, J.; Cufí, S.; et al. Polyphenols and the Modulation of Gene Expression Pathways: Can We Eat Our Way Out of the Danger of Chronic Disease? Crit. Rev. Food Sci. Nutr. 2014, 54, 985–1001. [Google Scholar] [CrossRef] [PubMed]
- Yessenkyzy, A.; Saliev, T.; Zhanaliyeva, M.; Masoud, A.-R.; Umbayev, B.; Sergazy, S.; Krivykh, E.; Gulyayev, A.; Nurgozhin, T. Polyphenols as Caloric-Restriction Mimetics and Autophagy Inducers in Aging Research. Nutrients 2020, 12, 1344. [Google Scholar] [CrossRef]
- Shabalala, S.; Muller, C.; Louw, J.; Johnson, R. Polyphenols, autophagy and doxorubicin-induced cardiotoxicity. Life Sci. 2017, 180, 160–170. [Google Scholar] [CrossRef]
- Zhou, H.; Chen, Y.; Huang, S.-W.; Hu, P.-F.; Tang, L.-J. Regulation of autophagy by tea polyphenols in diabetic cardiomyopathy. J. Zhejiang Univ. Sci. B 2018, 19, 333–341. [Google Scholar] [CrossRef]
- Sarubbo, F.; Moranta, D.; Asensio, V.J.; Miralles, A.; Esteban, S. Effects of Resveratrol and Other Polyphenols on the Most Common Brain Age-Related Diseases. Curr. Med. Chem. 2017, 24, 4245–4266. [Google Scholar] [CrossRef]
- Qin, N.; Wei, L.; Li, W.; Yang, W.; Cai, L.; Qian, Z.; Wu, S. Local intra-articular injection of resveratrol delays cartilage degeneration in C57BL/6 mice by inducing autophagy via AMPK/mTOR pathway. J. Pharmacol. Sci. 2017, 134, 166–174. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-H.; Zhu, L.; Hong, X.; Wang, Y.-T.; Wang, F.; Bao, J.-P.; Xie, X.-H.; Liu, L.; Wu, X.-T. Resveratrol attenuated TNF-α–induced MMP-3 expression in human nucleus pulposus cells by activating autophagy via AMPK/SIRT1 signaling pathway. Exp. Biol. Med. 2016, 241, 848–853. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Zhang, Q.; Song, L. Resveratrol enhances matrix biosynthesis of nucleus pulposus cells through activating autophagy via the PI3K/Akt pathway under oxidative damage. Biosci. Rep. 2018, 38. [Google Scholar] [CrossRef] [Green Version]
- Ansari, M.; Ahmad, N.; Haqqi, T.M. Butein Activates Autophagy Through AMPK/TSC2/ULK1/mTOR Pathway to Inhibit IL-6 Expression in IL-1β Stimulated Human Chondrocytes. Cell. Physiol. Biochem. 2018, 49, 932–946. [Google Scholar] [CrossRef]
- Chin, K.-Y.; Pang, K.-L. Therapeutic Effects of Olive and Its Derivatives on Osteoarthritis: From Bench to Bedside. Nutrients 2017, 9, 1060. [Google Scholar] [CrossRef] [PubMed]
- Bao, J.; Chen, Z.; Xu, L.; Wu, L.; Xiong, Y. Rapamycin protects chondrocytes against IL-18-induced apoptosis and ameliorates rat osteoarthritis. Aging 2020, 12, 5152–5167. [Google Scholar] [CrossRef] [PubMed]
- Moazedi-Fuerst, F.C.; Hofner, M.; Gruber, G.; Weinhaeusel, A.; Stradner, M.; Angerer, H.; Peischler, D.; Lohberger, B.; Glehr, M.; Leithner, A.; et al. Epigenetic differences in human cartilage between mild and severe OA. J. Orthop. Res. 2014, 32, 1636–1645. [Google Scholar] [CrossRef]
- Barter, M.J.; Young, D. Epigenetic Mechanisms and Non-coding RNAs in Osteoarthritis. Curr. Rheumatol. Rep. 2013, 15, 1–9. [Google Scholar] [CrossRef]
- Visconti, V.; Cariati, I.; Fittipaldi, S.; Iundusi, R.; Gasbarra, E.; Tarantino, U.; Botta, A. DNA Methylation Signatures of Bone Metabolism in Osteoporosis and Osteoarthritis Aging-Related Diseases: An Updated Review. Int. J. Mol. Sci. 2021, 22, 4244. [Google Scholar] [CrossRef] [PubMed]
- Fathollahi, A.; Aslani, S.; Jamshidi, A.; Mahmoudi, M. Epigenetics in osteoarthritis: Novel spotlight. J. Cell. Physiol. 2019, 234, 12309–12324. [Google Scholar] [CrossRef]
- Carthew, R.W.; Sontheimer, E.J. Origins and Mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642–655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gurău, F.; Baldoni, S.; Prattichizzo, F.; Espinosa, E.; Amenta, F.; Procopio, A.D.; Albertini, M.C.; Bonafè, M.; Olivieri, F. Anti-senescence compounds: A potential nutraceutical approach to healthy aging. Ageing Res. Rev. 2018, 46, 14–31. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Gong, L.; Zhang, P.; Li, Y.; Liu, B.; Zhang, L.; Zhuang, J.; Xiao, D. Epigenetic Down-Regulation of Sirt 1 via DNA Methylation and Oxidative Stress Signaling Contributes to the Gestational Diabetes Mellitus-Induced Fetal Programming of Heart Ischemia-Sensitive Phenotype in Late Life. Int. J. Biol. Sci. 2019, 15, 1240–1251. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.S.; Shin, H.R.; Kim, D.; Baek, S.A.; Choi, S.A.; Ahn, H.; Shamim, A.; Kim, J.; Kim, I.S.; Kim, K.K.; et al. Pontin arginine methylation by CARM1 is crucial for epigenetic regulation of autophagy. Nat. Commun. 2020, 11, 6297. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Wu, Z.; Xu, L.; Xu, K.; Chen, Z.; Ran, J.; Wu, L. The role of SIRT3-mediated mitochondrial homeostasis in osteoarthritis. Cell. Mol. Life Sci. 2020, 77, 3729–3743. [Google Scholar] [CrossRef]
- González-Rodríguez, P.; Cheray, M.; Füllgrabe, J.; Salli, M.; Engskog-Vlachos, P.; Keane, L.; Cunha, V.; Lupa, A.; Li, W.; Ma, Q.; et al. The DNA methyltransferase DNMT3A contributes to autophagy long-term memory. Autophagy 2020, 17, 1259–1277. [Google Scholar] [CrossRef]
- Füllgrabe, J.; Lynch-Day, M.A.; Heldring, N.; Li, W.; Struijk, R.B.; Ma, Q.; Hermanson, O.; Rosenfeld, M.G.; Klionsky, D.J.; Joseph, B. The histone H4 lysine 16 acetyltransferase hMOF regulates the outcome of autophagy. Nat. Cell Biol. 2013, 500, 468–471. [Google Scholar] [CrossRef]
- Wang, W.; Wang, Q.; Wan, D.; Sun, Y.; Wang, L.; Chen, H.; Liu, C.; Petersen, R.B.; Li, J.; Xue, W.; et al. Histone HIST1H1C/H1.2 regulates autophagy in the development of diabetic retinopathy. Autophagy 2017, 13, 941–954. [Google Scholar] [CrossRef]
- Weilner, S.; Grillari-Voglauer, R.; Redl, H.; Grillari, R.; Nau, T. The role of microRNAs in cellular senescence and age-related conditions of cartilage and bone: A review. Acta Orthop. 2014, 86, 92–99. [Google Scholar] [CrossRef]
- Cecconi, F. Autophagy regulation by miRNAs: When cleaning goes out of service. EMBO J. 2011, 30, 4517–4519. [Google Scholar] [CrossRef] [Green Version]
- Frankel, L.B.; Wen, J.; Lees, M.; Høyer-Hansen, M.; Farkas, T.; Krogh, A.; Jäättelä, M.; Lund, A.H. microRNA-101 is a potent inhibitor of autophagy. EMBO J. 2011, 30, 4628–4641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turei, D.; Földvári-Nagy, L.; Fazekas, D.; Modos, D.; Kubisch, J.; Kadlecsik, T.; Demeter, A.; Lenti, K.F.-N.L.; Csermely, P.; Vellai, T.; et al. Autophagy Regulatory Network—A systems-level bioinformatics resource for studying the mechanism and regulation of autophagy. Autophagy 2015, 11, 155–165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, W.; Fu, X.; Xie, J.; Meng, Z.; Gu, Y.; Wang, X.; Li, L.; Pan, H.; Huang, W. miR-26a enhances autophagy to protect against ethanol-induced acute liver injury. J. Mol. Med. 2015, 93, 1045–1055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, X.; Han, L.; Sun, T.; Zhang, F.; Ji, S.; Zhang, M.; Wang, X.; Yang, W. Silencing UHRF1 enhances cell autophagy to prevent articular chondrocytes from apoptosis in osteoarthritis through PI3K/AKT/mTOR signaling pathway. Biochem. Biophys. Res. Commun. 2020, 529, 1018–1024. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Liu, Y.; Xu, B.; Zhang, Y.; Yang, M. Noncoding RNAs: New regulatory code in chondrocyte apoptosis and autophagy. Wiley Interdiscip. Rev. RNA 2020, 11, e1584. [Google Scholar] [CrossRef]
- Cai, C.; Min, S.; Yan, B.; Liu, W.; Yang, X.; Li, L.; Wang, T.; Jin, A. MiR-27a promotes the autophagy and apoptosis of IL-1β treated-articular chondrocytes in osteoarthritis through PI3K/AKT/mTOR signaling. Aging 2019, 11, 6371–6384. [Google Scholar] [CrossRef]
- Li, H.; Li, Z.; Pi, Y.; Chen, Y.; Mei, L.; Luo, Y.; Xie, J.; Mao, X. MicroRNA-375 exacerbates knee osteoarthritis through repressing chondrocyte autophagy by targeting ATG2B. Aging 2020, 12, 7248–7261. [Google Scholar] [CrossRef]
- Wang, Z.; Hu, J.; Pan, Y.; Shan, Y.; Jiang, L.; Qi, X.; Jia, L. miR-140-5p/miR-149 Affects Chondrocyte Proliferation, Apoptosis, and Autophagy by Targeting FUT1 in Osteoarthritis. Inflammation 2017, 41, 959–971. [Google Scholar] [CrossRef]
- Li, H.; Miao, D.; Zhu, Q.; Huang, J.; Lu, G.; Xu, W. MicroRNA-17-5p contributes to osteoarthritis progression by binding p62/SQSTM1. Exp. Ther. Med. 2017, 15, 1789–1794. [Google Scholar] [CrossRef] [Green Version]
- He, W.; Cheng, Y. Inhibition of miR-20 promotes proliferation and autophagy in articular chondrocytes by PI3K/AKT/mTOR signaling pathway. Biomed. Pharmacother. 2018, 97, 607–615. [Google Scholar] [CrossRef]
- Yang, F.; Huang, R.; Ma, H.; Zhao, X.; Wang, G. miRNA-411 Regulates Chondrocyte Autophagy in Osteoarthritis by Targeting Hypoxia-Inducible Factor 1 alpha (HIF-1α). Med. Sci. Monit. 2020, 26, e921155-1. [Google Scholar] [CrossRef] [PubMed]
- Ayissi, V.B.O.; Ebrahimi, A.; Schluesenner, H. Epigenetic effects of natural polyphenols: A focus on SIRT1-mediated mechanisms. Mol. Nutr. Food Res. 2014, 58, 22–32. [Google Scholar] [CrossRef]
- Ferraresi, A.; Phadngam, S.; Morani, F.; Galetto, A.; Alabiso, O.; Chiorino, G.; Isidoro, C. Resveratrol inhibits IL-6-induced ovarian cancer cell migration through epigenetic up-regulation of autophagy. Mol. Carcinog. 2017, 56, 1164–1181. [Google Scholar] [CrossRef] [PubMed]
- Kornicka, K.; Szłapka-Kosarzewska, J.; Śmieszek, A.; Marycz, K. 5-Azacytydine and resveratrol reverse senescence and ageing of adipose stem cells via modulation of mitochondrial dynamics and autophagy. J. Cell. Mol. Med. 2019, 23, 237–259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ukai, T.; Sato, M.; Akutsu, H.; Umezawa, A.; Mochida, J. MicroRNA-199a-3p, microRNA-193b, and microRNA-320c are correlated to aging and regulate human cartilage metabolism. J. Orthop. Res. 2012, 30, 1915–1922. [Google Scholar] [CrossRef] [PubMed]
- Hollander, W.D.; Meulenbelt, I. DNA Methylation in Osteoarthritis. Curr. Genom. 2015, 16, 419–426. [Google Scholar] [CrossRef] [PubMed] [Green Version]

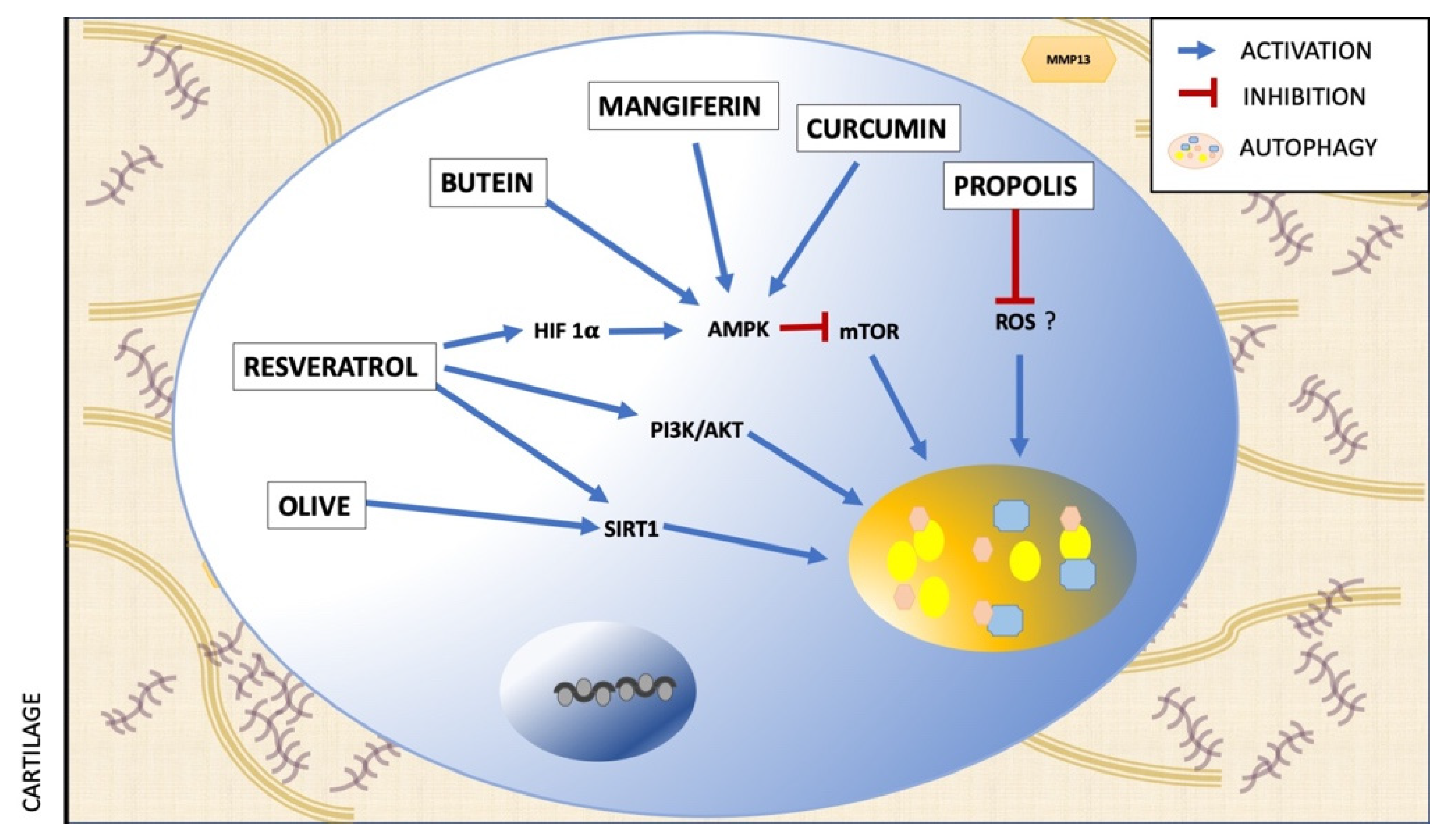
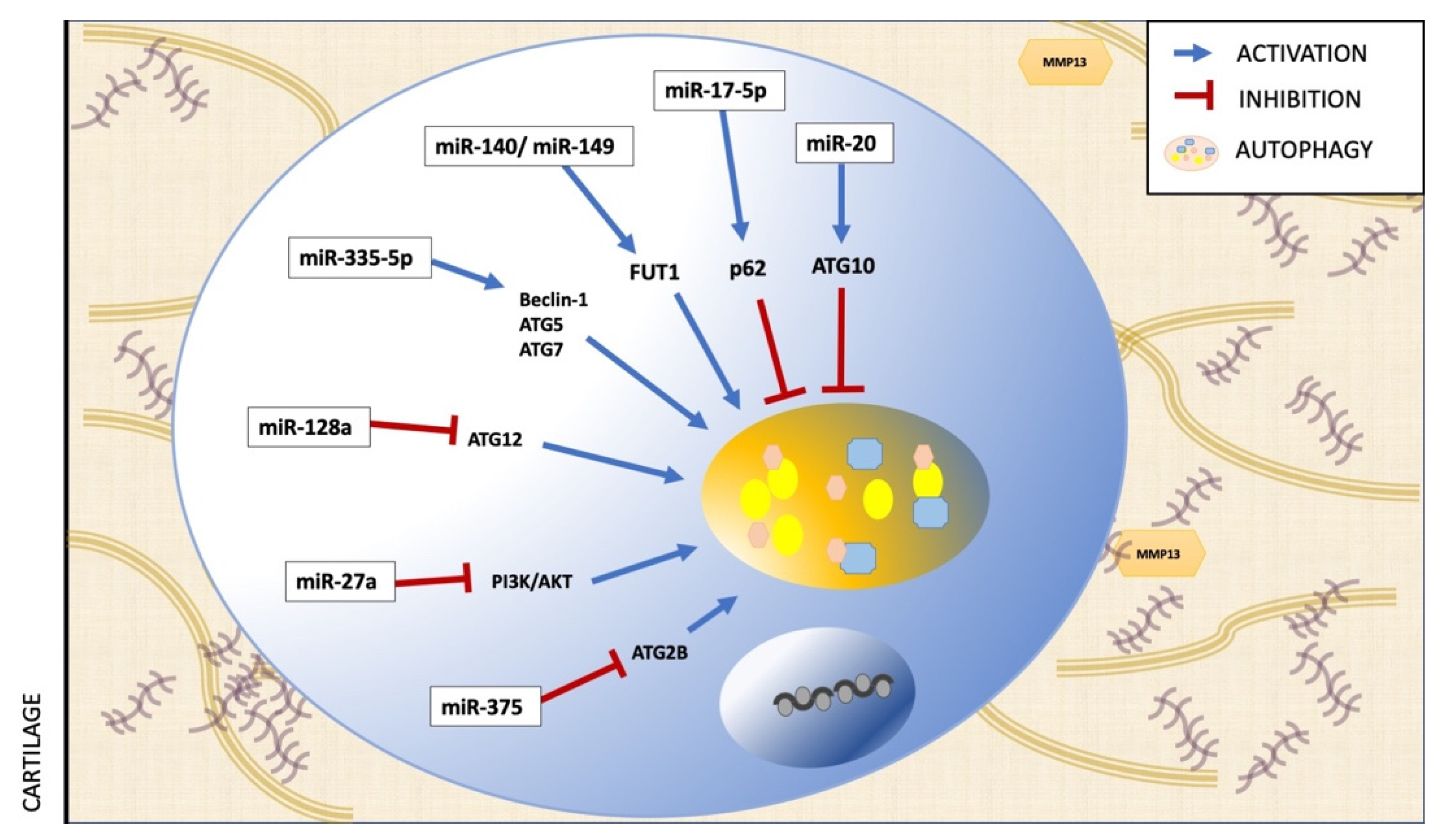
| Polyphenol | Assay | Effect on Autophagy | Mechanism | Ref. |
|---|---|---|---|---|
| Resveratrol | in vivo | Activation | AMPK/mTOR pathway | [63] |
| in vitro | Activation | AMK/SIRT1 pathway | [64] | |
| in vitro | Activation | PI3K/AKT pathway | [65] | |
| Butein | ex vivo | Increase of autophagic flux | AMPK/TSC2/ULK1/mTOR pathway | [66] |
| Olive polyphenols | in vitro | Activation | Suirtin-1 signaling | [67] |
| Mangiferin | in vitro | Increase of autophagic flux | AMPK signaling | [43] |
| Curcumin | in vitro | Activation | Akt/mTOR signaling pathway | [68] |
| Propolis polyphenols | in vitro | Reduction of protein autophagy | ROS signaling | [44] |
| miRNA | Assay | Model/Cell Line | Target in Autophagy | Effect on Autophagy | Ref. |
|---|---|---|---|---|---|
| miR-335-5p | in vitro | Chondrocytes from Human OA articular cartilage | Beclin-1, ATG5, ATG7 | Activation | [54] |
| miR-128a | in vitro in vivo | Chondrocytes from Human OA articular cartilage Rat anterior cruciate ligament transection (ACLT) | ATG12 | Inhibition | [55] |
| miR-27a | in vitro | Chondrocytes from OA human articular cartilage and traumatic amputees. | PI3K | Inhibition | [88] |
| miR-375 | in vitro in vivo | Chondrocytes from OA human articular cartilage. Destabilization of the medial meniscus | ATG2B | Inhibition | [89] |
| miR-140-5p/ miR-149 | in vitro | Chondrocytes from OA human articular cartilage and normal donors. | FUT1 | Inhibition | [90] |
| miR-17-5p | in vitro in vivo | SW1353 human chondrosarcoma cells Destabilization of the medial meniscus on C57BL/6J mice | p62 | Activation | [91] |
| miR-20 | in vitro in vivo | Chondrocytes from OA human articular cartilage and traumatic amputees. Sprague-Dawley (SD) rats | ATG10 | Inhibition | [92] |
| miR-411 | in vitro | Human chondrocyte C28/I2 line | Beclin-1, P62, ULK-1, LC3 | Inhibition | [93] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arias, C.; Salazar, L.A. Autophagy and Polyphenols in Osteoarthritis: A Focus on Epigenetic Regulation. Int. J. Mol. Sci. 2022, 23, 421. https://doi.org/10.3390/ijms23010421
Arias C, Salazar LA. Autophagy and Polyphenols in Osteoarthritis: A Focus on Epigenetic Regulation. International Journal of Molecular Sciences. 2022; 23(1):421. https://doi.org/10.3390/ijms23010421
Chicago/Turabian StyleArias, Consuelo, and Luis A. Salazar. 2022. "Autophagy and Polyphenols in Osteoarthritis: A Focus on Epigenetic Regulation" International Journal of Molecular Sciences 23, no. 1: 421. https://doi.org/10.3390/ijms23010421
APA StyleArias, C., & Salazar, L. A. (2022). Autophagy and Polyphenols in Osteoarthritis: A Focus on Epigenetic Regulation. International Journal of Molecular Sciences, 23(1), 421. https://doi.org/10.3390/ijms23010421







