Analysis of the Genome of the Heavy Metal Resistant and Hydrocarbon-Degrading Rhizospheric Pseudomonas qingdaonensis ZCR6 Strain and Assessment of Its Plant-Growth-Promoting Traits
Abstract
1. Introduction
2. Results and Discussion
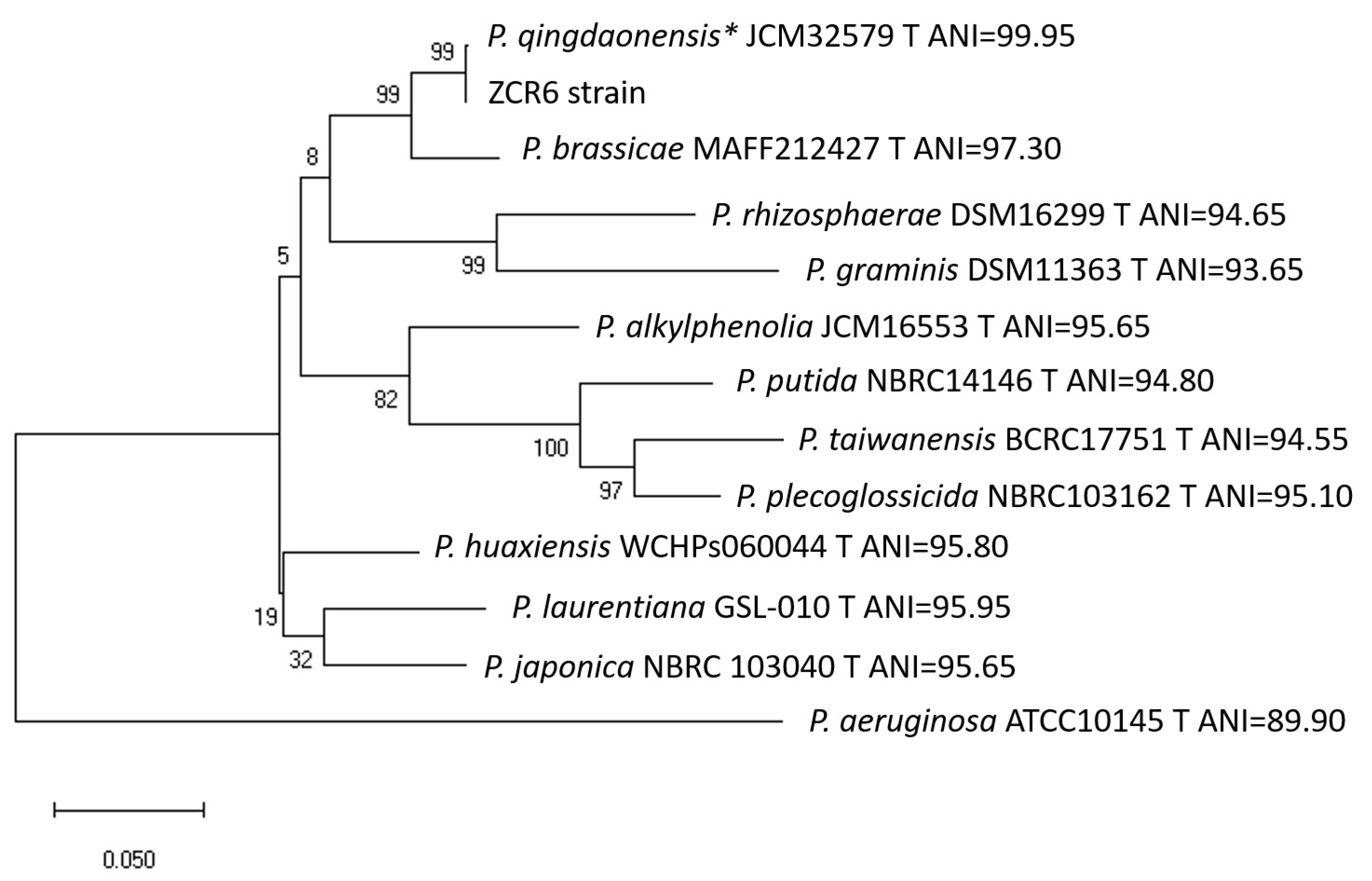
2.1. Isolation and Identification of the Isolate
2.2. Biochemical Characteristics and Cellular Fatty Acid Composition of the ZCR6 Strain
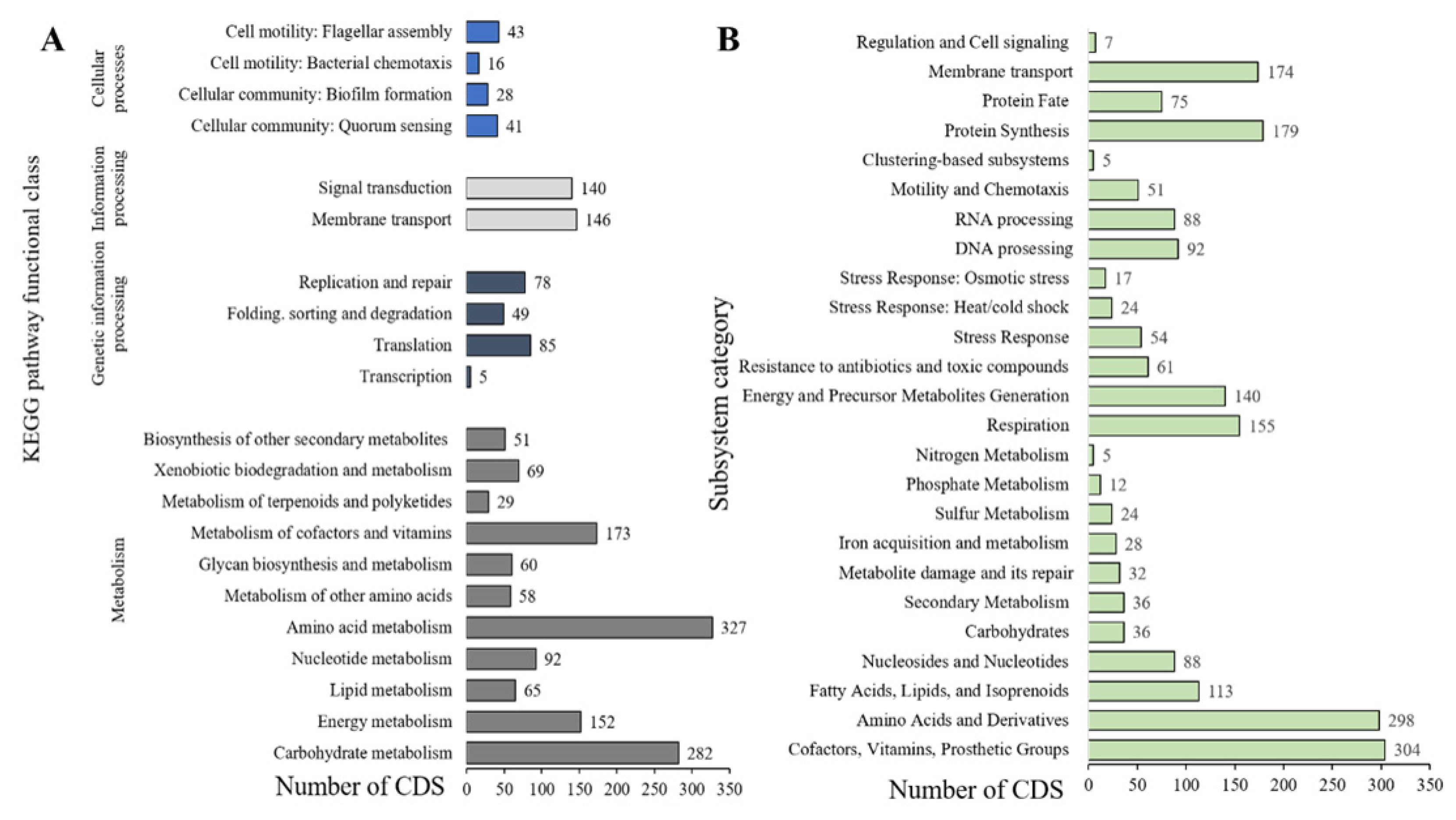
2.3. General Features of the Genome
2.4. Plant Growth Promotion Potential
2.4.1. Nutrient (N, P, S, Fe) Acquisition
2.4.2. Synthesis of the Phytohormone Indole3-Acetic Acid (IAA)
2.4.3. Ethylene Modulation
2.5. Heavy Metal Resistance
2.5.1. Cobalt/Zinc/Cadmium Resistance
2.5.2. Copper/Arsenate/Chromium/Mercury Resistance
2.5.3. Multidrug Resistance
2.6. Exopolysaccharide (EPS) Production
2.7. Response to Oxidative Stress
2.8. Organic Compound Degradation and Biosurfactant Production
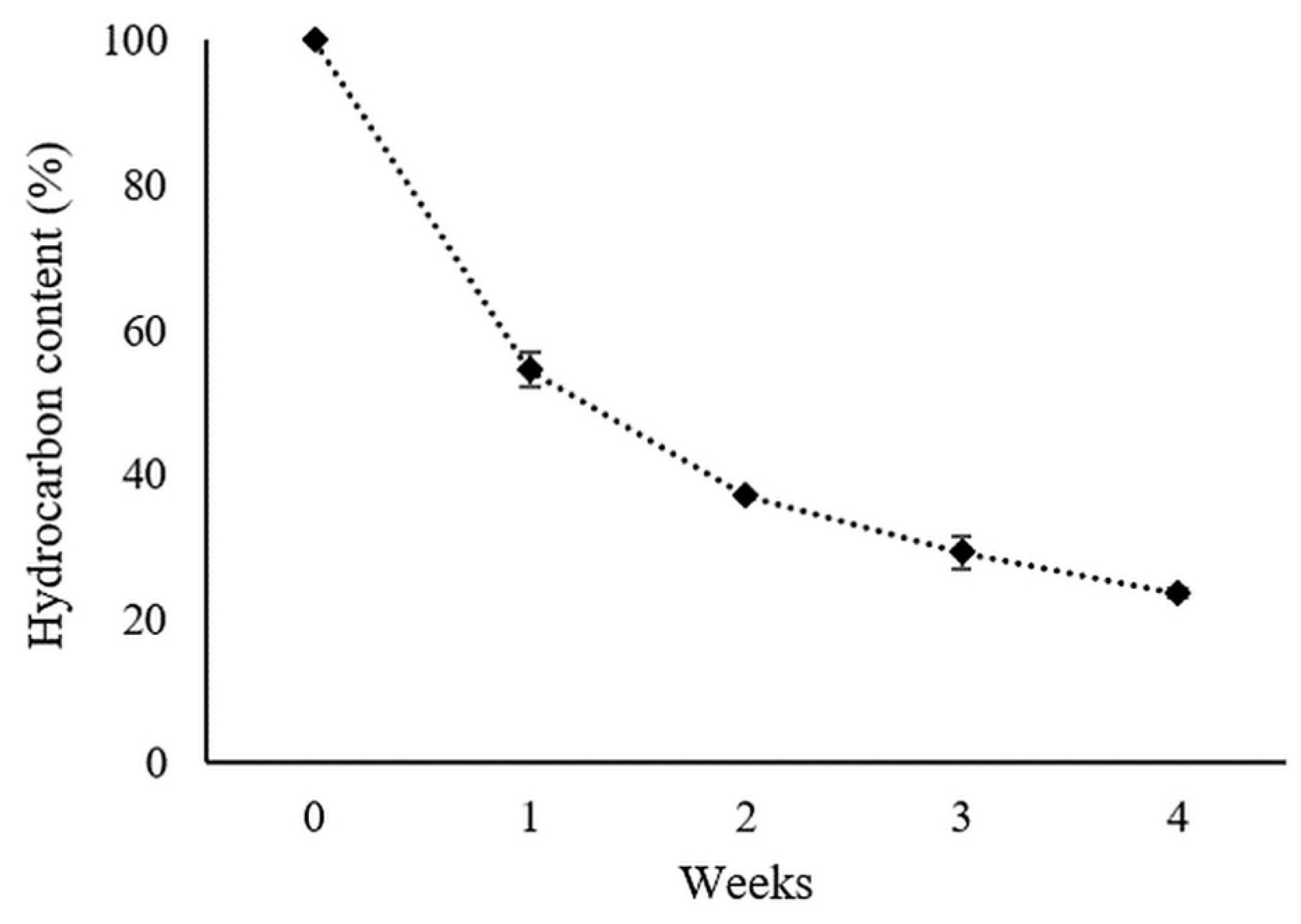
2.8.1. Degradation of Aliphatic Hydrocarbons
2.8.2. Degradation of Aromatic Hydrocarbons
3. Materials and Methods
3.1. Isolation of Hydrocarbon-Degrading and Metal-Resistant Bacterial Strains
3.2. Evaluation of Plant-Growth-Promoting Activities and Surface-Active Properties
3.3. Biochemical Characteristics and Cellular Fatty Acid Analysis of ZCR6 Strain
3.4. DNA Extraction, Whole Genome Sequencing, and Assembly
3.5. Genome Functional Annotation
3.6. Phylogenetic Analysis
3.7. Hydrocarbon Degradation Analysis
3.8. Minimal Inhibitory Concentration (MIC) Determination
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Germaine, K.J.; Keogh, E.; Ryan, D.; Dowling, D.N. Bacterial endophyte-mediated naphthalene phytoprotection and phytoremediation. FEMS Microbiol. Lett. 2009, 296, 226–234. [Google Scholar] [CrossRef] [PubMed]
- Glick, B.R. Using soil bacteria to facilitate phytoremediation. Biotechnol. Adv. 2010, 28, 367–374. [Google Scholar] [CrossRef]
- Afzal, M.; Khan, Q.M.; Sessitsch, A. Endophytic bacteria: Prospects and applications for the phytoremediation of organic pollutants. Chemosphere 2014, 117, 232–242. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Afzal, M.; Iqbal, S.; Khan, Q.M. Plant-bacteria partnerships for the remediation of hydrocarbon contaminated soils. Chemosphere 2013, 90, 1317–1332. [Google Scholar] [CrossRef] [PubMed]
- Manoj, S.R.; Karthik, C.; Kadirvelu, K.; Arulselvi, P.I.; Shanmugasundaram, T.; Bruno, B.; Rajkumar, M. Understanding the molecular mechanisms for the enhanced phytoremediation of heavy metals through plant growth promoting rhizobacteria: A review. J. Environ. Manag. 2020, 254, 109779. [Google Scholar] [CrossRef]
- Jeelani, N.; Yang, W.; Xu, L.; Qiao, Y.; An, S.; Leng, X. Phytoremediation potential of Acorus calamus in soils co-contaminated with cadmium and polycyclic aromatic hydrocarbons. Sci. Rep. 2017, 7, 8028. [Google Scholar] [CrossRef]
- Bello-Akinosho, M.; Makofane, R.; Adeleke, R.; Thantsha, M.; Pillay, M.; Johannes Chirima, G. Potential of Polycyclic Aromatic Hydrocarbon-Degrading Bacterial Isolates to Contribute to Soil Fertility. BioMed Res. Int. 2016, 2016, 5798593. [Google Scholar] [CrossRef]
- Das, K.; Abrol, S.; Verma, R.; Annapragada, H.; Katiyar, N.; Senthilkumar, M. Pseudomonas. Benef. Microbes Agro-Ecol. 2020, 133–148. [Google Scholar] [CrossRef]
- Sun, K.; Liu, J.; Gao, Y.; Li, J.; Gu, Y.; Wang, W. Isolation, plant colonization potential, and phenanthrene degradation performance of the endophytic bacterium Pseudomonas sp. Ph6-gfp. Sci. Rep. 2014, 4, 5462. [Google Scholar] [CrossRef]
- Zhu, X.; Ni, X.; Waigi, M.G.; Liu, J.; Sun, K.; Gao, Y. Biodegradation of Mixed PAHs by PAH-Degrading Endophytic Bacteria. Int. J. Environ. Res. Public Health 2016, 13, 805. [Google Scholar] [CrossRef]
- Wu, T.; Xu, J.; Xie, W.; Yao, Z.; Yang, H.; Sun, C.; Li, X. Pseudomonas aeruginosa L10: A hydrocarbon-degrading, biosurfactant-producing, and plant-growth-promoting endophytic bacterium isolated from a Reed (Phragmites australis). Front. Microbiol. 2018, 9, 1087. [Google Scholar] [CrossRef]
- Khan, Z.; Roman, D.; Kintz, T.; Delas Alas, M.; Yap, R.; Doty, S. Degradation, Phytoprotection and Phytoremediation of Phenanthrene by Endophyte Pseudomonas putida, PD1. Environ. Sci. Technol. 2014, 48, 12221–12228. [Google Scholar] [CrossRef] [PubMed]
- Germaine, K.J.; Liu, X.; Cabellos, G.G.; Hogan, J.P.; Ryan, D.; Dowling, D.N. Bacterial endophyte-enhanced phytoremediation of the organochlorine herbicide 2,4-dichlorophenoxyacetic acid. FEMS Microbiol. Ecol. 2006, 57, 302–310. [Google Scholar] [CrossRef] [PubMed]
- Bakaeva, M.; Kuzina, E.; Vysotskaya, L.; Kudoyarova, G.; Arkhipova, T.; Rafikova, G.; Chetverikov, S.; Korshunova, T.; Chetverikova, D.; Loginov, O. Capacity of Pseudomonas Strains to Degrade Hydrocarbons, Produce Auxins and Maintain Plant Growth under Normal Conditions and in the Presence of Petroleum Contaminants. Plants 2020, 9, 379. [Google Scholar] [CrossRef]
- Płociniczak, T.; Sinkkonen, A.; Romantschuk, M.; Piotrowska-Seget, Z. Characterization of Enterobacter intermedius MH8b and its use for the enhancement of heavy metals uptake by Sinapis alba L. Appl. Soil Ecol. 2013, 63, 1–7. [Google Scholar] [CrossRef]
- Chellaiah, E.R. Cadmium (heavy metals) bioremediation by Pseudomonas aeruginosa: A minireview. Appl. Water Sci. 2018, 8, 154. [Google Scholar] [CrossRef]
- Płociniczak, T.; Chodór, M.; Pacwa-Płociniczak, M.; Piotrowska-Seget, Z. Metal-tolerant endophytic bacteria associated with Silene vulgaris support the Cd and Zn phytoextraction in non-host plants. Chemosphere 2019, 219, 250–260. [Google Scholar] [CrossRef]
- Das, D.; Baruah, R.; Sarma Roy, A.; Singh, A.K.; Deka Boruah, H.P.; Kalita, J.; Bora, T.C. Complete genome sequence analysis of Pseudomonas aeruginosa N002 reveals its genetic adaptation for crude oil degradation. Genomics 2015, 105, 182–190. [Google Scholar] [CrossRef]
- Chen, S.; Chen, W.; Xingxing, P.; Ma, Y.; He, C.; Li, Y.; Huang, C.; Chen, F. Genome Sequence and Metabolic Analysis of a Fluoranthene-Degrading Strain Pseudomonas aeruginosa DN1. Front. Microbiol. 2018, 9, 2595. [Google Scholar] [CrossRef]
- Wang, M.Q.; Wang, Z.; Yu, L.N.; Zhang, C.S.; Bi, J.; Sun, J. Pseudomonas qingdaonensis sp. nov., an aflatoxin-degrading bacterium, isolated from peanut rhizospheric soil. Arch. Microbiol. 2019, 201, 673–678. [Google Scholar] [CrossRef]
- Mulet, M.; Lalucat, J.; García-Valdés, E. DNA sequence-based analysis of the Pseudomonas species. Environ. Microbiol. 2010, 12, 1513–1530. [Google Scholar] [CrossRef]
- Kang, S.-M.; Asaf, S.; Khan, A.L.; Lubna; Khan, A.; Mun, B.-G.; Khan, M.A.; Gul, H.; Lee, I.-J. Complete Genome Sequence of Pseudomonas psychrotolerans CS51, a Plant Growth-Promoting Bacterium, Under Heavy Metal Stress Conditions. Microorganisms 2020, 8, 382. [Google Scholar] [CrossRef]
- Khatri, S.; Sazinas, P.; Shivay, Y.S.; Sharma, S.; Jelsbak, L. Complete Genome Sequence of Pseudomonas sp. Strain SK, Isolated from Organic Wheat Rhizosphere. Microbiol. Resour. Announc. 2021, 9, e00510-20. [Google Scholar] [CrossRef]
- Andrés-Barrao, C.; Lafi, F.F.; Alam, I.; de Zélicourt, A.; Eida, A.A.; Bokhari, A.; Alzubaidy, H.; Bajic, V.B.; Hirt, H.; Saad, M.M. Complete genome sequence analysis of Enterobacter sp. SA187, a plant multi-stress tolerance promoting endophytic bacterium. Front. Microbiol. 2017, 8, 2023. [Google Scholar] [CrossRef] [PubMed]
- Szczerba, H.; Komoń-Janczara, E.; Krawczyk, M.; Dudziak, K.; Nowak, A.; Kuzdraliński, A.; Waśko, A.; Targoński, Z. Genome analysis of a wild rumen bacterium Enterobacter aerogenes LU2-a novel bio-based succinic acid producer. Sci. Rep. 2020, 10, 1986. [Google Scholar] [CrossRef]
- Eida, A.A.; Bougouffa, S.; L’Haridon, F.; Alam, I.; Weisskopf, L.; Bajic, V.B.; Saad, M.M.; Hirt, H. Genome Insights of the Plant-Growth Promoting Bacterium Cronobacter muytjensii JZ38 With Volatile-Mediated Antagonistic Activity Against Phytophthora infestans. Front. Microbiol. 2020, 11, 369. [Google Scholar] [CrossRef]
- Martínez-García, P.M.; Ruano-Rosa, D.; Schilirò, E.; Prieto, P.; Ramos, C.; Rodríguez-Palenzuela, P.; Mercado-Blanco, J. Complete genome sequence of Pseudomonas fluorescens strain PICF7, an indigenous root endophyte from olive (Olea europaea L.) and effective biocontrol agent against Verticillium dahliae. Stand. Genom. Sci. 2015, 10, 10. [Google Scholar] [CrossRef]
- Duan, J.; Jiang, W.; Cheng, Z.; Heikkila, J.J.; Glick, B.R. The Complete Genome Sequence of the Plant Growth-Promoting Bacterium pseudomonas sp. UW4. PLoS ONE 2013, 8, e58640. [Google Scholar] [CrossRef]
- Li, H.B.; Singh, R.K.; Singh, P.; Song, Q.Q.; Xing, Y.X.; Yang, L.T.; Li, Y.R. Genetic diversity of nitrogen-fixing and plant growth promoting Pseudomonas species isolated from sugarcane rhizosphere. Front. Microbiol. 2017, 8, 1–20. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, Q.L.; Yang, Z.M.; Li, Y.; Yan, Y.L.; Ping, S.Z.; Zhang, L.W.; Lin, M.; Lu, W. Identification of the nitrogen-fixing Pseudomonas stutzeri major flagellar gene regulator FleQ and its role in biofilm formation and root colonization. J. Integr. Agric. 2016, 15, 339–348. [Google Scholar] [CrossRef][Green Version]
- Yan, Y.; Yang, J.; Dou, Y.; Chen, M.; Ping, S.; Peng, J.; Lu, W.; Zhang, W.; Yao, Z.; Li, H.; et al. Nitrogen fixation island and rhizosphere competence traits in the genome of root-associated Pseudomonas stutzeri A1501. Proc. Natl. Acad. Sci. USA 2008, 105, 7564–7569. [Google Scholar] [CrossRef]
- Dos Santos, P.C.; Johnson, D.C.; Ragle, B.E.; Unciuleac, M.C.; Dean, D.R. Controlled expression of nif and isc iron-sulfur protein maturation components reveals target specificity and limited functional replacement between the two systems. J. Bacteriol. 2007, 189, 2854–2862. [Google Scholar] [CrossRef] [PubMed]
- Tokumoto, U.; Kitamura, S.; Fukuyama, K.; Takahashi, Y. Interchangeability and distinct properties of bacterial Fe-S cluster assembly systems: Functional replacement of the isc and suf operons in Escherichia coli with the nifSU-like operon from Helicobacter pylori. J. Biochem. 2004, 136, 199–209. [Google Scholar] [CrossRef] [PubMed]
- Dai, Z.; Liu, G.; Chen, H.; Chen, C.; Wang, J.; Ai, S.; Wei, D.; Li, D.; Ma, B.; Tang, C.; et al. Long-term nutrient inputs shift soil microbial functional profiles of phosphorus cycling in diverse agroecosystems. ISME J. 2020, 14, 757–770. [Google Scholar] [CrossRef]
- Hoelzle, K.; Peter, S.; Sidler, M.; Kramer, M.M.; Wittenbrink, M.M.; Felder, K.M.; Hoelzle, L.E. Inorganic pyrophosphatase in uncultivable hemotrophic mycoplasmas: Identification and properties of the enzyme from Mycoplasma suis. BMC Microbiol. 2010, 10. [Google Scholar] [CrossRef]
- Albi, T.; Serrano, A. Two exopolyphosphatases with distinct molecular architectures and substrate specificities from the thermophilic green-sulfur bacterium Chlorobium tepidum TLS. Microbiology 2014, 160, 2067–2078. [Google Scholar] [CrossRef] [PubMed]
- Gebhard, S.; Ekanayaka, N.; Cook, G.M. The low-affinity phosphate transporter PitA is dispensable for in vitro growth of Mycobacterium smegmatis. BMC Microbiol. 2009, 9, 1–7. [Google Scholar] [CrossRef]
- Hsieh, Y.-J.; Wanner, B.L. Global regulation by the seven-component Pi signaling system. Curr. Opin. Microbiol. 2010, 13, 198–203. [Google Scholar] [CrossRef]
- Sekine, M.; Tanikawa, S.; Omata, S.; Saito, M.; Fujisawa, T.; Tsukatani, N.; Tajima, T.; Sekigawa, T.; Kosugi, H.; Matsuo, Y.; et al. Sequence analysis of three plasmids harboured in Rhodococcus erythropolis strain PR4. Environ. Microbiol. 2006, 8, 334–346. [Google Scholar] [CrossRef]
- Coleman, N.V.; Wilson, N.L.; Barry, K.; Brettin, T.S.; Bruce, D.C.; Copeland, A.; Dalin, E.; Detter, J.C.; del Rio, T.G.; Goodwin, L.A.; et al. Genome sequence of the ethene-and vinyl chloride-oxidizing actinomycete Nocardioides sp. strain JS614. J. Bacteriol. 2011, 193, 3399–3400. [Google Scholar] [CrossRef][Green Version]
- Copeland, A.; Lapidus, A.; del Rio, T.G.; Nolan, M.; Lucas, S.; Chen, F.; Tice, H.; Cheng, J.F.; Bruce, D.; Goodwin, L.; et al. Complete genome sequence of Catenulispora acidiphila type strain (ID 139908T). Stand. Genom. Sci. 2009, 1, 119–125. [Google Scholar] [CrossRef][Green Version]
- Normand, P.; Lapierre, P.; Tisa, L.S.; Gogarten, J.P.; Alloisio, N.; Bagnarol, E.; Bassi, C.A.; Berry, A.M.; Bickhart, D.M.; Choisne, N.; et al. Genome characteristics of facultatively symbiotic Frankia sp. strains reflect host range and host plant biogeography. Genome Res. 2007, 17, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Marzan, L.W.; Shimizu, K. Metabolic regulation of Escherichia coli and its phoB and phoR genes knockout mutants under phosphate and nitrogen limitations as well as at acidic condition. Microb. Cell Fact. 2011, 10, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Lubin, E.A.; Henry, J.T.; Fiebig, A.; Crosson, S.; Laub, M.T. Identification of the PhoB regulon and role of PhoU in the phosphate starvation response of Caulobacter crescentus. J. Bacteriol. 2016, 198, 187–200. [Google Scholar] [CrossRef]
- Gigolashvili, T.; Kopriva, S. Transporters in plant sulfur metabolism. Front. Plant Sci. 2014, 5, 1–16. [Google Scholar] [CrossRef]
- Gahan, J.; Schmalenberger, A. The role of bacteria and mycorrhiza in plant sulfur supply. Front. Plant Sci. 2014, 5, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Asaf, S.; Khan, A.L.; Khan, M.A.; Al-Harrasi, A.; Lee, I.J. Complete genome sequencing and analysis of endophytic Sphingomonas sp. LK11 and its potential in plant growth. 3 Biotech 2018, 8, 389. [Google Scholar] [CrossRef] [PubMed]
- Zou, H.; Zhang, N.N.; Pan, Q.; Zhang, J.H.; Chen, J.; Wei, G.H. Hydrogen sulfide promotes nodulation and nitrogen fixation in soybean–rhizobia symbiotic system. Mol. Plant Microbe Interact. 2019, 32, 972–985. [Google Scholar] [CrossRef]
- Shariati, J.V.; Malboobi, M.A.; Tabrizi, Z.; Tavakol, E. Comprehensive genomic analysis of a plant growth-promoting rhizobacterium Pantoea agglomerans strain P5. Sci. Rep. 2017, 15610. [Google Scholar] [CrossRef]
- Rout, G.R.; Sahoo, S. Role of Iron in Plant Growth and Metabolism. Rev. Agric. Sci. 2015, 3, 1–24. [Google Scholar] [CrossRef]
- Mirus, O.; Strauss, S.; Nicolaisen, K.; von Haeseler, A.; Schleiff, E. TonB-dependent transporters and their occurrence in cyanobacteria. BMC Biol. 2009, 7, 68. [Google Scholar] [CrossRef]
- Johnson, G.V.; Lopez, A.; La Valle Foster, N. Reduction and transport of Fe from siderophores. Plant Soil 2002, 241, 27–33. [Google Scholar] [CrossRef]
- Fernàndez, V.; Ebert, G.; Winkelmann, G. The use of microbial siderophores for foliar iron application studies. Plant Soil 2005, 272, 245–252. [Google Scholar] [CrossRef]
- Crowley, D.E.; Römheld, V.; Marschner, H.; Szaniszlo, P.J. Root-microbial effects on plant iron uptake from siderophores and phytosiderophores. Plant Soil 1992, 142, 1–7. [Google Scholar] [CrossRef]
- Rajkumar, M.; Ae, N.; Prasad, M.N.V.; Freitas, H. Potential of siderophore-producing bacteria for improving heavy metal phytoextraction. Trends Biotechnol. 2010, 28, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.K.; Muhammad, H.; Lv, X.; Wei, T.; Ren, X.H.; Jia, H.L.; Atif, S.; Hua, L. Prospects and applications of plant growth promoting rhizobacteria to mitigate soil metal contamination: A review. Chemosphere 2020, 246, 125OK823. [Google Scholar] [CrossRef] [PubMed]
- Khalid, A.; Tahir, S.; Arshad, M.; Zahir, Z.A. Relative efficiency of rhizobacteria for auxin biosynthesis in rhizosphere and non-rhizosphere soils. Aust. J. Soil Res. 2004, 42, 921–926. [Google Scholar] [CrossRef]
- de Souza, R.; Beneduzi, A.; Ambrosini, A.; da Costa, P.B.; Meyer, J.; Vargas, L.K.; Schoenfeld, R.; Passaglia, L.M.P. The effect of plant growth-promoting rhizobacteria on the growth of rice (Oryza sativa L.) cropped in southern Brazilian fields. Plant Soil 2013, 366, 585–603. [Google Scholar] [CrossRef]
- Suarez, C.; Ratering, S.; Hain, T.; Fritzenwanker, M.; Goesmann, A.; Blom, J.; Chakraborty, T.; Bunk, B.; Spröer, C.; Overmann, J.; et al. Complete genome sequence of the plant growth-promoting bacterium hartmannibacter diazotrophicus strain E19T. Int. J. Genom. 2019, 2019. [Google Scholar] [CrossRef]
- McDonnell, L.; Plett, J.M.; Andersson-Gunneras, S.; Kozela, C.; Dugardeyn, J.; Van Der Straeten, D.; Glick, B.R.; Sundberg, B.; Regan, S. Ethylene levels are regulated by a plant encoded 1-aminocyclopropane-1- carboxylic acid deaminase. Physiol. Plant. 2009, 136, 94–109. [Google Scholar] [CrossRef]
- Todorovic, B.; Glick, B.R. The interconversion of ACC deaminase and D-cysteine desulfhydrase by directed mutagenesis. Planta 2008, 229, 193–205. [Google Scholar] [CrossRef]
- Penrose, D.M.; Glick, B.R. Methods for isolating and characterizing ACC deaminase-containing plant growth-promoting rhizobacteria. Physiol. Plant. 2003, 118, 10–15. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.H.; Ryu, H.W.; Kim, J.; Cho, K.S. Rhizoremediation of diesel-contaminated soil using the plant growth-promoting rhizobacterium Gordonia sp. S2RP-17. Biodegradation 2011, 22, 593–601. [Google Scholar] [CrossRef]
- Grobelak, A.; Kokot, P.; Świątek, J.; Jaskulak, M.; Rorat, A. Bacterial ACC Deaminase Activity in Promoting Plant Growth on Areas Contaminated with Heavy Metals. J. Ecol. Eng. 2018, 19, 150–157. [Google Scholar] [CrossRef]
- Kandel, S.L.; Joubert, P.M.; Doty, S.L. Bacterial endophyte colonization and distribution within plants. Microorganisms 2017, 5, 77. [Google Scholar] [CrossRef]
- Pérez-Montaño, F.; Alías-Villegas, C.; Bellogín, R.A.; Del Cerro, P.; Espuny, M.R.; Jiménez-Guerrero, I.; López-Baena, F.J.; Ollero, F.J.; Cubo, T. Plant growth promotion in cereal and leguminous agricultural important plants: From microorganism capacities to crop production. Microbiol. Res. 2014, 169, 325–336. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, M.; Munot, H.P.; Shouche, Y.; Meyer, J.M.; Goel, R.; Journal, A.I. Isolation and Functional Characterization of Siderophore-Producing Lead-and Cadmium-Resistant Pseudomonas putida KNP9 Current Microbiology. Curr. Microbiol. 2005, 50, 233–237. [Google Scholar] [CrossRef] [PubMed]
- Caracciolo, A.B.; Terenzi, V.; Saccà, L.; Manici, L.M. microorganisms Rhizosphere Microbial Communities and Heavy Metals. Microorganisms 2021, 9, 1462. [Google Scholar] [CrossRef]
- Ducret, V.; Gonzalez, M.R.; Leoni, S.; Valentini, M.; Perron, K. The CzcCBA Efflux System Requires the CadA P-Type ATPase for Timely Expression Upon Zinc Excess in Pseudomonas aeruginosa. Front. Microbiol. 2020, 11, 911. [Google Scholar] [CrossRef]
- Fernando, D.M.; Kumar, A. Resistance-Nodulation-Division multidrug efflux pumps in Gram-negative bacteria: Role in virulence. Antibiotics 2013, 2, 163–181. [Google Scholar] [CrossRef]
- Legatzki, A.; Grass, G.; Anton, A.; Rensing, C.; Nies, D.H. Interplay of the Czc system and two P-type ATPases in conferring metal resistance to Ralstonia metallidurans. J. Bacteriol. 2003, 185, 4354–4361. [Google Scholar] [CrossRef]
- Bondarczuk, K.; Piotrowska-Seget, Z. Molecular basis of active copper resistance mechanisms in Gram-negative bacteria. Cell Biol. Toxicol. 2013, 29, 397–405. [Google Scholar] [CrossRef]
- González-Guerrero, M.; Raimunda, D.; Cheng, X.; Argüello, J.M. Distinct functional roles of homologous Cu+ efflux ATPases in Pseudomonas aeruginosa. Mol. Microbiol. 2010, 78, 1246–1258. [Google Scholar] [CrossRef] [PubMed]
- Marrero, K.; Sánchez, A.; González, L.J.; Ledón, T.; Rodríguez-Ulloa, A.; Castellanos-Serra, L.; Pérez, C.; Fando, R. Periplasmic proteins encoded by VCA0261-0260 and VC2216 genes together with copA and cueR products are required for copper tolerance but not for virulence in Vibrio cholerae. Microbiology 2012, 158, 2005–2016. [Google Scholar] [CrossRef] [PubMed]
- Osman, D.; Waldron, K.J.; Denton, H.; Taylor, C.M.; Grant, A.J.; Mastroeni, P.; Robinson, N.J.; Cavet, J.S. Copper homeostasis in Salmonella is atypical and copper-CueP is a major periplasmic metal complex. J. Biol. Chem. 2010, 285, 25259–25268. [Google Scholar] [CrossRef]
- Outten, F.W.; Huffman, D.L.; Hale, J.A.; O’Halloran, T.V. The Independent cue and cus Systems Confer Copper Tolerance during Aerobic and Anaerobic Growth in Escherichia coli. J. Biol. Chem. 2001, 276, 30670–30677. [Google Scholar] [CrossRef] [PubMed]
- Ahemad, M. Bacterial mechanisms for Cr(VI) resistance and reduction: An overview and recent advances. Folia Microbiol. 2014, 59, 321–332. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, C.F.; Aekerley, D.F.; Lynch, S.V.; Matin, A. ChrR, a soluble quinone reductase of Pseudomonas putida that defends against H2O2. J. Biol. Chem. 2005, 280, 22590–22595. [Google Scholar] [CrossRef]
- Ben Fekih, I.; Zhang, C.; Li, Y.P.; Zhao, Y.; Alwathnani, H.A.; Saquib, Q.; Rensing, C.; Cervantes, C. Distribution of arsenic resistance genes in prokaryotes. Front. Microbiol. 2018, 9, 2473. [Google Scholar] [CrossRef]
- Chen, J.; Bhattacharjee, H.; Rosen, B.P. ArsH is an organoarsenical oxidase that confers resistance to trivalent forms of the herbicide monosodium methylarsenate and the poultry growth promoter roxarsone. Mol. Microbiol. 2015, 96, 1042–1052. [Google Scholar] [CrossRef]
- Kiyono, M.; Sone, Y.; Nakamura, R.; Pan-Hou, H.; Sakabe, K. The MerE protein encoded by transposon Tn21 is a broad mercury transporter in Escherichia coli. FEBS Lett. 2009, 583, 1127–1131. [Google Scholar] [CrossRef] [PubMed]
- Spaniol, V.; Bernhard, S.; Aebi, C. Moraxella catarrhalis AcrAB-OprM efflux pump contributes to antimicrobial resistance and is enhanced during cold shock response. Antimicrob. Agents Chemother. 2015, 59, 1886–1894. [Google Scholar] [CrossRef] [PubMed]
- Whitney, J.C.; Howell, P.L. Synthase-dependent exopolysaccharide secretion in Gram-negative bacteria. Trends Microbiol. 2013, 21, 63–72. [Google Scholar] [CrossRef]
- Nilsson, M.; Chiang, W.C.; Fazli, M.; Gjermansen, M.; Givskov, M.; Tolker-Nielsen, T. Influence of putative exopolysaccharide genes on Pseudomonas putida KT2440 biofilm stability. Environ. Microbiol. 2011, 13, 1357–1369. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Lim, E.J.; Kim, K.S.; Huang, S.L.; Veeranagouda, Y.; Rehm, B.H.A. An alginate-like exopolysaccharide biosynthesis gene cluster involved in biofilm aerial structure formation by Pseudomonas alkylphenolia. Appl. Microbiol. Biotechnol. 2014, 98, 4137–4148. [Google Scholar] [CrossRef]
- Gupta, P.; Diwan, B. Bacterial Exopolysaccharide mediated heavy metal removal: A Review on biosynthesis, mechanism and remediation strategies. Biotechnol. Rep. 2017, 13, 58–71. [Google Scholar] [CrossRef]
- Sönmez, M.; Ficai, A.; Ficai, D.; Trusca, R.; Andronescu, E. Alginate/cellulose composite beads for environmental applications. UPB Sci. Bull. Ser. B Chem. Mater. Sci. 2016, 78, 165–176. [Google Scholar]
- Liang, Y.; Chen, J.Q.; Mei, J.; Chang, J.J.; Wang, Q.Y.; Wan, G.S.; Yin, B.Y. Characterization of Cu and Cd biosorption by Pseudomonas sp. strain DC-B3 isolated from metal mine soil. Int. J. Environ. Sci. Technol. 2018, 16, 4035–4046. [Google Scholar] [CrossRef]
- Xu, S.; Xing, Y.; Liu, S.; Hao, X.; Chen, W.; Huang, Q. Characterization of Cd2+ biosorption by Pseudomonas sp. strain 375, a novel biosorbent isolated from soil polluted with heavy metals in Southern China. Chemosphere 2020, 240, 124893. [Google Scholar] [CrossRef]
- Taran, O.; Patel, V.; Lynn, D.G. Small molecule reaction networks that model the ROS dynamics of the rhizosphere. Chem. Commun. 2019, 55, 3602–3605. [Google Scholar] [CrossRef]
- Wu, X.; Monchy, S.; Taghavi, S.; Zhu, W.; Ramos, J.; Van Der Lelie, D. Comparative genomics and functional analysis of niche-specific adaptation in Pseudomonas putida. FEMS Microbiol. Rev. 2011, 35, 299–323. [Google Scholar] [CrossRef]
- Chen, Q.; Li, J.; Liu, M.; Sun, H.; Bao, M. Study on the biodegradation of crude oil by free and immobilized bacterial consortium in marine environment. PLoS ONE 2017, 12, e0174445. [Google Scholar] [CrossRef] [PubMed]
- Muriel-Millán, L.F.; Rodríguez-Mejía, J.L.; Godoy-Lozano, E.E.; Rivera-Gómez, N.; Gutierrez-Rios, R.M.; Morales-Guzmán, D.; Trejo-Hernández, M.R.; Estradas-Romero, A.; Pardo-López, L. Functional and Genomic Characterization of a Pseudomonas aeruginosa Strain Isolated From the Southwestern Gulf of Mexico Reveals an Enhanced Adaptation for Long-Chain Alkane Degradation. Front. Mar. Sci. 2019, 6, 572. [Google Scholar] [CrossRef]
- Mahjoubi, M.; Aliyu, H.; Neifar, M.; Cappello, S.; Chouchane, H.; Souissi, Y.; Masmoudi, A.S.; Cowan, D.A.; Cherif, A. Genomic characterization of a polyvalent hydrocarbonoclastic bacterium Pseudomonas sp. strain BUN14. Sci. Rep. 2021, 11, 8124. [Google Scholar] [CrossRef]
- Phillips, L.A.; Germida, J.J.; Farrell, R.E.; Greer, C.W. Hydrocarbon degradation potential and activity of endophytic bacteria associated with prairie plants. Soil Biol. Biochem. 2008, 40, 3054–3064. [Google Scholar] [CrossRef]
- Andria, V.; Reichenauer, T.G.; Sessitsch, A. Expression of alkane monooxygenase (alkB) genes by plant-associated bacteria in the rhizosphere and endosphere of Italian ryegrass (Lolium multiflorum L.) grown in diesel contaminated soil. Environ. Pollut. 2009, 157, 3347–3350. [Google Scholar] [CrossRef]
- Feng, L.; Wang, W.; Cheng, J.; Ren, Y.; Zhao, G.; Gao, C.; Tang, Y.; Liu, X.; Han, W.; Peng, X.; et al. Genome and proteome of long-chain alkane degrading Geobacillus thermodenitrificans NG80-2 isolated from a deep-subsurface oil reservoir. Proc. Natl. Acad. Sci. USA 2007, 104, 5602–5607. [Google Scholar] [CrossRef]
- Tourova, T.P.; Sokolova, D.S.; Semenova, E.M.; Shumkova, E.S.; Korshunova, A.V.; Babich, T.L.; Poltaraus, A.B.; Nazina, T.N. Detection of n-alkane biodegradation genes alkB and ladA in thermophilic hydrocarbon-oxidizing bacteria of the genera Aeribacillus and Geobacillus. Microbiology 2016, 85, 693–707. [Google Scholar] [CrossRef]
- Ji, Y.; Mao, G.; Wang, Y.; Bartlam, M. Structural insights into diversity and n-alkane biodegradation mechanisms of alkane hydroxylases. Front. Microbiol. 2013, 4, 58. [Google Scholar] [CrossRef]
- Meng, L.; Bao, M.; Sun, P. Construction of long-chain alkane degrading bacteria and its application in bioremediation of crude oil pollution. Int. J. Biol. Macromol. 2018, 119, 524–532. [Google Scholar] [CrossRef]
- Fuchs, G.; Boll, M.; Heider, J. Microbial degradation of aromatic compounds—from one strategy to four. Nat. Rev. Microbiol. 2011, 9, 803–816. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Yan, Y.; Ping, S.; Chen, M.; Zhang, W.; Li, L.; Lin, W.; Geng, L.; Liu, W.; Lu, W.; et al. Genome-wide investigation and functional characterization of the β-ketoadipate pathway in the nitrogen-fixing and root-associated bacterium Pseudomonas stutzeriA1501. BMC Microbiol. 2010, 10, 36. [Google Scholar] [CrossRef][Green Version]
- Nogales, J.; García, J.L.; Díaz, E. Degradation of Aromatic Compounds in Pseudomonas: A Systems Biology View. In Aerobic Utilization of Hydrocarbons, Oils and Lipids; Rojo, F., Ed.; Springer International Publishing: Cham, Switzerland, 2017; pp. 1–49. ISBN 978-3-319-39782-5. [Google Scholar]
- Alejandro-Marín, C.M.; Bosch, R.; Nogales, B. Comparative genomics of the protocatechuate branch of the β-ketoadipate pathway in the Roseobacter lineage. Mar. Genom. 2014, 17, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Tikariha, H.; Pal, R.R.; Qureshi, A.; Kapley, A.; Purohit, H.J. In silico analysis for prediction of degradative capacity of Pseudomonas putida SF1. Gene 2016, 591, 382–392. [Google Scholar] [CrossRef] [PubMed]
- Méndez, V.; Agulló, L.; González, M.; Seeger, M. The homogentisate and homoprotocatechuate central pathways are involved in 3- and 4-hydroxyphenylacetate degradation by Burkholderia xenovorans LB400. PLoS ONE 2011, 6. [Google Scholar] [CrossRef]
- Pacwa-Płociniczak, M.; Płaza, G.A.; Poliwoda, A.; Piotrowska-Seget, Z. Characterization of hydrocarbon-degrading and biosurfactant-producing Pseudomonas sp. P-1 strain as a potential tool for bioremediation of petroleum-contaminated soil. Environ. Sci. Pollut. Res. 2014, 21. [Google Scholar] [CrossRef]
- Guo, G.; Tian, F.; Ding, K.; Wang, L.; Liu, T.; Yang, F. Effect of a bacterial consortium on the degradation of polycyclic aromatic hydrocarbons and bacterial community composition in Chinese soils. Int. Biodeterior. Biodegradation 2017, 123, 56–62. [Google Scholar] [CrossRef]
- Chlebek, D.; Pinski, A.; Żur, J.; Michalska, J.; Hupert-Kocurek, K. Genome Mining and Evaluation of the Biocontrol Potential of Pseudomonas fluorescens BRZ63, a New Endophyte of Oilseed Rape (Brassica napus L.) against Fungal Pathogens. Int. J. Mol. Sci. 2020, 21, 8740. [Google Scholar] [CrossRef] [PubMed]
- Cheng, T.; Liang, J.; He, J.; Hu, X.; Ge, Z.; Liu, J. A novel rhamnolipid-producing pseudomonas aeruginosa zs1 isolate derived from petroleum sludge suitable for bioremediation. AMB Express 2017, 7, 1–14. [Google Scholar] [CrossRef]
- Omoboye, O.O.; Geudens, N.; Duban, M.; Chevalier, M.; Flahaut, C.; Martins, J.C.; Leclère, V.; Oni, F.E.; Höfte, M. Pseudomonas sp. COW3 produces new bananamide-type cyclic lipopeptides with antimicrobial activity against Pythium myriotylum and Pyricularia oryzae. Molecules 2019, 24, 4170. [Google Scholar] [CrossRef]
- Zhang, J.; Xue, Q.; Gao, H.; Lai, H.; Wang, P. Production of lipopeptide biosurfactants by Bacillus atrophaeus 5-2a and their potential use in microbial enhanced oil recovery. Microb. Cell Fact. 2016, 15, 168. [Google Scholar] [CrossRef]
- Gutiérrez-Chávez, C.; Benaud, N.; Ferrari, B.C. The ecological roles of microbial lipopeptides: Where are we going? Comput. Struct. Biotechnol. J. 2021, 19, 1400–1413. [Google Scholar] [CrossRef] [PubMed]
- Viramontes-Ramos, S.; Portillo-Ruiz, M.C.; de Lourdes Ballinas-Casarrubias, M.; Torres-Muñoz, J.V.; Rivera-Chavira, B.E.; Nevárez-Moorillón, G.V. Selection of biosurfactan/bioemulsifier-producing bacteria from hydrocarbon-contaminated soil. Braz. J. Microbiol. 2010, 41, 668–675. [Google Scholar] [CrossRef]
- Saleh, S.S.; Glick, B.R. Involvement of gacS and rpoS in enhancement of the plant growth-promoting capabilities of Enterobacter cloacae CAL2 and UW4. Can. J. Microbiol. 2001, 47, 698–705. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Pointing, S.B. Qualitative methods for the determination of lignocellulolytic enzyme production by tropical fungi. Fungal Divers. 1999, 2, 17–33. [Google Scholar]
- Schwyn, B.; Neilands, J.B. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Bric, J.M.; Bostock, R.M.; Silverstone, S.E. Rapid in situ assay for indoleacetic acid production by bacteria immobilized on a nitrocellulose membrane. Appl. Environ. Microbiol. 1991, 57, 535–538. [Google Scholar] [CrossRef] [PubMed]
- Cappuccino, J.C.; Sherman, N. Microbiology: A Laboratory Manual, 3rd ed.; Benjamin Cummings Publisher: New York, NY, USA, 1992. [Google Scholar]
- Nautiyal, C.S. An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol. Lett. 1999, 170, 265–270. [Google Scholar] [CrossRef]
- Lorck, H. Production of Hydrocyanic Acid by Bacteria. Physiol. Plant. 1948, 1, 142–146. [Google Scholar] [CrossRef]
- Pacwa-Płociniczak, M.; Płociniczak, T.; Iwan, J.; Zarska, M.; Chorazewski, M.; Dzida, M.; Piotrowska-Seget, Z. Isolation of hydrocarbon-degrading and biosurfactant-producing bacteria and assessment their plant growth-promoting traits. J. Environ. Manag. 2016, 168, 175–184. [Google Scholar] [CrossRef]
- Ptaszek, N.; Pacwa-Płociniczak, M.; Noszczyńska, M.; Płociniczak, T. Comparative Study on Multiway Enhanced Bio- and Phytoremediation of Aged Petroleum-Contaminated Soil. Agronomy 2020, 10, 947. [Google Scholar] [CrossRef]
- Iverson, W.G.; Millis, N.F. A Method for the Detection of Starch Hydrolysis by Bacteria. J. Appl. Bacteriol. 1974, 37, 443–446. [Google Scholar] [CrossRef]
- Hidayat, I. Characteristics of the peat soil Bacilllus sp. Berk. Penelit. Hayati 2004, 10, 31–35. [Google Scholar] [CrossRef]
- Peck, H.D.; Gest, H. Formic dehydrogenase and the hydrogenlyase enzyme complex in coli-aerogenes bacteria. J. Bacteriol. 1957, 73, 706–721. [Google Scholar] [CrossRef]
- Taylor, J.J.; Whitby, J.L. Pseudomonas Pyocyanea and the Arginine Dihydrolase System. J. Clin. Pathol. 1964, 17, 122–125. [Google Scholar] [CrossRef]
- Christensen, W.B. Urea Decomposition as a Means of Differentiating Proteus and Paracolon Cultures from Each Other and from Salmonella and Shigella Types. J. Bacteriol. 1946, 52, 461–466. [Google Scholar] [CrossRef]
- King, E.O.; Ward, M.K.; Raney, D.E. Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 1954, 44, 301–307. [Google Scholar] [PubMed]
- Piotrowska-Seget, Z.; Cycoń, M.; Kozdrój, J. Metal-tolerant bacteria occurring in heavily polluted soil and mine spoil. Appl. Soil Ecol. 2005, 28, 237–246. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. EggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef]
- Davis, J.J.; Wattam, A.R.; Aziz, R.K.; Brettin, T.; Butler, R.; Butler, R.M.; Chlenski, P.; Conrad, N.; Dickerman, A.; Dietrich, E.M.; et al. The PATRIC Bioinformatics Resource Center: Expanding data and analysis capabilities. Nucleic Acids Res. 2020, 48. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Zhu, Z.; Fu, L.; Niu, B.; Li, W. WebMGA: A customizable web server for fast metagenomic sequence analysis. BMC Genomics 2011, 12, 444. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. AntiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef]
- Zhou, Y.; Liang, Y.; Lynch, K.H.; Dennis, J.J.; Wishart, D.S. PHAST: A Fast Phage Search Tool. Nucleic Acids Res. 2011, 39, 347–352. [Google Scholar] [CrossRef]
- Furmanczyk, E.M.; Kaminski, M.A.; Lipinski, L.; Dziembowski, A.; Sobczak, A. Pseudomonas laurylsulfatovorans sp. nov., sodium dodecyl sulfate degrading bacteria, isolated from the peaty soil of a wastewater treatment plant. Syst. Appl. Microbiol. 2018, 41, 348–354. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.; Lim, J.; Kwon, S.; Chun, J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie van Leeuwenhoek/Int. J. Gen. Mol. Microbiol. 2017, 110, 1281–1286. [Google Scholar] [CrossRef] [PubMed]
- Ha, S.M.; Kim, C.K.; Roh, J.; Byun, J.H.; Yang, S.J.; Bin Choi, S.; Chun, J.; Yong, D. Application of the whole genome-based bacterial identification system, TRUEBAC ID, using clinical isolates that were not identified with three matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) systems. Ann. Lab. Med. 2019, 39, 530–536. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R.; Glöckner, F.O.; Rg Peplies, J. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef]
- Rathnayake, I.V.N.; Megharaj, M.; Krishnamurti, G.S.R.; Bolan, N.S.; Naidu, R. Heavy metal toxicity to bacteria—Are the existing growth media accurate enough to determine heavy metal toxicity? Chemosphere 2013, 90, 1195–1200. [Google Scholar] [CrossRef] [PubMed]
- Manogaran, M.; Shukor, M.Y.; Yasid, N.A.; Khalil, K.A.; Ahmad, S.A. Optimisation of culture composition for glyphosate degradation by Burkholderia vietnamiensis strain AQ5-12. 3 Biotech 2018, 8, 108. [Google Scholar] [CrossRef] [PubMed]


| PGP Mechanisms | Plant Colonisation | Surface-Active Properties | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ACCD a | IAA b | Ca3(PO4)2 c | Siderophore d | HCN e | NH3 f | CMC g | Surface Tension h | Oil-Spreading i | Emulsification Index j | ||
| H k | X l | D m | |||||||||
| 296.73 ± 1.27 | 3.59 ± 0.11 | 6.5 ± 0.53 | + | - | + | + | 68.1 ± 0.55 | 4.0 ± 0.2 | 25.71 ± 0.57 | 71.43 ± 1.43 | 14.71 ± 0.29 |
| Percentage (%) | ||
|---|---|---|
| 24 h | 72 h | |
| C16:0 | 30.95 | 22.52 |
| C16:1ω7c and/or C16:1ω6c | 26.35 | 11.42 |
| C18:1ω7c and/or C18:1ω6c | 15.44 | 11.68 |
| C17:0 cyclo | 10.22 | 23.07 |
| C12:0 2OH | 4.63 | 8.74 |
| C12:0 3OH | 4.11 | 7.57 |
| C10:0 3OH | 3.34 | 4.35 |
| C12:0 | 2.22 | 4.07 |
| C 19:0 cyclo ω8c | <1 | 5.14 |
| Others | <2 | <2 |
| Feature | Value |
|---|---|
| Assembly information | |
| Genome size (bp) | 5,507,067 |
| Contigs | 52 |
| Conting N50 (bp) | 292,270 |
| Conting L50 | 7 |
| G + C content (%) | 64.5 |
| Annotation information | |
| Genes (total) | 5055 |
| CDSs (total) | 4979 |
| Genes (coding) | 4943 |
| CDSs (with protein) | 4943 |
| Pseudogenes | 36 |
| RNA genes | 76 |
| rRNA | 3, 3, 2 (5S, 16S, 23S) |
| tRNA | 64 |
| ncRNA | 4 |
| Prophage regions | 2 |
| Genes assigned to KEGG pathways | 2945 |
| Genes assigned to KEGG Orthology (KO) | 2209 |
| Genes assigned to COGs | 4877 |
| Genes connected to RASTtk | 2319 |
| BioProject ID | PRJNA681194 |
| BioSample ID | SAMN16935590 |
| GeneBank accession number | JADWSX000000000 |
| Gene | Accession Number | KO Number | Gene Product | Activity |
|---|---|---|---|---|
| iscS | MBG8561789.1 | K04487 | Cysteine desulfurase | Nitrogen fixation |
| iscU | MBG8560681.1 | K04488 | Nitrogen fixation protein nifu | |
| gcd | MBG8562075.1 | K00117 | Quinoprotein glucose dehydrogenase (EC:1.1.5.2) | Phosphate solubilization |
| pqqA | MBG8558041.1 | K06135 | Pyrroloquinoline quinone biosynthesis protein A | |
| pqqB | MBG8558042.1 | K06136 | Pyrroloquinoline quinone biosynthesis protein B | |
| pqqC | MBG8558043.1 | K06137 | Pyrroloquinoline-quinone synthase (EC:1.3.3.11) | |
| pqqD | MBG8558044.1 | K06138 | Pyrroloquinoline quinone biosynthesis protein D | |
| pqqE | MBG8558045.1 | K06139 | Pqqa peptide cyclase (EC:1.21.98.4) | |
| ppa | MBG8560327.1 | K01507 | Inorganic pyrophosphatase (EC:3.6.1.1) | |
| ppx-gppA | MBG8558276.1 | K01524 | Exopolyphosphatase/guanosine-5′-triphosphate,3′-diphosphate pyrophosphatase (EC:3.6.1.11 3.6.1.40) | |
| pstB | MBG8558408.1 | K02036 | Phosphate transport system ATP-binding protein (EC:7.3.2.1) | |
| pstA | MBG8558409.1 | K02038 | Phosphate transport system permease protein | |
| pstC | MBG8558410.1 | K02037 | Phosphate transport system permease protein | |
| pstS | MBG8558411.1 | K02040 | Phosphate transport system substrate-binding protein | |
| pit | MBG8560129.1 | K16322 | Low-affinity inorganic phosphate transporter | |
| phoB | MBG8558402.1 | K07657 | Two-component system. Ompr family. Phosphate regulon response regulator phob | |
| phoR | MBG8558403.1 | K07636 | Two-component system. Ompr family. Phosphate regulon sensor histidine kinase phor (EC:2.7.13.3) | |
| phoU | MBG8558407.1 | K02039 | Phosphate transport system protein | |
| ssuA | MBG8561374.1 | K15553 | Sulfonate transport system substrate-binding protein | Sulfur metabolism |
| ssuB | MBG8560999.1 | K15555 | Sulfonate transport system ATP-binding protein (EC:7.6.2.14) | |
| ssuC | MBG8561372.1 | K15554 | Sulfonate transport system permease protein | |
| ssuD | MBG8561373.1 | K04091 | Alkanesulfonate monooxygenase (EC:1.14.14.5) | |
| ssuE | MBG8561375.1 | K00299 | FMN reductase (EC:1.5.1.38) | |
| cysP | MBG8561508.1 | K02048 | Sulfate/thiosulfate transport system substrate-binding protein | |
| cysU | MBG8561415.1 | K02046 | Sulfate/thiosulfate transport system permease protein | |
| cysW | MBG8561414.1 | K02047 | Sulfate/thiosulfate transport system permease protein | |
| cysA | MBG8561413.1 | K02045 | Sulfate/thiosulfate transport system ATP-binding protein (EC:7.3.2.3) | |
| cysI | MBG8558903.1 | K00381 | Sulfite reductase (NADPH) hemoprotein beta-component (EC:1.8.1.2) | |
| cysJ | MBG8560698.1 | K00380 | Sulfite reductase (NADPH) flavoprotein alpha-component (EC:1.8.1.2) | |
| CTH | MBG8560530.1 | K01758 | Cystathionine γ-lyase | |
| CBS | MBG8560531.1 | K01697 | Cystathionine β-lyase | |
| TST | MBG8560836.1 | K01011 | Thiosulfate/3-mercaptopyruvate sulfurtransferase | |
| bfr | MBG8560256.1 | K03594 | Bacterioferritin (EC:1.16.3.1) | Siderophore biosynthesis |
| entD | MBG8562760.1 | K02362 | Enterobactin synthetase component D (EC:6.3.2.14 2.7.8.-) | |
| fiu | MBG8560699.1 | K16090 | Catecholate siderophore receptor | Siderophore uptake |
| tonB | MBG8561215.1 | K03832 | Periplasmic protein tonb | |
| exbB | MBG8561408.1 | K03561 | Biopolymer transport protein exbb | |
| exbD | MBG8558382.1 | K03559 | Biopolymer transport protein exbd | |
| fepA | MBG8559634.1 | K19611 | Ferric enterobactin receptor | |
| pvdF | MBG8561616.1 | K06160 | Putative pyoverdin transport system ATP-binding/permease protein | |
| trpC | MBG8557998.1 | K01609 | Indole-3-glycerol phosphate synthase (EC:4.1.1.48) | Synthesis of the phytohormone: |
| trpD | MBG8557999.1 | K00766 | Anthranilate phosphoribosyltransferase (EC:2.4.2.18) | indole acetic acid (IAA) |
| trpG | MBG8558000.1 | K01658 | Anthranilate synthase component II (EC:4.1.3.27) | |
| trpE | MBG8558001.1 | K01657 | Anthranilate synthase component I (EC:4.1.3.27) | |
| trpF | MBG8560957.1 | K01817 | Phosphoribosylanthranilate isomerase (EC:5.3.1.24) | |
| trpA | MBG8562725.1 | K01695 | Tryptophan synthase alpha chain (EC:4.2.1.20) | |
| trpB | MBG8562726.1 | K01696 | Tryptophan synthase beta chain (EC:4.2.1.20) | |
| ipdC | MBG8559993.1 | K04103 | Indolepyruvate decarboxylase (EC:4.1.1.74) | |
| aspC | MBG8558823.1 | K00813 | Aspartate aminotransferase (EC:2.6.1.1) | |
| aldA | MBG8559624.1 | K07248 | Lactaldehyde dehydrogenase/glycolaldehyde dehydrogenase (EC:1.2.1.22 1.2.1.21) | |
| aldB | MBG8560334.1 | K00138 | Aldehyde dehydrogenase (EC:1.2.1.-) | |
| iaaM | MBG8558038.1 | K00466 | Amine oxidase | |
| acdS | MBG8559878.1 | K01505 | 1-aminocyclopropane-1-carboxylate deaminase (EC:3.5.99.7) | Ethylene modulation |
| dcyD | MBG8561384.1 | K05396 | D-cysteine desulfhydrase (EC:4.4.1.15) |
| Gene | Accession Number | KO Numbers | Gene Product | Metal Resistance |
|---|---|---|---|---|
| czcA | MBG8562084.1 | K15726 | Cobalt-zinc-cadmium resistance protein czca | Cobalt/zinc/cadmium resistance |
| czcB | MBG8562085.1 | K15727 | Membrane fusion protein. Cobalt-zinc-cadmium efflux system | |
| czcC | MBG8562086.1 | K15725 | Outer membrane protein. Cobalt-zinc-cadmium efflux system | |
| copB | MBG8561457.1 | K01533 | P-type Cu2+ transporter (EC:7.2.2.9) | Cooper resistance |
| copA | MBG8562586.1 | K17686 | P-type Cu+ transporter (EC:7.2.2.8) | |
| cusR | MBG8559773.1 | K07665 | Two-component system. Ompr family. Copper resistance phosphate regulon response regulator cusr | |
| cusS | MBG8559774.1 | K07644 | Two-component system. Ompr family. Heavy metal sensor histidine kinase cuss (EC:2.7.13.3) | |
| cueR | MBG8562587.1 | K19591 | Merr family transcriptional regulator. Copper efflux regulator | |
| arsR | MBG8562772.1 | K03892 | Arsr family transcriptional regulator. Arsenate/arsenite/antimonite-responsive transcriptional repressor | Arsenate resistance |
| arsB | MBG8559938.1 | K03893 | Arsenical pump membrane protein | |
| arsC | MBG8559939.1 | K03741 | Arsenate reductase (thioredoxin) (EC:1.20.4.4) | |
| arsC | MBG8562772.1 | K00537 | Arsenate reductase (glutaredoxin) (EC:1.20.4.1) | |
| arsH | MBG8559940.1 | K11811 | Arsenical resistance protein arsh | |
| chrA | MBG8559121.1 | K07240 | Chromate transporter | Chromium resistance |
| chrR | MBG8561757.1 | K19784 | Chromate reductase. Nad(p)h dehydrogenase (quinone) | |
| oprM | MBG8560149.1 | K18139 | Outer membrane protein. Efflux system | Multidrug resistance |
| acrB | MBG8560150.1 | K18138 | Multidrug efflux pump | |
| acrA | MBG8560151.1 | K03585 | Membrane fusion protein. Multidrug efflux system | |
| acrR | MBG8560152.1 | K03577 | Tetr/acrr family transcriptional regulator. Acrab operon repressor | |
| merE | MBG8560549.1 | K00549 | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase (EC:2.1.1.14) | Mercury resistance |
| Gene | Accession Number | KO Numbers | Gene Product | Activity |
|---|---|---|---|---|
| bcsA | MBG8561468.1 | K00694 | Cellulose synthase (UDP-forming) (EC:2.4.1.12) | Exopolysaccharides biosynthesis |
| bcsB | MBG8561469.1 | K20541 | Cellulose synthase operon protein B | |
| bcsZ | MBG8561470.1 | K20542 | Endoglucanase (EC:3.2.1.4) | |
| bcsC | MBG8561471.1 | K20543 | Cellulose synthase operon protein C | |
| algF | MBG8561473.1 | K19296 | Alginate O-acetyltransferase complex protein algf | |
| algI | MBG8561474.1 | K19294 | Alginate O-acetyltransferase complex protein algi | |
| algJ | MBG8561475.1 | K19295 | Alginate O-acetyltransferase complex protein algj | |
| algB | MBG8562662.1 | K11384 | Two-component system. Ntrc family. Response regulator algb | |
| algA | MBG8559808.1 | K16011 | Mannose-1-phosphate guanylyltransferase/mannose-6-phosphate isomerase (EC:2.7.7.13 5.3.1.8) | |
| algL | MBG8560007.1 | K01729 | Poly(beta-D-mannuronate) lyase (EC:4.2.2.3) | |
| algX | MBG8560008.1 | K19293 | Alginate biosynthesis protein algx | |
| algG | MBG8560009.1 | K01795 | Mannuronan 5-epimerase (EC:5.1.3.37) | |
| algE | MBG8560010.1 | K16081 | Alginate production protein | |
| algK | MBG8560011.1 | K19292 | Alginate biosynthesis protein algk | |
| alg44 | MBG8560012.1 | K19291 | Mannuronan synthase (EC:2.4.1.33) | |
| alg8 | MBG8560013.1 | K19290 | Mannuronan synthase (EC:2.4.1.33) | |
| algD | MBG8560014.1 | K00066 | GDP-mannose 6-dehydrogenase (EC:1.1.1.132) | |
| algH | MBG8561216.1 | K07735 | Putative transcriptional regulator | |
| algR | MBG8558298.1 | K08083 | Two-component system. Lyttr family. Response regulator algr |
| Gene | Accession Number | KO Numbers | Gene Product | Activity |
|---|---|---|---|---|
| katE | MBG8562679.1 | K03781 | Catalase (EC:1.11.1.6) | Oxidative stress response |
| katG | MBG8559566.1 | K03782 | Catalase-peroxidase (EC:1.11.1.21) | |
| sod2 | MBG8560099.1 | K04564 | SOD2; superoxide dismutase. Fe-Mn family (EC:1.15.1.1) | |
| ggt | MBG8558271.1 | K00681 | Gamma-glutamyltranspeptidase/glutathione hydrolase (EC:2.3.2.2 3.4.19.13) | |
| gst | MBG8558300.1 | K00799 | Glutathione S-transferase (EC:2.5.1.18) | |
| gpx | MBG8560587.1 | K00432 | Glutathione peroxidase (EC:1.11.1.9) | |
| gsr | MBG8561128.1 | K00383 | GSR; glutathione reductase (NADPH) (EC:1.8.1.7) | |
| oxyR | MBG8558139.1 | K04761 | Lysr family transcriptional regulator. Hydrogen peroxide-inducible gene activator |
| Gene | Accession Number | KO Numbers | Gene Product | Activity |
|---|---|---|---|---|
| ladA | MBG8560652.1 | K20938 | long-chain alkane monooxygenase (EC:1.14.14.28) | Aliphatic compounds’ degradation |
| pcaG | MBG8562545.1 | K00448 | Protocatechuate 3.4-dioxygenase. Alpha subunit (EC:1.13.11.3) | Aromatic compounds’ degradation |
| pcaH | MBG8562546.1 | K00449 | Protocatechuate 3.4-dioxygenase. Beta subunit (EC:1.13.11.3) | |
| pcaQ | MBG8560766.1 | K02623 | Lysr family transcriptional regulator. Pca operon transcriptional activator | |
| pcaC | MBG8560139.1 | K01607 | 4-carboxymuconolactone decarboxylase (EC:4.1.1.44) | |
| pcaD | MBG8560138.1 | K01055 | 3-oxoadipate enol-lactonase (EC:3.1.1.24) | |
| pcaR | MBG8560131.1 | K02624 | Iclr family transcriptional regulator. Pca regulon regulatory protein | |
| pcaK | MBG8560132.1 | K08195 | MFS transporter. AAHS family. 4-hydroxybenzoate transporter | |
| pcaF | MBG8560135.1 | K07823 | 3-oxoadipyl-coa thiolase (EC:2.3.1.174) | |
| pcaT | MBG8560136.1 | K02625 | MFS transporter. MHS family. Dicarboxylic acid transporter pcat | |
| pcaB | MBG8560137.1 | K01857 | 3-carboxy-cis.cis-muconate cycloisomerase (EC:5.5.1.2) | |
| hpaF | MBG8558716.1 | K01826 | 5-carboxymethyl-2-hydroxymuconate isomerase (EC:5.3.3.10) | |
| hpaI | MBG8558890.1 | K02510 | 4-hydroxy-2-oxoheptanedioate aldolase (EC:4.1.2.52) | |
| hpaH | MBG8558891.1 | K02509 | 2-oxo-hept-3-ene-1.7-dioate hydratase (EC:4.2.1.-) | |
| hpaF | MBG8558893.1 | K01826 | 5-carboxymethyl-2-hydroxymuconate isomerase (EC:5.3.3.10) | |
| hpaD | MBG8558894.1 | K00455 | 3.4-dihydroxyphenylacetate 2.3-dioxygenase (EC:1.13.11.15) | |
| hpaE | MBG8558895.1 | K00151 | 5-carboxymethyl-2-hydroxymuconic-semialdehyde dehydrogenase (EC:1.2.1.60) | |
| hpaG | MBG8558896.1 | K05921 | 5-oxopent-3-ene-1.2.5-tricarboxylate decarboxylase/2-hydroxyhepta-2.4-diene-1.7-dioate isomerase (EC:4.1.1.68 5.3.3.-) | |
| hpaA | MBG8558898.1 | K02508 | Arac family transcriptional regulator. 4-hydroxyphenylacetate 3-monooxygenase operon regulatory protein | |
| paaF | MBG8559324.1 | K01692 | Enoyl-coa hydratase (EC:4.2.1.17) | |
| paaX | MBG8559346.1 | K02616 | Phenylacetic acid degradation operon negative regulatory protein | |
| paaY | MBG8559347.1 | K02617 | Phenylacetic acid degradation protein | |
| paaG | MBG8559349.1 | K15866 | 2-(1.2-epoxy-1.2-dihydrophenyl)acetyl-coa isomerase (EC:5.3.3.18) | |
| paaH | MBG8559350.1 | K00074 | 3-hydroxybutyryl-coa dehydrogenase (EC:1.1.1.157) | |
| paaI | MBG8559351.1 | K02614 | Acyl-coa thioesterase (EC:3.1.2.-) | |
| paaK | MBG8559353.1 | K01912 | Phenylacetate-coa ligase (EC:6.2.1.30) | |
| paaA | MBG8559354.1 | K02609 | Ring-1.2-phenylacetyl-coa epoxidase subunit paaa (EC:1.14.13.149) | |
| paaB | MBG8559355.1 | K02610 | Ring-1.2-phenylacetyl-coa epoxidase subunit paab | |
| paaC | MBG8559356.1 | K02611 | Ring-1.2-phenylacetyl-coa epoxidase subunit paac (EC:1.14.13.149) | |
| paaD | MBG8559357.1 | K02612 | Ring-1.2-phenylacetyl-coa epoxidase subunit paad | |
| paaE | MBG8559358.1 | K02613 | Ring-1.2-phenylacetyl-coa epoxidase subunit paae | |
| paaZ | MBG8559362.1 | K02618 | Oxepin-coa hydrolase/3-oxo-5.6-dehydrosuberyl-coa semialdehyde dehydrogenase (EC:3.3.2.12 1.2.1.91) | |
| etbAa; | MBG8559252.1 | K14748 | Ethylbenzene dioxygenase subunit alpha (EC:1.14.12.-) | |
| etbAb; | MBG8559253.1 | K14749 | Ethylbenzene dioxygenase subunit beta (EC:1.14.12.-) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chlebek, D.; Płociniczak, T.; Gobetti, S.; Kumor, A.; Hupert-Kocurek, K.; Pacwa-Płociniczak, M. Analysis of the Genome of the Heavy Metal Resistant and Hydrocarbon-Degrading Rhizospheric Pseudomonas qingdaonensis ZCR6 Strain and Assessment of Its Plant-Growth-Promoting Traits. Int. J. Mol. Sci. 2022, 23, 214. https://doi.org/10.3390/ijms23010214
Chlebek D, Płociniczak T, Gobetti S, Kumor A, Hupert-Kocurek K, Pacwa-Płociniczak M. Analysis of the Genome of the Heavy Metal Resistant and Hydrocarbon-Degrading Rhizospheric Pseudomonas qingdaonensis ZCR6 Strain and Assessment of Its Plant-Growth-Promoting Traits. International Journal of Molecular Sciences. 2022; 23(1):214. https://doi.org/10.3390/ijms23010214
Chicago/Turabian StyleChlebek, Daria, Tomasz Płociniczak, Sara Gobetti, Agata Kumor, Katarzyna Hupert-Kocurek, and Magdalena Pacwa-Płociniczak. 2022. "Analysis of the Genome of the Heavy Metal Resistant and Hydrocarbon-Degrading Rhizospheric Pseudomonas qingdaonensis ZCR6 Strain and Assessment of Its Plant-Growth-Promoting Traits" International Journal of Molecular Sciences 23, no. 1: 214. https://doi.org/10.3390/ijms23010214
APA StyleChlebek, D., Płociniczak, T., Gobetti, S., Kumor, A., Hupert-Kocurek, K., & Pacwa-Płociniczak, M. (2022). Analysis of the Genome of the Heavy Metal Resistant and Hydrocarbon-Degrading Rhizospheric Pseudomonas qingdaonensis ZCR6 Strain and Assessment of Its Plant-Growth-Promoting Traits. International Journal of Molecular Sciences, 23(1), 214. https://doi.org/10.3390/ijms23010214







