Localization and Absolute Quantification of Dopamine in Discrete Intravesicular Compartments Using NanoSIMS Imaging
Abstract
:1. Introduction
2. Results and Discussion
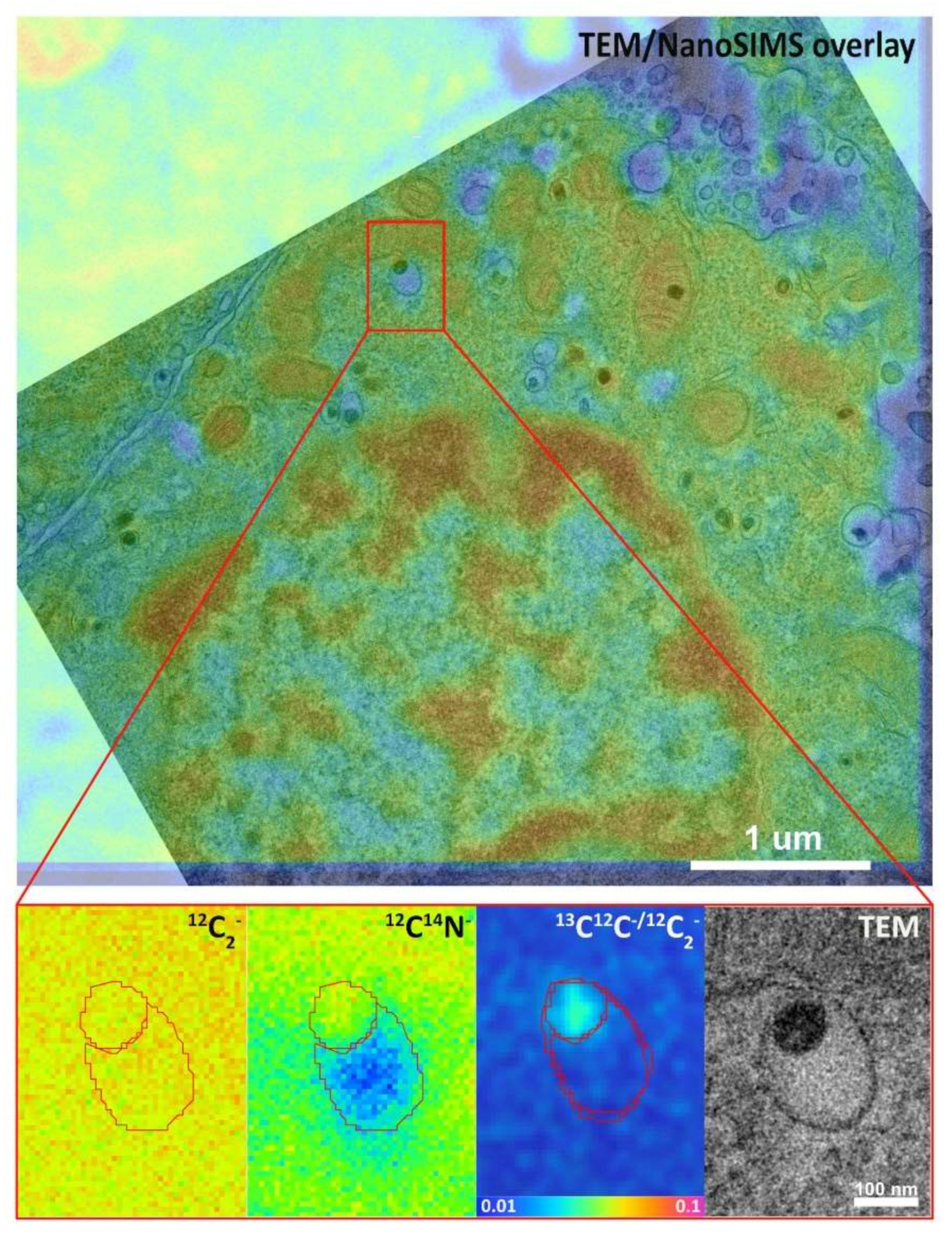
2.1. Imaging Workflow and NanoSIMS Parameters
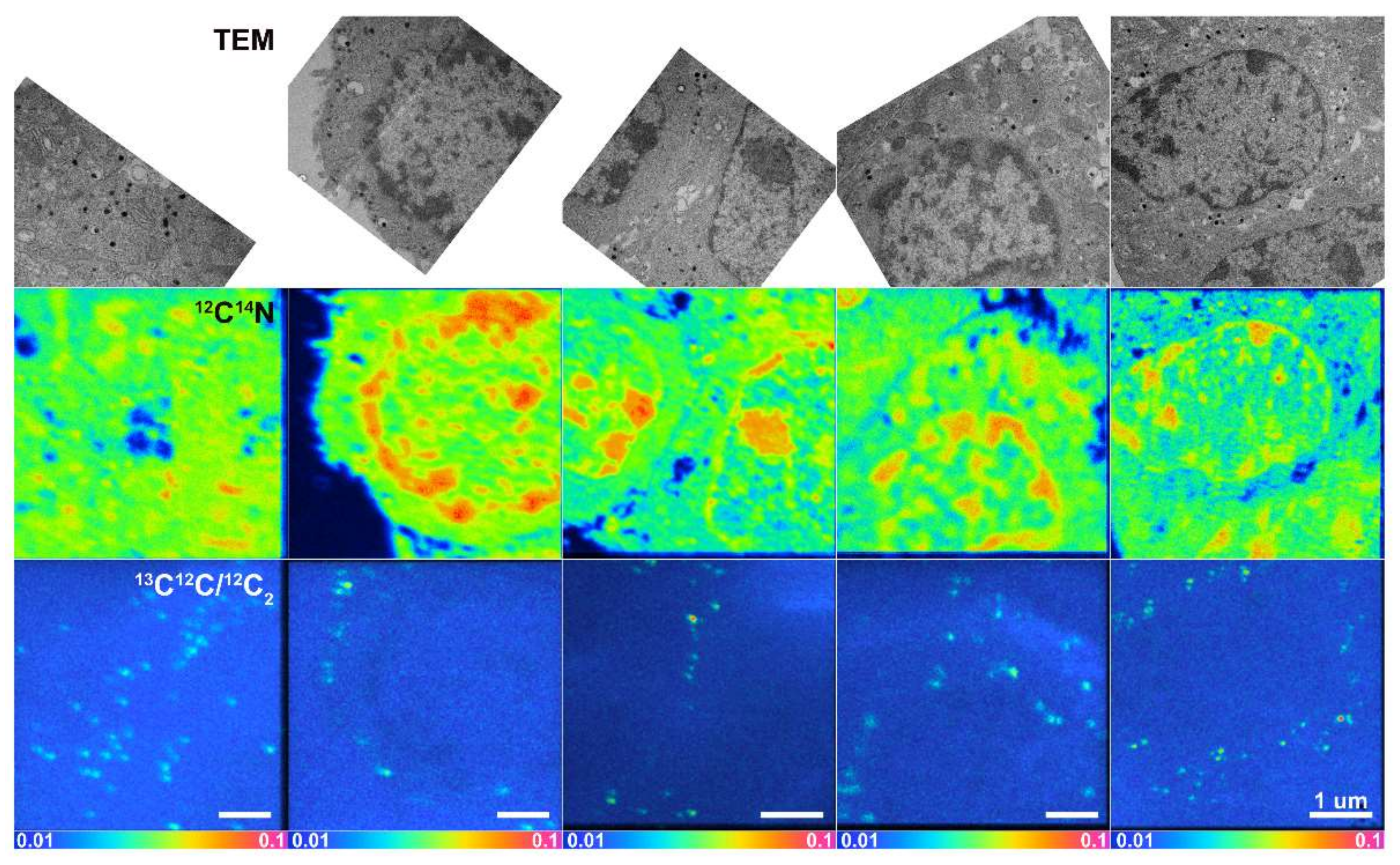
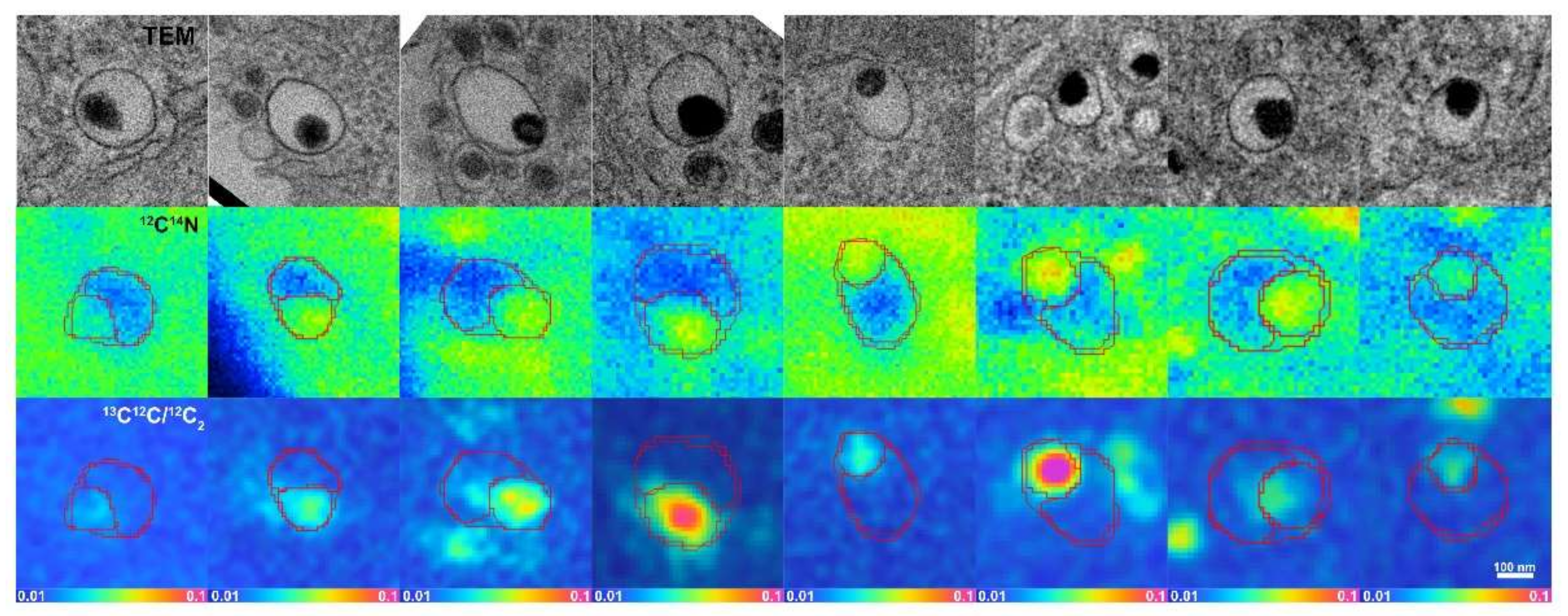
2.2. 12C14N Compartmentalization in Relation to Distinct Vesicular Subdomains
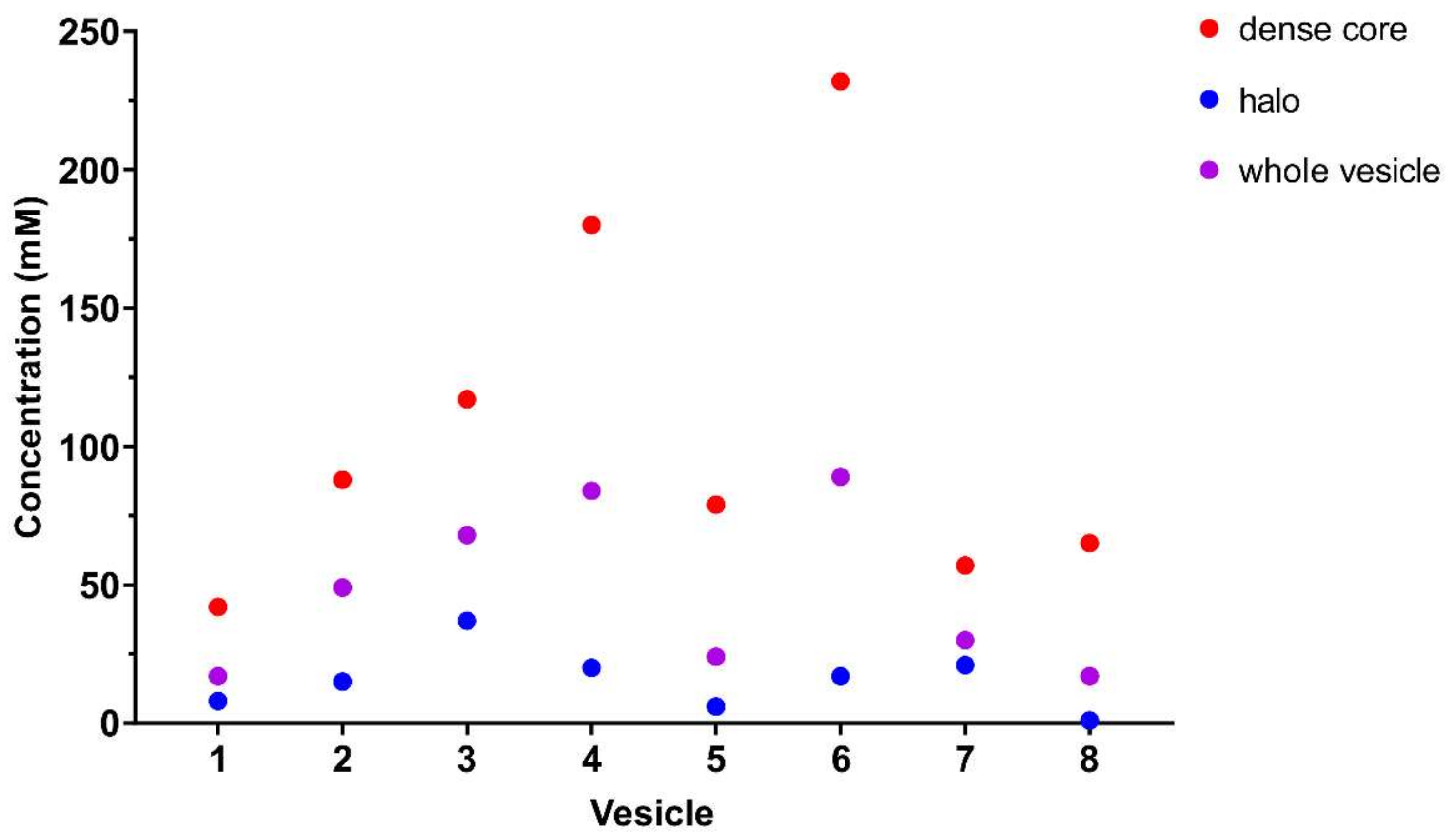
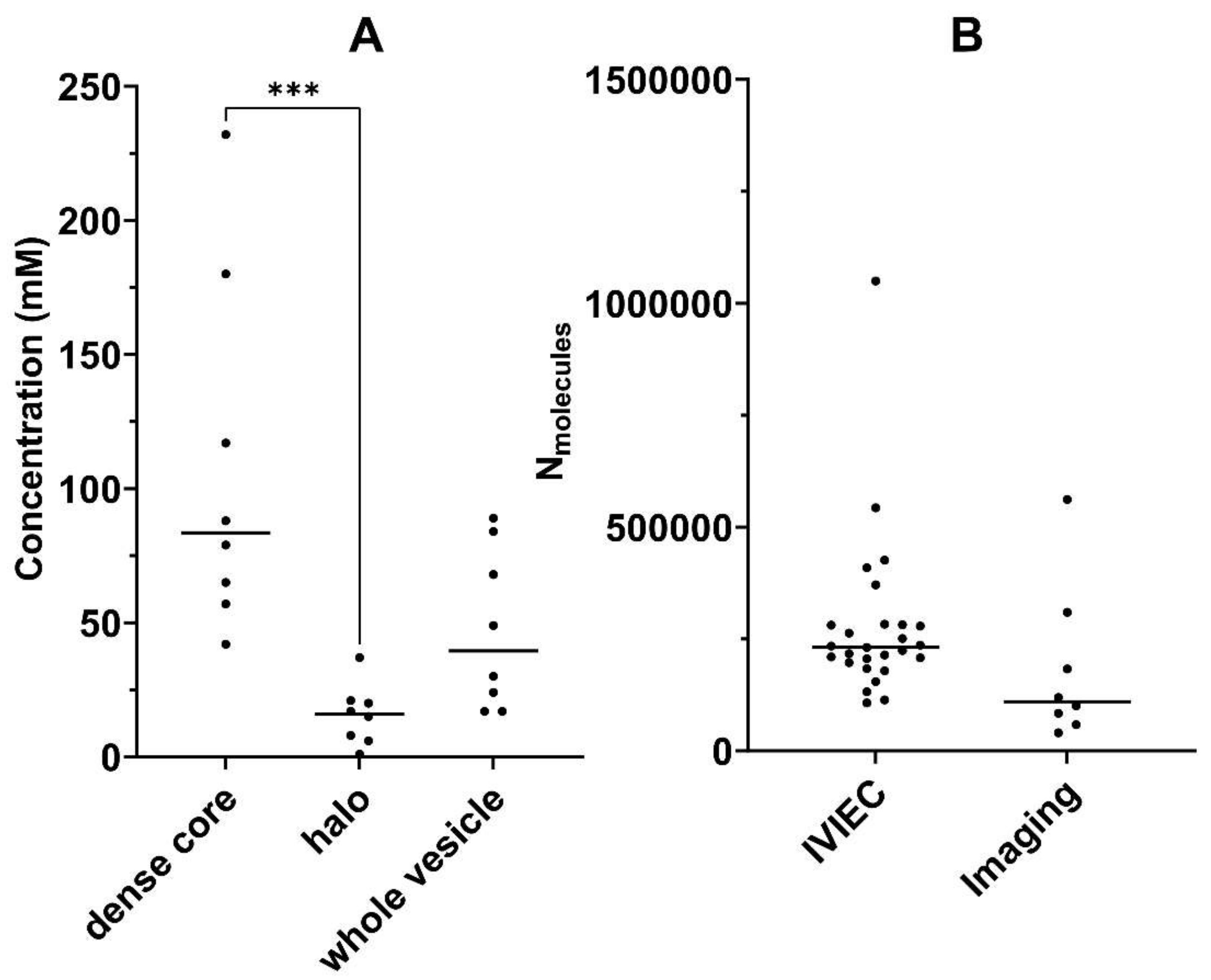
2.3. Dopamine Concentration: Dense Core versus Halo
2.4. Total Dopamine Concentration: Electrochemistry versus Imaging
3. Materials and Methods
3.1. Cell Culture
3.2. Sample Preparation
3.3. TEM
3.4. Electrode Fabrication and IVIEC Measurements
3.5. NanoSIMS Measurements
3.6. Data Analysis and Statistics for IVIEC Experiments
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Maurer, T.S.; Smith, D.; Beaumont, K.; Di, L. Dose Predictions for Drug Design. J. Med. Chem. 2020, 63, 6423–6435. [Google Scholar] [CrossRef] [PubMed]
- Hann, M.M.; Simpson, G.L. Intracellular drug concentration and disposition—The missing link? Methods 2014, 68, 283–285. [Google Scholar] [CrossRef]
- Rawat, A.; Vaidya, B.; Khatri, K. Targeted intracellular delivery of therapeutics: An overview. Pharmazie 2007, 62, 643–658. [Google Scholar]
- Harisa, G.I.; Faris, T.M. Direct Drug Targeting into Intracellular Compartments: Issues, Limitations, and Future Outlook. J. Membr. Biol. 2019, 252, 527–539. [Google Scholar] [CrossRef]
- Rosenkranz, A.A.; Ulasov, A.v.; Slastnikova, T.A.; Khramtsov, Y.v.; Sobolev, A.S. Use of intracellular transport processes for targeted drug delivery into a specified cellular compartment. Biochemistry 2014, 79, 928–946. [Google Scholar] [CrossRef] [PubMed]
- Benhamou, R.I.; Bibi, M.; Berman, J.; Fridman, M. Localizing Antifungal Drugs to the Correct Organelle Can Markedly Enhance their Efficacy. Angew. Chem. Int. Ed. 2018, 57, 6230–6235. [Google Scholar] [CrossRef]
- Yin, Q.; Pan, A.; Chen, B.; Wang, Z.; Tang, M.; Yan, Y.; Wang, Y.; Xia, H.; Chen, W.; Du, H.; et al. Quantitative Imaging of Intracellular Nanoparticle Exposure Enables Prediction of Nanotherapeutic Efficacy. Nat. Commun. 2021, 12, 2385. [Google Scholar] [CrossRef]
- Mateus, A.; Gordon, L.J.; Wayne, G.J.; Almqvist, H.; Axelsson, H.; Seashore-Ludlow, B.; Treyer, A.; Matsson, P.; Lundbäck, T.; West, A.; et al. Prediction of intracellular exposure bridges the gap between target- and cell-based drug discovery. Proc. Natl. Acad. Sci. USA 2017, 114, E6231–E6239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, R.; Lillard, J.W. Nanoparticle-based targeted drug delivery. Exp. Mol. Pathol. 2009, 86, 215–223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reinholz, J.; Diesler, C.; Schöttler, S.; Kokkinopoulou, M.; Ritz, S.; Landfester, K.; Mailänder, V. Protein machineries defining pathways of nanocarrier exocytosis and transcytosis. Acta Biomater. 2018, 71, 432–443. [Google Scholar] [CrossRef]
- Majumder, J.; Taratula, O.; Minko, T. Nanocarrier-based systems for targeted and site specific therapeutic delivery. Adv. Drug Deliv. Rev. 2019, 144, 57–77. [Google Scholar] [CrossRef]
- Westerink, R.H.S.; Ewing, A.G. The PC12 cell as model for neurosecretion. Acta Physiol. 2008, 192, 273–285. [Google Scholar] [CrossRef] [Green Version]
- Colliver, T.L.; Pyott, S.J.; Achalabun, M.; Ewing, A.G. VMAT-Mediated Changes in Quantal Size and Vesicular Volume. J. Neurosci. 2000, 20, 5276–5282. [Google Scholar] [CrossRef] [Green Version]
- Ren, L.; Oleinick, A.; Svir, I.; Amatore, C.; Ewing, A.G. Amperometric Measurements and Dynamic Models Reveal a Mechanism for How Zinc Alters Neurotransmitter Release. Angew. Chem. Int. Ed. 2020, 59, 3083–3087. [Google Scholar] [CrossRef] [PubMed]
- Fathali, H.; Dunevall, J.; Majdi, S.; Cans, A.-S. Extracellular Osmotic Stress Reduces the Vesicle Size while Keeping a Constant Neurotransmitter Concentration. ACS Chem. Neurosci. 2017, 8, 368–375. [Google Scholar] [CrossRef]
- Machado, J.D.; Camacho, M.; Alvarez, J.; Borges, R. On the role of intravesicular calcium in the motion and exocytosis of secretory organelles. Commun. Integr. Biol. 2009, 2, 71–73. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.Z.; Wen, J.; Raid, A.G.; Chuang, V. Studies of the Rate Constant of L-DOPA Oxidation and Decarboxylation by HPLC. Chromatographia 2012, 75, 597–606. [Google Scholar] [CrossRef]
- Jin, C.M.; Yang, Y.J.; Huang, H.S.; Lim, S.C.; Kai, M.; Lee, M.K. Induction of dopamine biosynthesis by l-DOPA in PC12 cells: Implications of l-DOPA influx and cyclic AMP. Eur. J. Pharmacol. 2008, 591, 88–95. [Google Scholar] [CrossRef]
- El-Kordy, E.A.; Alshahrani, A.M. Effect of genistein, a natural soy isoflavone, on pancreatic β-cells of streptozotocin-induced diabetic rats: Histological and immunohistochemical study. J. Microsc. Ultrastruct. 2015, 3, 108–119. [Google Scholar] [CrossRef] [Green Version]
- Winey, M.; Meehl, J.B.; O’Toole, E.T.; Giddings, T.H., Jr. Conventional transmission electron microscopy. Mol. Biol. Cell. 2014, 25, 319–323. [Google Scholar] [CrossRef]
- Hoppe, P.; Cohen, S.; Meibom, A. NanoSIMS: Technical Aspects and Applications in Cosmochemistry and Biological Geochemistry. Geostand. Geoanal. Res. 2013, 37, 111–154. [Google Scholar] [CrossRef]
- Thomen, A.; Najafinobar, N.; Penen, F.; Kay, E.; Upadhyay, P.P.; Li, X.; Phan, N.T.N.; Malmberg, P.; Klarqvist, M.; Andersson, S.; et al. Subcellular Mass Spectrometry Imaging and Absolute Quantitative Analysis across Organelles. ACS Nano 2020, 14, 4316–4325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jurchen, J.C.; Rubakhin, S.S.; Sweedler, J.v. MALDI-MS Imaging of Features Smaller than the size of the laser beam. J. Am. Soc. Mass Spectrom. 2005, 16, 1654–1659. [Google Scholar] [CrossRef] [Green Version]
- Nazari, M.; Muddiman, D.C. Cellular-level mass spectrometry imaging using infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI) by oversampling. Anal. Bioanal. Chem. 2014, 407, 2265–2271. [Google Scholar] [CrossRef]
- Yue, L.; Bindesh, S.; Vertes, A. Atmospheric Pressure Molecular Imaging by Infrared MALDI Mass Spectrometry. Anal. Chem. 2006, 79, 523–532. [Google Scholar]
- Duncan, K.D.; Lanekoff, I. Oversampling to Improve Spatial Resolution for Liquid Extraction Mass Spectrometry Imaging. Anal. Chem. 2018, 90, 2451–2455. [Google Scholar] [CrossRef] [PubMed]
- Maimó-Barceló, A.; Garate, J.; Bestard-Escalas, J.; Fernández, R.A.; Berthold, L.; Lopez, D.H.; Fernandez, J.; Barceló-Coblijn, G. Confirmation of sub-cellular resolution using oversampling imaging mass spectrometry. Anal. Bioanal. Chem. 2019, 411, 7935–7941. [Google Scholar] [CrossRef]
- Lechene, C.; Hillion, F.; McMahon, G.; Benson, D.; Kleinfeld, A.M.; Kampf, J.P.; Distel, D.; Luyten, Y.; Bonventre, J.; Hentschel, D.; et al. High-resolution quantitative imaging of mammalian and bacterial cells using stable isotope mass spectrometry. J. Biol. 2006, 5, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Craig, H. Isotopic standards for carbon and oxygen and correction factors for mass-spectrometric analysis of carbon dioxide. Geochim. Cosmochim. Acta 1957, 12, 133–149. [Google Scholar] [CrossRef]
- Lovrić, J.; Dunevall, J.; Larsson, A.; Ren, L.; Andersson, S.; Meibom, A.; Malmberg, P.; Kurczy, M.E.; Ewing, A.G. Nano Secondary Ion Mass Spectrometry Imaging of Dopamine Distribution Across Nanometer Vesicles. ACS Nano 2017, 11, 3446–3455. [Google Scholar] [CrossRef]
- Li, X.; Dunevall, J.; Ewing, A.G. Electrochemical quantification of transmitter concentration in single nanoscale vesicles isolated from PC12 cells. Faraday Discuss. 2018, 210, 353–364. [Google Scholar] [CrossRef]
- Li, X.; Majdi, S.; Dunevall, J.; Fathali, H.; Ewing, A.G. Quantitative Measurement of Transmitters in Individual Vesicles in the Cytoplasm of Single Cells with Nanotip Electrodes. Angew. Chem. Int. Ed. 2015, 54, 11978–11982. [Google Scholar] [CrossRef] [PubMed]
- Phan, N.T.N.; Li, X.; Ewing, A.G. Measuring synaptic vesicles using cellular electrochemistry and nanoscale molecular imaging. Nat. Rev. Chem. 2017, 1, 48. [Google Scholar] [CrossRef]
- Wang, Y.; Ewing, A.G. Electrochemical quantification of neurotransmitters in single live cell vesicles shows exocytosis is predominantly partial. ChemBioChem 2021, 22, 807. [Google Scholar] [CrossRef] [PubMed]
- Banker, G.; Goslin, K. Culturing Nerve Cells; MIT Press: Boston, MA, USA, 1998. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rabasco, S.; Nguyen, T.D.K.; Gu, C.; Kurczy, M.E.; Phan, N.T.N.; Ewing, A.G. Localization and Absolute Quantification of Dopamine in Discrete Intravesicular Compartments Using NanoSIMS Imaging. Int. J. Mol. Sci. 2022, 23, 160. https://doi.org/10.3390/ijms23010160
Rabasco S, Nguyen TDK, Gu C, Kurczy ME, Phan NTN, Ewing AG. Localization and Absolute Quantification of Dopamine in Discrete Intravesicular Compartments Using NanoSIMS Imaging. International Journal of Molecular Sciences. 2022; 23(1):160. https://doi.org/10.3390/ijms23010160
Chicago/Turabian StyleRabasco, Stefania, Tho D. K. Nguyen, Chaoyi Gu, Michael E. Kurczy, Nhu T. N. Phan, and Andrew G. Ewing. 2022. "Localization and Absolute Quantification of Dopamine in Discrete Intravesicular Compartments Using NanoSIMS Imaging" International Journal of Molecular Sciences 23, no. 1: 160. https://doi.org/10.3390/ijms23010160
APA StyleRabasco, S., Nguyen, T. D. K., Gu, C., Kurczy, M. E., Phan, N. T. N., & Ewing, A. G. (2022). Localization and Absolute Quantification of Dopamine in Discrete Intravesicular Compartments Using NanoSIMS Imaging. International Journal of Molecular Sciences, 23(1), 160. https://doi.org/10.3390/ijms23010160







