Candida Administration in Bilateral Nephrectomy Mice Elevates Serum (1→3)-β-D-glucan That Enhances Systemic Inflammation Through Energy Augmentation in Macrophages
Abstract
1. Introduction
2. Results
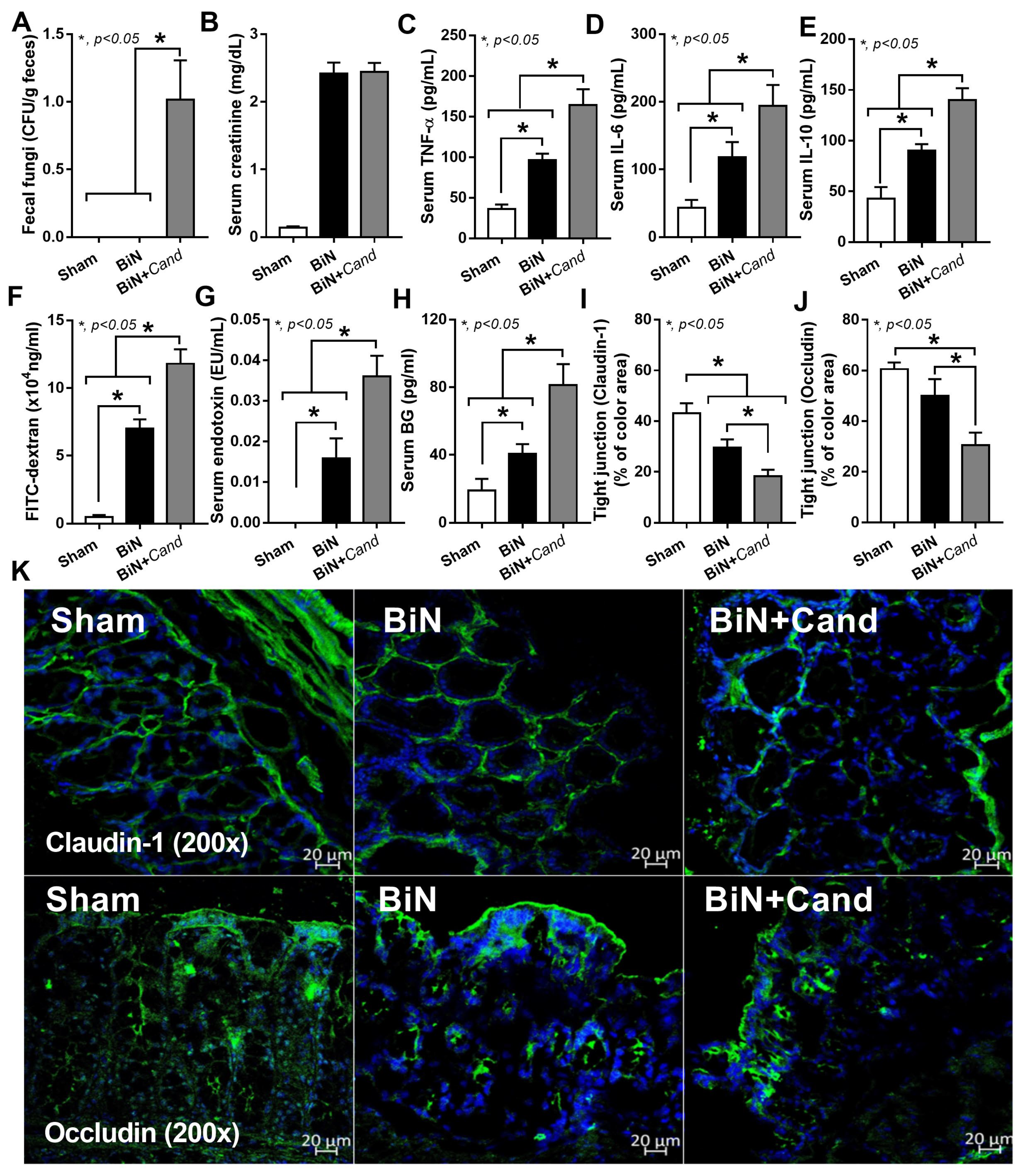
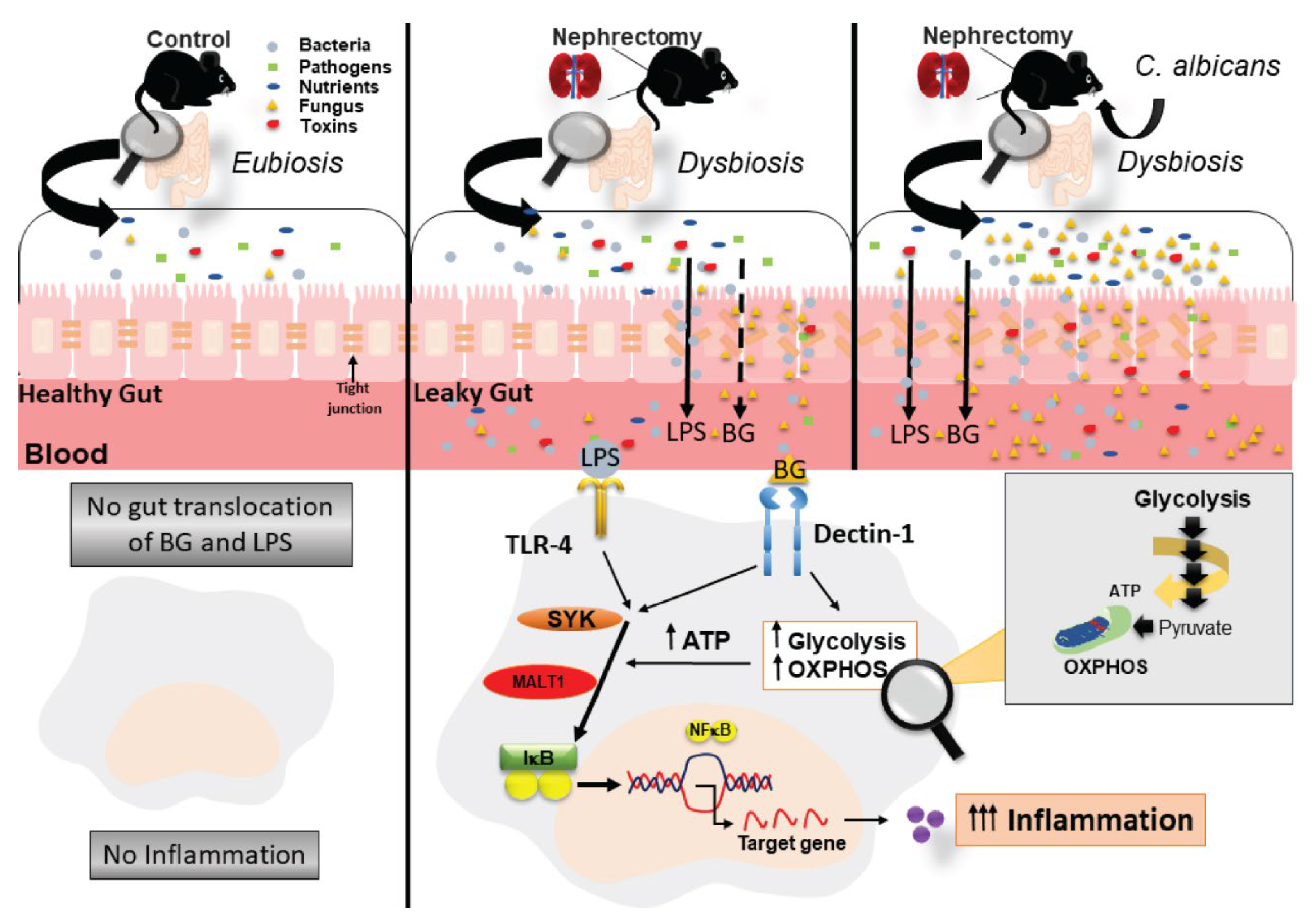
2.1. Candida Administration Worsened Uremia-Induced Systemic Inflammation through the Induction of Gut Dysbiosis and Gut Translocation of LPS and BG in Bilateral Nephrectomy Mice
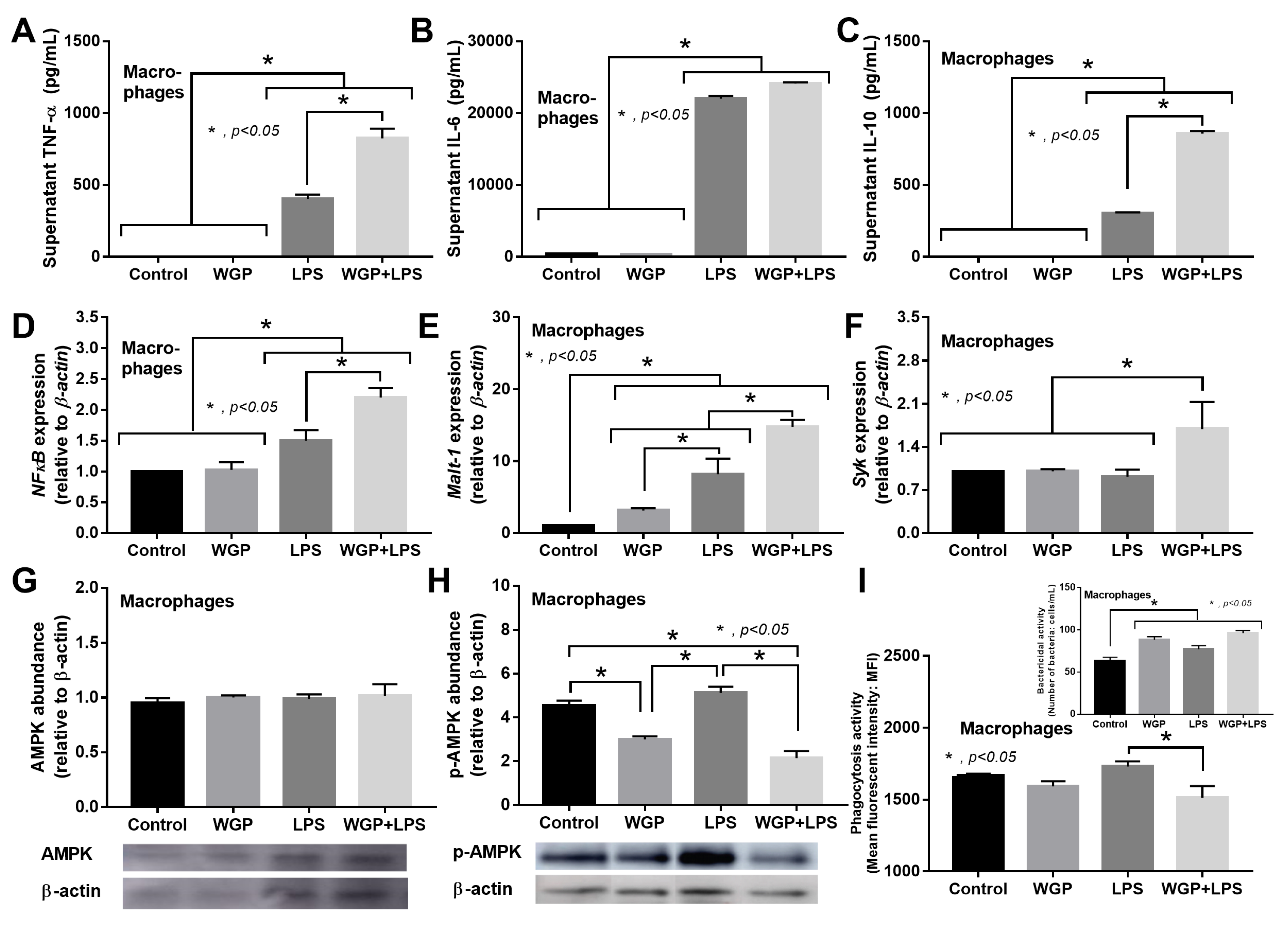
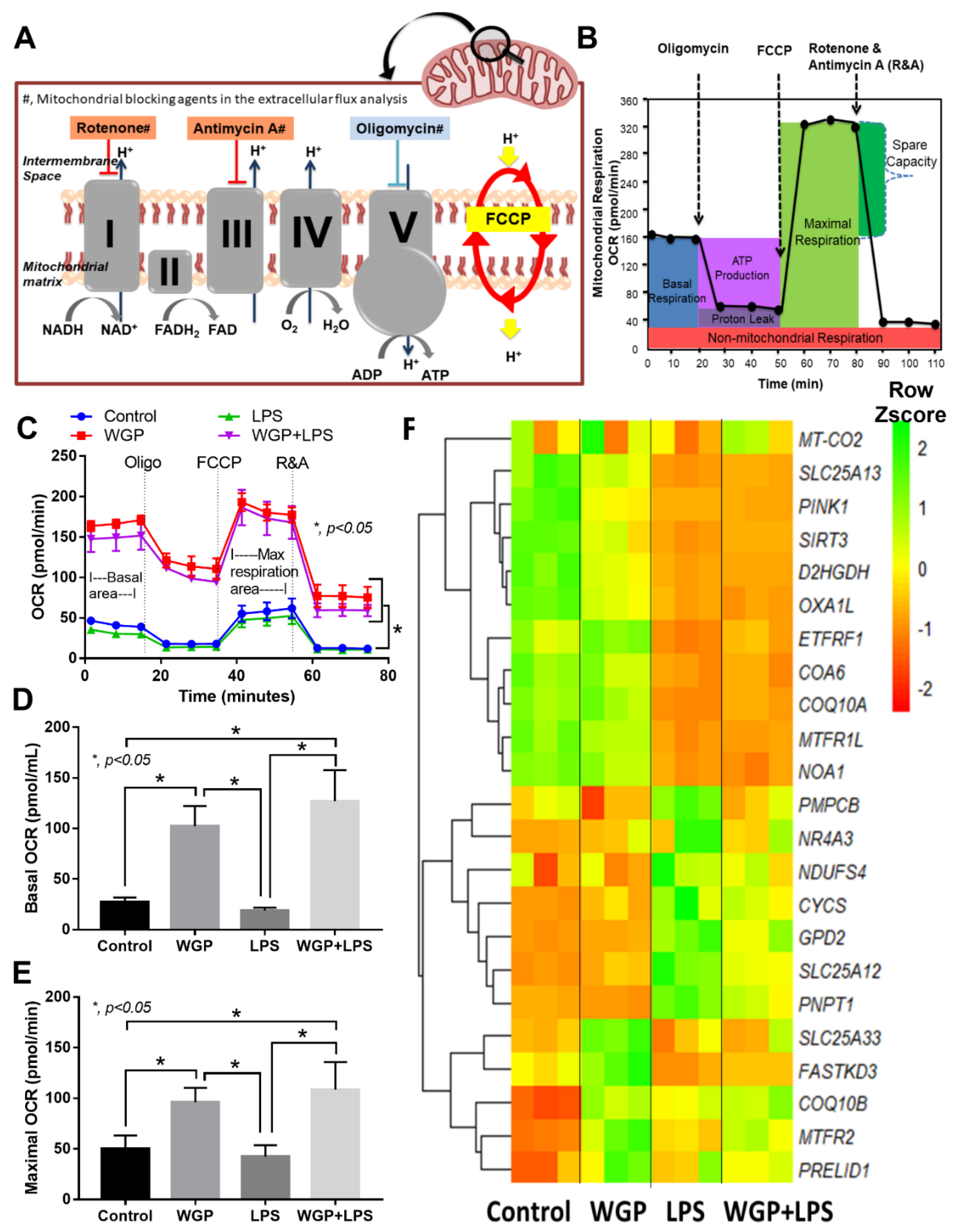
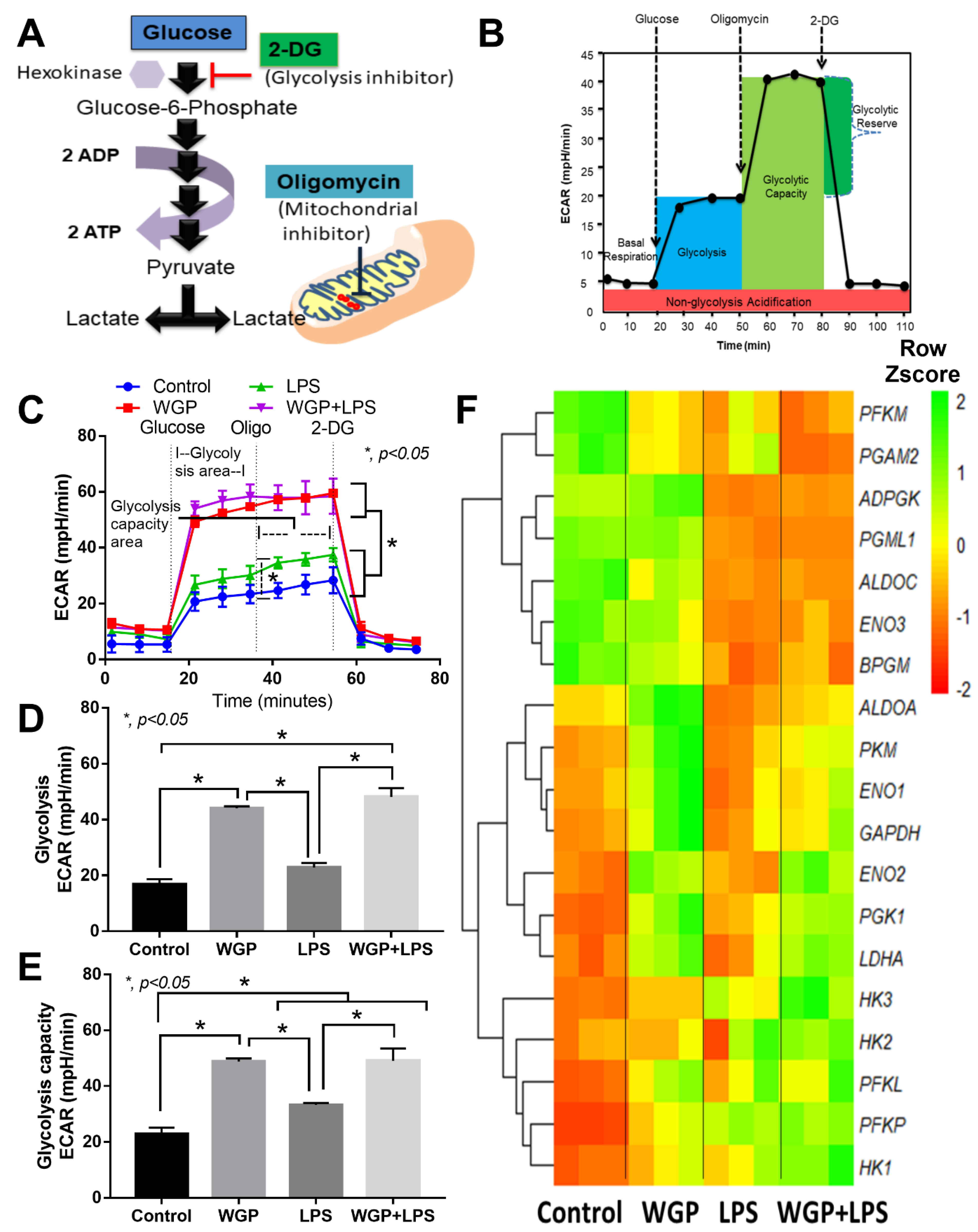
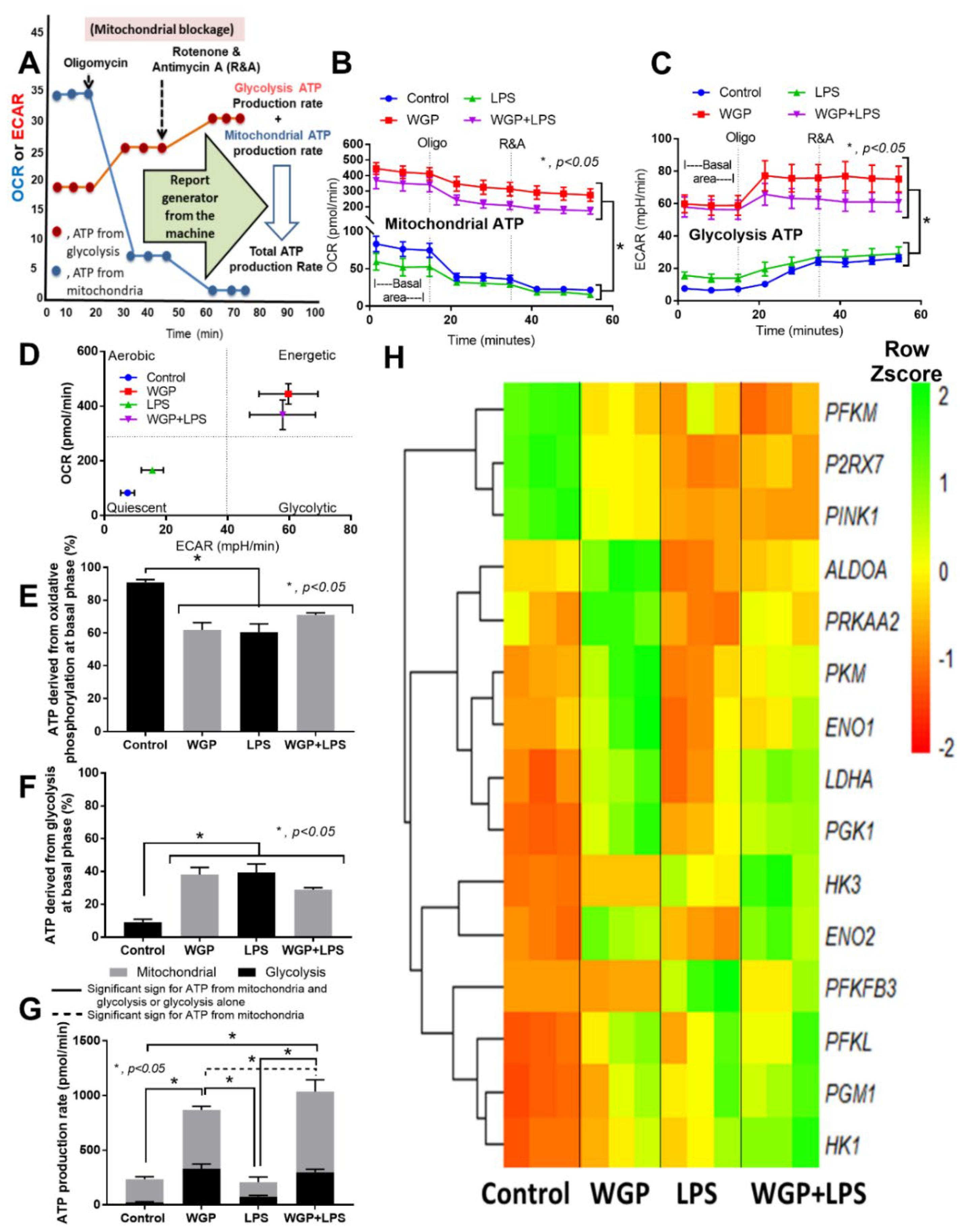
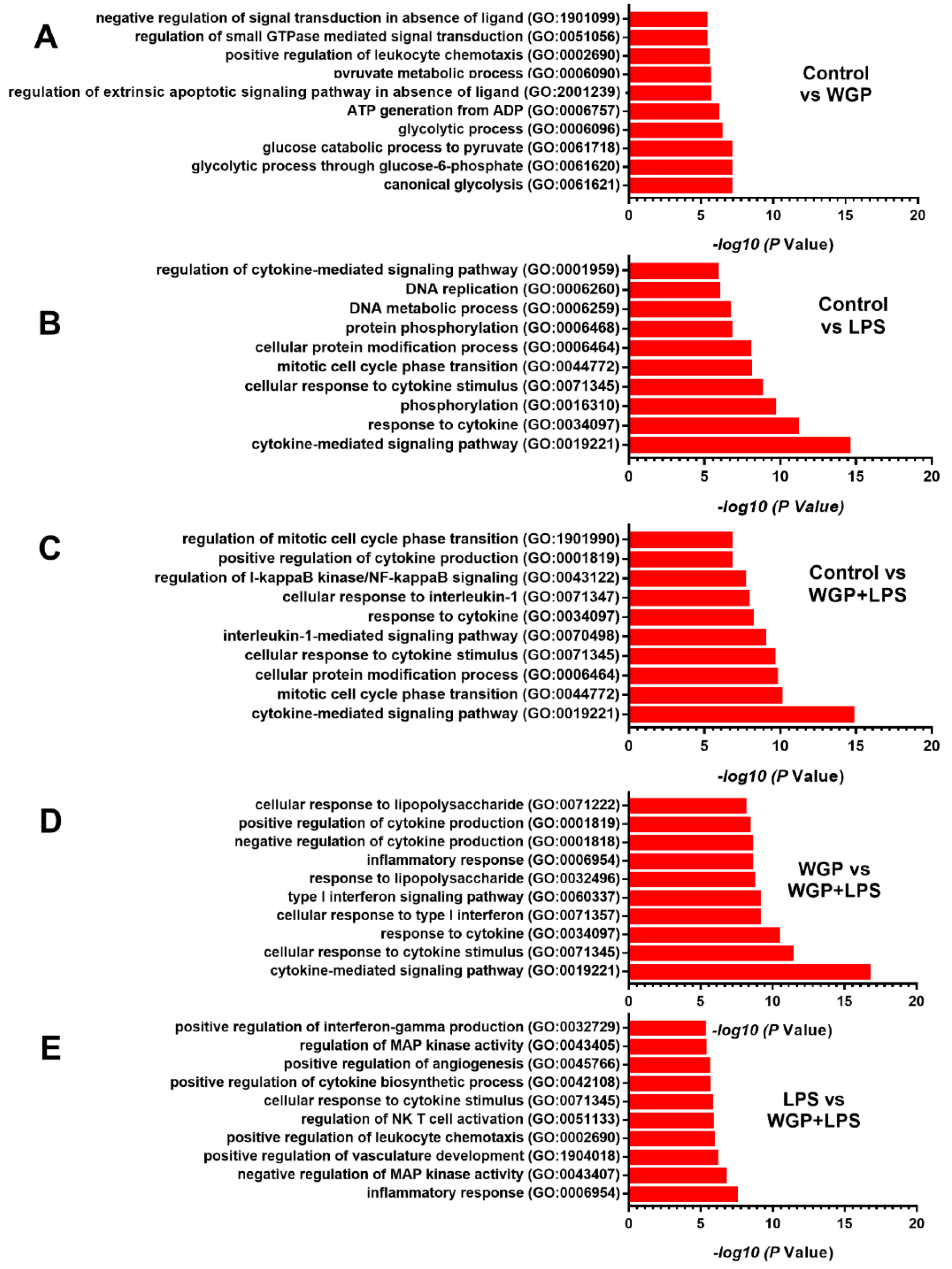
2.2. The Acceleration of Cell Energy in Macrophages by BG, an Impact on the Enhanced Pro-Inflammatory Responses
3. Discussion
3.1. Gut Dysbiosis in Candida Administered Mice with Bilateral Nephrectomy Induced Gut Permeability Defect and the Translocation of Organismal Molecules from Intestinal Contents
3.2. Prominent Macrophage Inflammatory Responses to LPS with BG, When Compared with LPS Alone, an Impact of BG-Induced Cell Energy Augmentation
3.3. Clinical Translation and Utilization of BG for Macrophage Manipulation
4. Materials and Methods
4.1. Animals and Animal Model
4.2. Mouse Sample Analysis and Immunofluorescence
4.3. Fecal Microbiome Analysis
4.4. Macrophage Cytokines and Western Blot Analysis
4.5. Real-Time Polymerase Chain Reaction and RNA Sequencing Analysis
4.6. Macrophage Phagocytosis and Bactericidal Activity
4.7. Extracellular Flux Analysis
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wang, H.E.; Muntner, P.; Chertow, G.M.; Warnock, D.G. Acute kidney injury and mortality in hospitalized patients. Am. J. Nephrol. 2012, 35, 349–355. [Google Scholar] [CrossRef]
- Cohen, G.; Horl, W.H. Immune dysfunction in uremia—An update. Toxins 2012, 4, 962–990. [Google Scholar] [CrossRef] [PubMed]
- Leelahavanichkul, A.; Huang, Y.; Hu, X.; Zhou, H.; Tsuji, T.; Chen, R.; Kopp, J.B.; Schnermann, J.; Yuen, P.S.; Star, R.A. Chronic kidney disease worsens sepsis and sepsis—induced acute kidney injury by releasing High Mobility Group Box Protein-1. Kidney Int. 2011, 80, 1198–1211. [Google Scholar] [CrossRef]
- Leelahavanichkul, A.; Somparn, P.; Issara-Amphorn, J.; Eiam-ong, S.; Avihingsanon, Y.; Hirankarn, N.; Srisawat, N. Serum Neutrophil Gelatinase Associated Lipocalin (NGAL) Outperforms Serum Creatinine in Detecting Sepsis-Induced Acute Kidney Injury, Experiments on Bilateral Nephrectomy and Bilateral Ureter Obstruction Mouse Models. Shock 2016, 45, 570–576. [Google Scholar] [CrossRef] [PubMed]
- Amornphimoltham, P.; Yuen, P.S.T.; Star, R.A.; Leelahavanichkul, A. Gut Leakage of Fungal-Derived Inflammatory Mediators: Part of a Gut-Liver-Kidney Axis in Bacterial Sepsis. Dig. Dis. Sci. 2019, 64, 2416–2428. [Google Scholar] [CrossRef] [PubMed]
- Helander, H.F.; Fandriks, L. Surface area of the digestive tract—Revisited. Scand. J. Gastroenterol. 2014, 49, 681–689. [Google Scholar] [CrossRef]
- Visitchanakun, P.; Saisorn, W.; Wongphoom, J.; Chatthanathon, P.; Somboonna, N.; Svasti, S.; Fucharoen, S.; Leelahavanichkul, A. Gut leakage enhances sepsis susceptibility in iron-overloaded beta-thalassemia mice through macrophage hyperinflammatory responses. Am. J. Physiol. Gastrointest. Liver Physiol. 2020, 318, G966–G979. [Google Scholar] [CrossRef]
- Sae-Khow, K.; Charoensappakit, A.; Visitchanakun, P.; Saisorn, W.; Svasti, S.; Fucharoen, S.; Leelahavanichkul, A. Pathogen-Associated Molecules from Gut Translocation Enhance Severity of Cecal Ligation and Puncture Sepsis in Iron-Overload beta-Thalassemia Mice. J. Inflamm. Res. 2020, 13, 719–735. [Google Scholar] [CrossRef]
- Thim-Uam, A.; Surawut, S.; Issara-Amphorn, J.; Jaroonwitchawan, T.; Hiengrach, P.; Chatthanathon, P.; Wilantho, A.; Somboonna, N.; Palaga, T.; Pisitkun, P.; et al. Leaky-gut enhanced lupus progression in the Fc gamma receptor-IIb deficient and pristane-induced mouse models of lupus. Sci. Rep. 2020, 10, 777. [Google Scholar] [CrossRef] [PubMed]
- Bhunyakarnjanarat, T.; Udompornpitak, K.; Saisorn, W.; Chantraprapawat, B.; Visitchanakun, P.; Dang, C.P.; Issara-Amphorn, J.; Leelahavanichkul, A. Prominent Indomethacin-Induced Enteropathy in Fcgriib Defi-cient lupus Mice: An Impact of Macrophage Responses and Immune Deposition in Gut. Int. J. Mol. Sci. 2021, 22, 1377. [Google Scholar] [CrossRef] [PubMed]
- Panpetch, W.; Sawaswong, V.; Chanchaem, P.; Ondee, T.; Dang, C.P.; Payungporn, S.; Leelahavanichkul, A. Candida Administration Worsens Cecal Ligation and Puncture-Induced Sepsis in Obese Mice Through Gut Dysbiosis Enhanced Systemic Inflammation, Impact of Pathogen-Associated Molecules From Gut Translocation and Saturated Fatty Acid. Front. Immunol. 2020, 11, 561652. [Google Scholar] [CrossRef]
- Nisbeth, U.; Hallgren, R.; Eriksson, O.; Danielson, B.G. Endotoxemia in chronic renal failure. Nephron 1987, 45, 93–97. [Google Scholar] [CrossRef]
- Thongprayoon, C.; Kaewput, W.; Hatch, S.T.; Bathini, T.; Sharma, K.; Wijarnpreecha, K.; Ungprasert, P.; D’Costa, M.; Mao, M.A.; Cheungpasitporn, W. Effects of Probiotics on Inflammation and Uremic Toxins Among Patients on Dialysis: A Systematic Review and Meta-Analysis. Dig. Dis. Sci. 2019, 64, 469–479. [Google Scholar] [CrossRef]
- Samonis, G.; Gikas, A.; Toloudis, P.; Maraki, S.; Vrentzos, G.; Tselentis, Y.; Tsaparas, N.; Bodey, G. Prospective study of the impact of broad-spectrum antibiotics on the yeast flora of the human gut. Eur. J. Clin. Microbiol. Infect. Dis. 1994, 13, 665–667. [Google Scholar] [CrossRef]
- Eggimann, P.; Pittet, D. Candida colonization index and subsequent infection in critically ill surgical patients: 20 years later. Intensive Care Med. 2014, 40, 1429–1448. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.M.; Inamine, T.; Hochrath, K.; Chen, P.; Wang, L.; Llorente, C.; Bluemel, S.; Hartmann, P.; Xu, J.; Koyama, Y.; et al. Intestinal fungi contribute to development of alcoholic liver disease. J. Clin. Investig. 2017, 127, 2829–2841. [Google Scholar] [CrossRef]
- Saltzman, E.T.; Palacios, T.; Thomsen, M.; Vitetta, L. Intestinal Microbiome Shifts, Dysbiosis, Inflammation, and Non-alcoholic Fatty Liver Disease. Front. Microbiol. 2018, 9, 61. [Google Scholar] [CrossRef]
- Lang, S.; Duan, Y.; Liu, J.; Torralba, M.G.; Kuelbs, C.; Ventura-Cots, M.; Abraldes, J.G.; Bosques-Padilla, F.; Verna, E.C.; Brown, R.S., Jr.; et al. Intestinal Fungal Dysbiosis and Systemic Immune Response to Fungi in Patients With Alcoholic Hepatitis. Hepatology 2020, 71, 522–538. [Google Scholar] [CrossRef] [PubMed]
- Panpetch, W.; Somboonna, N.; Bulan, D.E.; Issara-Amphorn, J.; Finkelman, M.; Worasilchai, N.; Chindamporn, A.; Palaga, T.; Tumwasorn, S.; Leelahavanichkul, A. Oral administration of live- or heat-killed Candida albicans worsened cecal ligation and puncture sepsis in a murine model possibly due to an increased serum (1→3)-beta-D-glucan. PLoS ONE 2017, 12, e0181439. [Google Scholar] [CrossRef]
- Panpetch, W.; Hiengrach, P.; Nilgate, S.; Tumwasorn, S.; Somboonna, N.; Wilantho, A.; Chatthanathon, P.; Prueksapanich, P.; Leelahavanichkul, A. Additional Candida albicans administration enhances the severity of dextran sulfate solution induced colitis mouse model through leaky gut-enhanced systemic inflammation and gut-dysbiosis but attenuated by Lactobacillus rhamnosus L34. Gut Microbes 2020, 11, 465–480. [Google Scholar] [CrossRef]
- Dennehy, K.M.; Ferwerda, G.; Faro-Trindade, I.; Pyz, E.; Willment, J.A.; Taylor, P.R.; Kerrigan, A.; Tsoni, S.V.; Gordon, S.; Meyer-Wentrup, F.; et al. Syk kinase is required for collaborative cytokine production induced through Dectin-1 and Toll-like receptors. Eur. J. Immunol. 2008, 38, 500–506. [Google Scholar] [CrossRef]
- Koh, A.Y. Murine models of Candida gastrointestinal colonization and dissemination. Eukaryot. Cell 2013, 12, 1416–1422. [Google Scholar] [CrossRef]
- Borges, F.M.; de Paula, T.O.; Sarmiento, M.R.A.; de Oliveira, M.G.; Pereira, M.L.M.; Toledo, I.V.; Nascimento, T.C.; Ferreira-Machado, A.B.; Silva, V.L.; Diniz, C.G. Fungal Diversity of Human Gut Microbiota Among Eutrophic, Overweight, and Obese Individuals Based on Aerobic Culture-Dependent Approach. Curr. Microbiol. 2018, 75, 726–735. [Google Scholar] [CrossRef]
- Meijers, B.; Farre, R.; Dejongh, S.; Vicario, M.; Evenepoel, P. Intestinal Barrier Function in Chronic Kidney Disease. Toxins 2018, 10, 298. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, N.; Kobayashi, K. Macrophages in inflammation. Curr. Drug Targets. Inflamm. Allergy 2005, 4, 281–286. [Google Scholar] [CrossRef]
- Sheikh, S.Z.; Plevy, S.E. The role of the macrophage in sentinel responses in intestinal immunity. Curr. Opin. Gastroenterol. 2010, 26, 578–582. [Google Scholar] [CrossRef] [PubMed]
- Franken, L.; Schiwon, M.; Kurts, C. Macrophages: Sentinels and regulators of the immune system. Cell. Microbiol. 2016, 18, 475–487. [Google Scholar] [CrossRef] [PubMed]
- Gordon, S.; Pluddemann, A. Tissue macrophages: Heterogeneity and functions. BMC Biol. 2017, 15, 53. [Google Scholar] [CrossRef]
- Taratummarat, S.; Sangphech, N.; Vu, C.T.B.; Palaga, T.; Ondee, T.; Surawut, S.; Sereemaspun, A.; Ritprajak, P.; Leelahavanichkul, A. Gold nanoparticles attenuates bacterial sepsis in cecal ligation and puncture mouse model through the induction of M2 macrophage polarization. BMC Microbiol. 2018, 18, 85. [Google Scholar] [CrossRef]
- Dang, C.P.; Leelahavanichkul, A. Over-expression of miR-223 induces M2 macrophage through glycolysis alteration and attenuates LPS-induced sepsis mouse model, the cell-based therapy in sepsis. PLoS ONE 2020, 15, e0236038. [Google Scholar] [CrossRef]
- Murray, P.J. Macrophage Polarization. Annu. Rev. Physiol. 2017, 79, 541–566. [Google Scholar] [CrossRef]
- O’Neill, L.A.; Kishton, R.J.; Rathmell, J. A guide to immunometabolism for immunologists. Nat. Rev. Immunol. 2016, 16, 553–565. [Google Scholar] [CrossRef]
- Domblides, C.; Lartigue, L.; Faustin, B. Metabolic Stress in the Immune Function of T Cells, Macrophages and Dendritic Cells. Cells 2018, 7, 68. [Google Scholar] [CrossRef]
- Thapa, B.; Lee, K. Metabolic influence on macrophage polarization and pathogenesis. Bmb Rep. 2019, 52, 360–372. [Google Scholar] [CrossRef]
- Ondee, T.; Gillen, J.; Visitchanakun, P.; Somparn, P.; Issara-Amphorn, J.; Dang Phi, C.; Chancharoenthana, W.; Gurusamy, D.; Nita-Lazar, A.; Leelahavanichkul, A. Lipocalin-2 (Lcn-2) Attenuates Polymicrobial Sepsis with LPS Preconditioning (LPS Tolerance) in FcGRIIb Deficient Lupus Mice. Cells 2019, 8, 1064. [Google Scholar] [CrossRef]
- Panpetch, W.; Kullapanich, C.; Dang, C.P.; Visitchanakun, P.; Saisorn, W.; Wongphoom, J.; Wannigama, D.L.; Thim-Uam, A.; Patarakul, K.; Somboonna, N.; et al. Candida Administration Worsens Uremia-Induced Gut Leakage in Bilateral Nephrectomy Mice, an Impact of Gut Fungi and Organismal Molecules in Uremia. mSystems 2021, 6, e01187. [Google Scholar] [CrossRef] [PubMed]
- Issara-Amphorn, J.; Surawut, S.; Worasilchai, N.; Thim-Uam, A.; Finkelman, M.; Chindamporn, A.; Palaga, T.; Hirankarn, N.; Pisitkun, P.; Leelahavanichkul, A. The Synergy of Endotoxin and (1→3)-beta-D-Glucan, from Gut Translocation, Worsens Sepsis Severity in a Lupus Model of Fc Gamma Receptor IIb-Deficient Mice. J. Innate Immun. 2018, 10, 189–201. [Google Scholar] [CrossRef]
- Issara-Amphorn, J.; Somboonna, N.; Pisitkun, P.; Hirankarn, N.; Leelahavanichkul, A. Syk inhibitor attenuates inflammation in lupus mice from FcgRIIb deficiency but not in pristane induction: The influence of lupus pathogenesis on the therapeutic effect. Lupus 2020, 29, 1248–1262. [Google Scholar] [CrossRef]
- Issara-Amphorn, J.; Chancharoenthana, W.; Visitchanakun, P.; Leelahavanichkul, A. Syk Inhibitor Attenuates Polymicrobial Sepsis in FcgRIIb-Deficient Lupus Mouse Model, the Impact of Lupus Characteristics in Sepsis. J. Innate Immun. 2020, 12, 461–479. [Google Scholar] [CrossRef]
- Moorlag, S.; Khan, N.; Novakovic, B.; Kaufmann, E.; Jansen, T.; van Crevel, R.; Divangahi, M.; Netea, M.G. Beta-Glucan Induces Protective Trained Immunity against Mycobacterium tuberculosis Infection: A Key Role for IL-1. Cell Rep. 2020, 31, 107634. [Google Scholar] [CrossRef]
- Bien, J.; Palagani, V.; Bozko, P. The intestinal microbiota dysbiosis and Clostridium difficile infection: Is there a relationship with inflammatory bowel disease? Ther. Adv. Gastroenterol. 2013, 6, 53–68. [Google Scholar] [CrossRef] [PubMed]
- Shin, N.R.; Whon, T.W.; Bae, J.W. Proteobacteria: Microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015, 33, 496–503. [Google Scholar] [CrossRef] [PubMed]
- Fan, K.; Lin, L.; Ai, Q.; Wan, J.; Dai, J.; Liu, G.; Tang, L.; Yang, Y.; Ge, P.; Jiang, R.; et al. Lipopolysaccharide-Induced Dephosphorylation of AMPK-Activated Protein Kinase Potentiates Inflammatory Injury via Repression of ULK1-Dependent Autophagy. Front. Immunol. 2018, 9, 1464. [Google Scholar] [CrossRef]
- Youle, R.J.; van der Bliek, A.M. Mitochondrial fission, fusion, and stress. Science 2012, 337, 1062–1065. [Google Scholar] [CrossRef]
- Cali, C.; Tauffenberger, A.; Magistretti, P. The Strategic Location of Glycogen and Lactate: From Body Energy Reserve to Brain Plasticity. Front. Cell. Neurosci. 2019, 13, 82. [Google Scholar] [CrossRef]
- Galvan-Pena, S.; O’Neill, L.A. Metabolic reprograming in macrophage polarization. Front. Immunol. 2014, 5, 420. [Google Scholar]
- Wang, N.; Zhu, P.; Huang, R.; Wang, C.; Sun, L.; Lan, B.; He, Y.; Zhao, H.; Gao, Y. PINK1: The guard of mitochondria. Life Sci. 2020, 259, 118247. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Huang, K.; Chen, Z.; Hu, M.; Bai, Y.; Lin, S.; Du, H. Characterization of Glycolysis-Associated Molecules in the Tumor Microenvironment Revealed by Pan-Cancer Tissues and Lung Cancer Single Cell Data. Cancers 2020, 12, 1788. [Google Scholar] [CrossRef]
- Tian, Y.; Huo, M.; Li, G.; Li, Y.; Wang, J. Regulation of LPS-induced mRNA expression of pro-inflammatory cytokines via alteration of NF-kappaB activity in mouse peritoneal macrophages exposed to fluoride. Chemosphere 2016, 161, 89–95. [Google Scholar] [CrossRef]
- Konieczna, I.; Zarnowiec, P.; Kwinkowski, M.; Kolesinska, B.; Fraczyk, J.; Kaminski, Z.; Kaca, W. Bacterial urease and its role in long-lasting human diseases. Curr. Protein Pept. Sci. 2012, 13, 789–806. [Google Scholar] [CrossRef] [PubMed]
- Mitch, W.E. Effects of intestinal flora on nitrogen metabolism in patients with chronic renal failure. Am. J. Clin. Nutr. 1978, 31, 1594–1600. [Google Scholar] [CrossRef]
- Wong, J.; Piceno, Y.M.; DeSantis, T.Z.; Pahl, M.; Andersen, G.L.; Vaziri, N.D. Expansion of urease- and uricase-containing, indole- and p-cresol-forming and contraction of short-chain fatty acid-producing intestinal microbiota in ESRD. Am. J. Nephrol. 2014, 39, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Vaziri, N.D.; Wong, J.; Pahl, M.; Piceno, Y.M.; Yuan, J.; DeSantis, T.Z.; Ni, Z.; Nguyen, T.H.; Andersen, G.L. Chronic kidney disease alters intestinal microbial flora. Kidney Int. 2013, 83, 308–315. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Ma, L.; Fu, P. Gut microbiota-derived short-chain fatty acids and kidney diseases. Drug Des. Dev. Ther. 2017, 11, 3531–3542. [Google Scholar] [CrossRef] [PubMed]
- Vaziri, N.D.; Goshtasbi, N.; Yuan, J.; Jellbauer, S.; Moradi, H.; Raffatellu, M.; Kalantar-Zadeh, K. Uremic plasma impairs barrier function and depletes the tight junction protein constituents of intestinal epithelium. Am. J. Nephrol. 2012, 36, 438–443. [Google Scholar] [CrossRef]
- Gandhi, B.V.; Bahadur, M.M.; Dodeja, H.; Aggrwal, V.; Thamba, A.; Mali, M. Systemic fungal infections in renal diseases. J. Postgrad. Med. 2005, 51, S30–S36. [Google Scholar]
- Yu, C.; Wang, Z.; Tan, S.; Wang, Q.; Zhou, C.; Kang, X.; Zhao, S.; Liu, S.; Fu, H.; Yu, Z.; et al. Chronic Kidney Disease Induced Intestinal Mucosal Barrier Damage Associated with Intestinal Oxidative Stress Injury. Gastroenterol. Res. Pract. 2016, 2016, 6720575. [Google Scholar] [CrossRef] [PubMed]
- Sokol, H.; Leducq, V.; Aschard, H.; Pham, H.P.; Jegou, S.; Landman, C.; Cohen, D.; Liguori, G.; Bourrier, A.; Nion-Larmurier, I.; et al. Fungal microbiota dysbiosis in IBD. Gut 2017, 66, 1039–1048. [Google Scholar] [CrossRef]
- Heisel, T.; Montassier, E.; Johnson, A.; Al-Ghalith, G.; Lin, Y.W.; Wei, L.N.; Knights, D.; Gale, C.A. High-Fat Diet Changes Fungal Microbiomes and Interkingdom Relationships in the Murine Gut. mSphere 2017, 2, e00351. [Google Scholar] [CrossRef]
- Hiengrach, P.; Panpetch, W.; Worasilchai, N.; Chindamporn, A.; Tumwasorn, S.; Jaroonwitchawan, T.; Wilantho, A.; Chatthanathon, P.; Somboonna, N.; Leelahavanichkul, A. Administration of Candida Albicans to Dextran Sulfate Solution Treated Mice Causes Intestinal Dysbiosis, Emergence and Dissemination of Intestinal Pseudomonas Aeruginosa and Lethal Sepsis. Shock 2020, 53, 189–198. [Google Scholar] [CrossRef]
- Panpetch, W.; Somboonna, N.; Palasuk, M.; Hiengrach, P.; Finkelman, M.; Tumwasorn, S.; Leelahavanichkul, A. Oral Candida administration in a Clostridium difficile mouse model worsens disease severity but is attenuated by Bifidobacterium. PLoS ONE 2019, 14, e0210798. [Google Scholar] [CrossRef]
- Bloom, S.M.; Bijanki, V.N.; Nava, G.M.; Sun, L.; Malvin, N.P.; Donermeyer, D.L.; Dunne, W.M., Jr.; Allen, P.M.; Stappenbeck, T.S. Commensal Bacteroides species induce colitis in host-genotype-specific fashion in a mouse model of inflammatory bowel disease. Cell Host Microbe 2011, 9, 390–403. [Google Scholar] [CrossRef]
- Falgier, C.; Kegley, S.; Podgorski, H.; Heisel, T.; Storey, K.; Bendel, C.M.; Gale, C.A. Candida species differ in their interactions with immature human gastrointestinal epithelial cells. Pediatric Res. 2011, 69, 384–389. [Google Scholar] [CrossRef] [PubMed]
- Allert, S.; Forster, T.M.; Svensson, C.M.; Richardson, J.P.; Pawlik, T.; Hebecker, B.; Rudolphi, S.; Juraschitz, M.; Schaller, M.; Blagojevic, M.; et al. Candida albicans-Induced Epithelial Damage Mediates Translocation through Intestinal Barriers. mBio 2018, 9, e00915–e00918. [Google Scholar] [CrossRef] [PubMed]
- Vaziri, N.D.; Zhao, Y.Y.; Pahl, M.V. Altered intestinal microbial flora and impaired epithelial barrier structure and function in CKD: The nature, mechanisms, consequences and potential treatment. Nephrol. Dial. Transplant. 2016, 31, 737–746. [Google Scholar] [CrossRef]
- Li, B.; Cramer, D.; Wagner, S.; Hansen, R.; King, C.; Kakar, S.; Ding, C.; Yan, J. Yeast glucan particles activate murine resident macrophages to secrete proinflammatory cytokines via MyD88- and Syk kinase-dependent pathways. Clin. Immunol. 2007, 124, 170–181. [Google Scholar] [CrossRef]
- Goodridge, H.S.; Reyes, C.N.; Becker, C.A.; Katsumoto, T.R.; Ma, J.; Wolf, A.J.; Bose, N.; Chan, A.S.; Magee, A.S.; Danielson, M.E.; et al. Activation of the innate immune receptor Dectin-1 upon formation of a ‘phagocytic synapse’. Nature 2011, 472, 471–475. [Google Scholar] [CrossRef] [PubMed]
- Ferwerda, G.; Meyer-Wentrup, F.; Kullberg, B.J.; Netea, M.G.; Adema, G.J. Dectin-1 synergizes with TLR2 and TLR4 for cytokine production in human primary monocytes and macrophages. Cell. Microbiol. 2008, 10, 2058–2066. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.W.; Hoffman, S.; Beal, A.M.; Dykon, A.; Ringenberg, M.A.; Hughes, A.C.; Dare, L.; Anderson, A.D.; Finger, J.; Kasparcova, V.; et al. MALT1 Protease Activity Is Required for Innate and Adaptive Immune Responses. PLoS ONE 2015, 10, e0127083. [Google Scholar] [CrossRef] [PubMed]
- Schorey, J.S.; Lawrence, C. The pattern recognition receptor Dectin-1: From fungi to mycobacteria. Curr. Drug Targets 2008, 9, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Escobar, D.A.; Botero-Quintero, A.M.; Kautza, B.C.; Luciano, J.; Loughran, P.; Darwiche, S.; Rosengart, M.R.; Zuckerbraun, B.S.; Gomez, H. Adenosine monophosphate-activated protein kinase activation protects against sepsis-induced organ injury and inflammation. J. Surg. Res. 2015, 194, 262–272. [Google Scholar] [CrossRef]
- Zhang, M.; Yang, D.; Gong, X.; Ge, P.; Dai, J.; Lin, L.; Zhang, L. Protective benefits of AMP-activated protein kinase in hepatic ischemia-reperfusion injury. Am. J. Transl. Res. 2017, 9, 823–829. [Google Scholar] [PubMed]
- Zhao, W.; Zhang, L.; Chen, R.; Lu, H.; Sui, M.; Zhu, Y.; Zeng, L. SIRT3 Protects Against Acute Kidney Injury via AMPK/mTOR-Regulated Autophagy. Front. Physiol. 2018, 9, 1526. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Zmijewski, J.W.; Lorne, E.; Liu, G.; Park, Y.J.; Tsuruta, Y.; Abraham, E. Activation of AMPK attenuates neutrophil proinflammatory activity and decreases the severity of acute lung injury. Am. J. Physiol. Lung Cell. Mol. Physiol. 2008, 295, L497–L504. [Google Scholar] [CrossRef] [PubMed]
- Alba, G.; El Bekay, R.; Alvarez-Maqueda, M.; Chacon, P.; Vega, A.; Monteseirin, J.; Santa Maria, C.; Pintado, E.; Bedoya, F.J.; Bartrons, R.; et al. Stimulators of AMP-activated protein kinase inhibit the respiratory burst in human neutrophils. FEBS Lett. 2004, 573, 219–225. [Google Scholar] [CrossRef]
- O’Neill, L.A.; Pearce, E.J. Immunometabolism governs dendritic cell and macrophage function. J. Exp. Med. 2016, 213, 15–23. [Google Scholar] [CrossRef]
- Odegaard, J.I.; Ricardo-Gonzalez, R.R.; Red Eagle, A.; Vats, D.; Morel, C.R.; Goforth, M.H.; Subramanian, V.; Mukundan, L.; Ferrante, A.W.; Chawla, A. Alternative M2 activation of Kupffer cells by PPARdelta ameliorates obesity-induced insulin resistance. Cell Metab. 2008, 7, 496–507. [Google Scholar] [CrossRef] [PubMed]
- Odegaard, J.I.; Chawla, A. Alternative macrophage activation and metabolism. Annu. Rev. Pathol. 2011, 6, 275–297. [Google Scholar] [CrossRef]
- Viola, A.; Munari, F.; Sanchez-Rodriguez, R.; Scolaro, T.; Castegna, A. The Metabolic Signature of Macrophage Responses. Front. Immunol. 2019, 10, 1462. [Google Scholar] [CrossRef]
- De Simone, R.; Ajmone-Cat, M.A.; Pandolfi, M.; Bernardo, A.; De Nuccio, C.; Minghetti, L.; Visentin, S. The mitochondrial uncoupling protein-2 is a master regulator of both M1 and M2 microglial responses. J. Neurochem. 2015, 135, 147–156. [Google Scholar] [CrossRef]
- Sanz, J.M.; Di Virgilio, F. Kinetics and mechanism of ATP-dependent IL-1 beta release from microglial cells. J. Immunol. 2000, 164, 4893–4898. [Google Scholar] [CrossRef]
- Tian, Y.; Pate, C.; Andreolotti, A.; Wang, L.; Tuomanen, E.; Boyd, K.; Claro, E.; Jackowski, S. Cytokine secretion requires phosphatidylcholine synthesis. J. Cell Biol. 2008, 181, 945–957. [Google Scholar] [CrossRef]
- Leonhardt, J.; Grosse, S.; Marx, C.; Siwczak, F.; Stengel, S.; Bruns, T.; Bauer, R.; Kiehntopf, M.; Williams, D.L.; Wang, Z.Q.; et al. Candida albicans beta-Glucan Differentiates Human Monocytes Into a Specific Subset of Macrophages. Front. Immunol. 2018, 9, 2818. [Google Scholar] [CrossRef] [PubMed]
- Marzocco, S.; Fazeli, G.; Di Micco, L.; Autore, G.; Adesso, S.; Dal Piaz, F.; Heidland, A.; Di Iorio, B. Supplementation of Short-Chain Fatty Acid, Sodium Propionate, in Patients on Maintenance Hemodialysis: Beneficial Effects on Inflammatory Parameters and Gut-Derived Uremic Toxins, A Pilot Study (PLAN Study). J. Clin. Med. 2018, 7, 315. [Google Scholar] [CrossRef] [PubMed]
- Allavena, P.; Anfray, C.; Ummarino, A.; Andon, F.T. Therapeutic Manipulation of Tumor-associated Macrophages: Facts and Hopes from a Clinical and Translational Perspective. Clin. Cancer Res. 2021, 20, 1679. [Google Scholar]
- Ono, S.; Tsujimoto, H.; Hiraki, S.; Aosasa, S. Mechanisms of sepsis-induced immunosuppression and immunological modification therapies for sepsis. Ann. Gastroenterol. Surg. 2018, 2, 351–358. [Google Scholar] [CrossRef] [PubMed]
- Leelahavanichkul, A.; Somparn, P.; Panich, T.; Chancharoenthana, W.; Wongphom, J.; Pisitkun, T.; Hirankarn, N.; Eiam-Ong, S. Serum miRNA-122 in acute liver injury induced by kidney injury and sepsis in CD-1 mouse models. Hepatol. Res. 2015, 45, 1341–1352. [Google Scholar] [CrossRef] [PubMed]
- Scheithauer, T.P.; Dallinga-Thie, G.M.; de Vos, W.M.; Nieuwdorp, M.; van Raalte, D.H. Causality of small and large intestinal microbiota in weight regulation and insulin resistance. Mol. Metab. 2016, 5, 759–770. [Google Scholar] [CrossRef] [PubMed]
- Panpetch, W.; Somboonna, N.; Bulan, D.E.; Issara-Amphorn, J.; Worasilchai, N.; Finkelman, M.; Chindamporn, A.; Palaga, T.; Tumwasorn, S.; Leelahavanichkul, A. Gastrointestinal Colonization of Candida Albicans Increases Serum (1→3)-beta-D-Glucan, without Candidemia, and Worsens Cecal Ligation and Puncture Sepsis in Murine Model. Shock 2018, 49, 62–70. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- Bailey, J.D.; Shaw, A.; McNeill, E.; Nicol, T.; Diotallevi, M.; Chuaiphichai, S.; Patel, J.; Hale, A.; Channon, K.M.; Crabtree, M.J. Isolation and culture of murine bone marrow-derived macrophages for nitric oxide and redox biology. Nitric Oxide Biol. Chem. 2020, 100–101, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Liu, C.; Zhou, R.; Lu, A.; Huang, B.; Liu, L.; Chen, L.; Luo, B.; Huang, J.; Tian, Z. SEQdata-BEACON: A comprehensive database of sequencing performance and statistical tools for performance evaluation and yield simulation in BGISEQ-500. Biodata Min. 2019, 12, 21. [Google Scholar] [CrossRef] [PubMed]
- Jansen, W.T.; Vakevainen-Anttila, M.; Kayhty, H.; Nahm, M.; Bakker, N.; Verhoef, J.; Snippe, H.; Verheul, A.F. Comparison of a classical phagocytosis assay and a flow cytometry assay for assessment of the phagocytic capacity of sera from adults vaccinated with a pneumococcal conjugate vaccine. Clin. Diagn. Lab. Immunol. 2001, 8, 245–250. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Stevens, M.G.; Olsen, S.C. Comparative analysis of using MTT and XTT in colorimetric assays for quantitating bovine neutrophil bactericidal activity. J. Immunol. Methods 1993, 157, 225–231. [Google Scholar] [CrossRef]
- Jaroonwitchawan, T.; Visitchanakun, P.; Dang, P.C.; Ritprajak, P.; Palaga, T.; Leelahavanichkul, A. Dysregulation of Lipid Metabolism in Macrophages Is Responsible for Severe Endotoxin Tolerance in FcgRIIB-Deficient Lupus Mice. Front. Immunol. 2020, 11, 959. [Google Scholar] [CrossRef]

| Primers | Forward | Reverse |
|---|---|---|
| NFκB | 5’-CTTCCTCAGCCATGGTACCTCT-3’ | 5’-CAAGTCTTCATCACATCAAACTG-3’ |
| Malt-1 | 5’-CACACTGAGGTTCTTCCGCT-3’ | 5’-TGATGCATTCGGGGGCGTAC-3’ |
| Syk | 5’-CTACTACAAGGCCCAGACCC-3’ | 5’-TGATGCATTCGGGGGCGTAC-3’ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Issara-Amphorn, J.; Dang, C.P.; Saisorn, W.; Limbutara, K.; Leelahavanichkul, A. Candida Administration in Bilateral Nephrectomy Mice Elevates Serum (1→3)-β-D-glucan That Enhances Systemic Inflammation Through Energy Augmentation in Macrophages. Int. J. Mol. Sci. 2021, 22, 5031. https://doi.org/10.3390/ijms22095031
Issara-Amphorn J, Dang CP, Saisorn W, Limbutara K, Leelahavanichkul A. Candida Administration in Bilateral Nephrectomy Mice Elevates Serum (1→3)-β-D-glucan That Enhances Systemic Inflammation Through Energy Augmentation in Macrophages. International Journal of Molecular Sciences. 2021; 22(9):5031. https://doi.org/10.3390/ijms22095031
Chicago/Turabian StyleIssara-Amphorn, Jiraphorn, Cong Phi Dang, Wilasinee Saisorn, Kavee Limbutara, and Asada Leelahavanichkul. 2021. "Candida Administration in Bilateral Nephrectomy Mice Elevates Serum (1→3)-β-D-glucan That Enhances Systemic Inflammation Through Energy Augmentation in Macrophages" International Journal of Molecular Sciences 22, no. 9: 5031. https://doi.org/10.3390/ijms22095031
APA StyleIssara-Amphorn, J., Dang, C. P., Saisorn, W., Limbutara, K., & Leelahavanichkul, A. (2021). Candida Administration in Bilateral Nephrectomy Mice Elevates Serum (1→3)-β-D-glucan That Enhances Systemic Inflammation Through Energy Augmentation in Macrophages. International Journal of Molecular Sciences, 22(9), 5031. https://doi.org/10.3390/ijms22095031







