Abstract
Diabetic neuropathy (DN), the most common chronic and progressive complication of diabetes mellitus (DM), strongly affects patients’ quality of life. DN could be present as peripheral, autonomous or, clinically also relevant, uremic neuropathy. The etiopathogenesis of DN is multifactorial, and genetic components play a role both in its occurrence and clinical course. A number of gene polymorphisms in candidate genes have been assessed as susceptibility factors for DN, and most of them are linked to mechanisms such as reactive oxygen species production, neurovascular impairments and modified protein glycosylation, as well as immunomodulation and inflammation. Different epigenomic mechanisms such as DNA methylation, histone modifications and non-coding RNA action have been studied in DN, which also underline the importance of “metabolic memory” in DN appearance and progression. In this review, we summarize most of the relevant data in the field of genetics and epigenomics of DN, hoping they will become significant for diagnosis, therapy and prevention of DN.
1. Introduction
Diabetes mellitus (DM) presents a worldwide public health burden. It is associated with numerous complications, including diabetic neuropathy (DN), which occurs in around 60–70% of affected individuals [1]. Furthermore, it was stated that the prevalence of DN in newly diagnosed patients with DM is around 8% [2]. The DN can be presented as peripheral diabetic neuropathy (PDN) as well as diabetic autonomic neuropathy (DAN) [3]. Additionally, from the clinical perspective, DN can be presented as uremic neuropathy (UN) [1].
The etiopathogenesis of DN is multifactorial and involves environmental, external factors and internal factors [1,2,4,5]. In the meta-analysis of Wu et al., it was further stated that genetic factors might influence the course of PDN [4]. Moreover, in the study of Kallinikou et al., it was stressed that subclinical DN can be present in the pediatric population even with good metabolic control and short DM duration, which might suggest potential genetic susceptibility [3].
The role of heritability in DN has been assumed based on the results of twin studies and observed familial aggregation [1]. In contemporary studies, medical genetics main tools for assessment of genetic predisposition to multifactorial diseases are association studies. Such studies analyze variants known as DNA/gene polymorphisms with the aim to detect if a particular variant is significantly linked to disease occurrence, representing a genetic risk factor. Protective variants which are less frequent in affected persons could be detected also. Common association studies analyze variants in a small number of candidate genes, which are selected by known patho-physiological mechanisms, by results in animal models or in segregation studies. In the last two decades, Genome Wide Association Studies (GWAS) are available, which allow simultaneous analysis of thousands of (unselected) DNA variants scattered throughout the entire genome. More recently, development of Next Generation Sequencing technology enabled rapid detection of DNA/gene variants contributing to genetics of multifactorial diseases also.
In the field of genetics of DN, a number of candidate gene studies have been published so far, but often with inconsistent results and with substantial missing heritability. Several issues, well known for such type of studies in general, are different sample size and structure, population and ethnic specificity, complex interaction between genes and environment, etc. [1,2,6].
In this article, first will be presented the main patho-physiological mechanisms important in DN, and then based on that, major results of association studies for candidate genes. Finally, epigenetic concepts important in DN will be discussed.
Besides the multidimensional role of different factors that could be associated with the DN, including but not limited to: reactive oxygens species production, neurovascular impairments and modified protein glycosylation, there are not sufficient findings regarding the role of inflammatory mediators in the DN etiopathogenesis [7].
2. Anatomical, Histological and Clinical Considerations
Peripheral diabetic neuropathy can be described as a common late complication of diabetes and is significant contributor to the increased morbidity and mortality [8,9]. It is presented with a wide plethora of abnormalities affecting motor, sensory and autonomic nerves that can be clinically expressed as isolated or combined.
PDN is usually painful and despite the fact that several structural and functional differences were noticed in central nervous system (CNS) between painful and painless PDN, in the study of Groener et al. [9] no specific distinguishing features in peripheral nervous system (PNS) between these two groups of PDN were not noticed. Diabetic polyneuropathy is often presented as mixed fiber polyneuropathy that involves Aβ, Aδ and C-fibers, however neuropathic pain is mostly associated with a small-fiber affection, but in the study of Galosi et al. [10], neuropathic pain was described in pure large-fiber neuropathy as well. In Table 1, characteristics of different types of neuropathies are presented [11,12,13,14,15,16,17,18,19,20].

Table 1.
Characteristics of different types of neuropathies in diabetic patients.
The complexity of diabetic autonomic neuropathy (DAN) can be found in numerous studies [12,13,14,15,16,17,18,19,20]. It was noticed that patients with DAN have changes in CNS including neuron loss in intermediolateral nuclei of the spinal cord, presence of degeneration along with the loss of sympathetic neurons of prevertebral and paravertebral autonomic ganglia, as well as vagal nerve axons degeneration, and loss of autonomic axons in somatic nerves [21].
In DN, reduction of unmyelinated and small myelinated axons was shown to appear earlier than decrease in large myelinated fibers, with assumption that changes in Schwann cells precede axonal degeneration [22]. These changes that are described in Schwann cells include: cytoplasm edema, glycogen particles aggregates as well as hyperplasia of surrounding basal lamina [22].
In the study of De Gregorio et al., it was noticed how complex anatomopathological changes can be in subjects with DN [23]. There is significant reduction in intraepidermal nerve fiber (IENF) density in diabetic mice particularly from 18 weeks of age compared to normal littermates, while in a later period from 26 weeks of age in diabetic mice, a significant increase in TUNEL+ cells in sciatic nerve was observed [23]. The increase of such cells suggests to the greater susceptibility of Schwann cells to apoptosis. Furthermore, at the same age (26 weeks) in diabetic mice, an increase of T lymphocytes in sciatic nerves was also noticed, indicating the increase of the inflammation process [23]. Finally, besides the fact that there was no difference in nerve fiber diameters of sciatic nerve between diabetic and non-diabetic mice, there was an increase in the fibers density particularly in diabetic mice age of 32 weeks [23]. It was shown that there was a decrease in large fibers and increase in small fibers in diabetic mice [23].
In the study of Younger, on sural nerve in human subjects with DN, it was noticed that there was perivasculitis in larger proportion than microvasculitis and necrotizing arteritis (NA) [24]. The presence of microvasculitis and NA might suggest the theory of ischemic axonopathy. However, it seems that microvasculitis and severe perivasculitis have more influence in epineurial ischemia than NA does [24]. Additionally, the study pointed to more frequent primary axonopathy than primary myelinopathy, and the presence of deposits of C3 and C5b-9 complements in the walls of endoneurial microvessels from around two-thirds of tested nerves [24].
3. Peripheral Nervous System Dysfunction in Diabetic Neuropathy
The peripheral nervous system has its own specific characteristics from other tissues that are characterized in structure, function and metabolic needs [25]. It should be pointed out that axons and Schwann cells are interdependent [26]. The significant components in saltatory impulse conduction along the myelinated nerve are nodes. Thus, the processes that affects structural and functional parts of these elements including Schwannopathy, axonopathy and nodal degeneration that could be associated with the onset and the progression of PDN. Zenker et al. stressed that nodal degeneration, disjunctions on the axonal-glial levels and altered ion-channel localization might be associated with PDN [25]. Robertson and Sima, in their study on mutant mouse models, described the presence of temporal discrepancy between the occurrence of functional and structural deficits in nerves, suggesting that the origin of the DN might be primarily metabolic [27]. However, it should be noticed that so far, no animal model was produced to be completely comparable with the human DN onset and the progression.
In DM patients with present neuropathy, metabolic disturbances alter the synthesis of neurotrophic factors both in neurons and Schwann cells that affect the processes of regeneration [28]. Reduced production of nerve growth factor (NGF) and neurotrophin-3 (NT-3) were observed in cultured Schwann cells of diabetic mice [22]. Modifications of Schwann cells functions that might be altered by diabetes, including the changes in the production of signaling molecules as well as impaired receptor-mediated interactions between these cells and basal lamina and the endoneurium, which could affect PNS structure and function [28].
Another important aspect is the mitochondrial role in neuron and Schwann cells dysfunctions in DM subjects. In diabetic rats there is downregulation of mitochondrial proteins [29]. Furthermore, changes in mitochondrial membrane potential in sensory neurons in diabetic rats were also described [30] as well as downregulation of mitochondrial respiratory chain complex proteins in sensory neurons [31]. Additionally, fragmented mitochondria and an increase in the expression of Drp1 were noticed in sensory neurons of DM subjects [32]. Finally, exposure of mitochondria to the increased concentration of glucose could lead to the apoptosis by releasing into cytosol some apoptosis-induced factors, thus reducing the mitochondrial number in cells [33] (Figure 1).
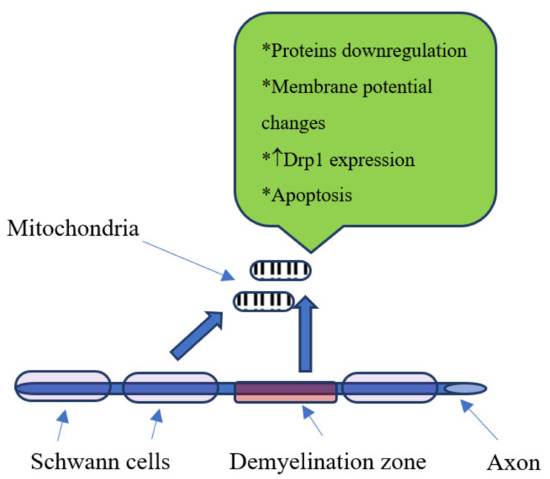
Figure 1.
Mitochondrial changes in diabetic neuropathy, ↑-increase.
In previous studies on animal models with DM type 1 it was stressed that induced hyperglycemia is associated with changes in calcium (Ca++) homeostasis/signaling pathway and mitochondrial function [34]. The impaired expression and function of Ca++ channels are associated with PDN, and Ca++ is found to be associated as well with dysfunctions of mitochondria in subjects with sensory DN [34].
4. Biochemical Changes in Diabetic Neuropathy
Diabetic neuropathy in subjects with DM could be associated with several conditions: excessive polyol pathway flux, reactive oxygen species formation (ROS), advanced glycosylation end products (AGE) production and inadequate neurotrophic support [26,35]. Furthermore, hyperglycemia was associated also with activation of mitogen-activated protein kinases (MAPK) and upregulation of hexosamine pathway [35,36].
The flux of polyol pathway increases in hyperglycemia in order to metabolize elevated glucose levels and such an increase of the flux is associated with diabetic neuropathy [37]. In diabetes, the polyol pathway is catalyzed by the two enzymes: aldose reductase (AR) and sorbitol dehydrogenase (SD). The localization of AR is noticed in Schwann cells of nerve fibers that are myelinated and in the satellite cells of the dorsal root ganglia [26]. Increased activity of these two enzymes is associated with elevated levels of sorbitol, fructose, NADP+ and NADH along with the lower levels of NADPH and NAD+ [37]. The conversion of glucose to sorbitol is done by the AR, while the SD‘s role is in the oxidation of the sorbitol to fructose [36]. Additional AR function in DM is the up-regulation of MAPK activity in nerve and dorsal root ganglia [36]. Polyol pathway activation as well as altered protein kinase C activity trigger oxidative stress, thus these mechanisms are associated with the altered cell‘s redox state, leading to the cell injury [35]. Furthermore, previously it was pointed that the lipid peroxidation in nerve cells, as a consequence of polyol pathway flux, can be prevented by inhibiting the AR [26].
In diabetic patients, the glucose via nonenzymatic processes forms Amadori products, which are further converted to a more stable substrate called AGE [38]. Further, the AGE attaches to the receptor (RAGE) on the cell surface, initiating a cascade of signal transduction events that lead to the ROS production [38].
The role of oxidative stress in pathogenesis of DN was broadly evaluated previously. Increased ROS is considered as one of the factors associated with programmed cell death [39]. This is of particular importance sine ROS were observed in dorsal root ganglia and Schwann cells; thus they might play important role in the DN pathogenesis [39]. Additionally, the oxidative stress affects the mitochondria via several mechanisms that include inhibition of ATP synthesis as well as essential proteins in to mitochondria, and affects mitochondrial membrane permeability by damaging the inner membrane proteins [38]. In a hyperglycemic state, the activation of Nuclear factor-κB (NF-κB), cyclooxigenase-2 (COX-2) mRNA induction as well as COX-2 protein expression are described [40]. The changes associated with the COX-2 upregulation lead to the complex processes resulting in the vasoconstriction and the onset of ischemic conditions in affected tissues as well as ROS production [40].
5. Inflammation, Cytokines and Neurotrophins in Diabetic Neuropathy
It is noticed that chronic low-grade inflammation plays an important role in the development and progression of DN [41]. In individuals with DN, increased inflammatory cytokines as well as different levels of certain growth factors were observed [42]. However, it should be mentioned that the inflammation has a more complex role in DN pathogenesis, since it is not only involved in processes that results in nerve damage, but also in process associated with regeneration [43,44].
Hypoxia or ischemia that are associated with the endothelial injury and or microvascular dysfunctions that lead to the increase in the levels of the certain cytokines including tumor necrosis factor (TNF)-α and interleukin (IL)-6, while such states lower nerve growth factor (NGF) [35]. However, NGF is more closely referred as neurotrophin and plays important roles in the axonal generation stimulation after a neuron injury [45]. Other cytokines could play a role in the pathogenesis of DN, including elevated levels of IL-1β, IL-13 and IL-17 [36] (Figure 2). Previous reports stated that the reduction of IL-1β, IL-6 and TNF-α by genetic deletion or pharmacological inhibition has favorable effects on changes in nerve conduction velocities [43]. As it was stressed in the study of Bönhof et al., IL-6 might have pro-and anti-inflammatory characteristics that could depend on immunological context [43], thus its exact role in the pathogenesis of DN should be studied further.
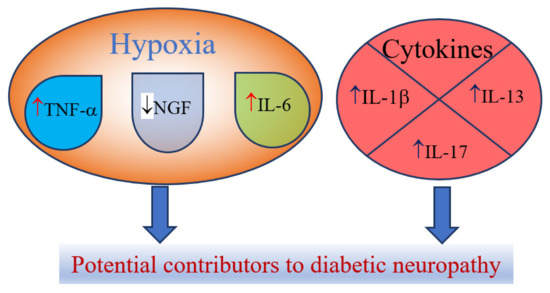
Figure 2.
Cytokines and neurotrophins as potential contributors to diabetic neuropathy, ↑-increase; ↓-decrease
6. The Role of Dyslipidemia in Diabetic Neuropathy
Previous studies investigated the complex interactions between dyslipidemia and inflammation in diabetic neuropathy [46,47]. In the study of Vincent et al. [47], it was noticed that sorbitol, oxidized lipids as well as poly ADP-ribose polymerase are increased in animal models that were on a high-fat diet, along with the lipoxygenases activation before diabetes development in peripheral nerves. Furthermore, it was stressed that fatty acids with long chains could penetrate the blood-nerve barrier, thus inducing neurogenic inflammation that might lead to the increased expression of TNF-α and IL-6 [46]. Additionally, free fatty acids are shown to have lipotoxicity potential that is mediated via lysosomal dysfunction in cultured neuronal and Schwann cell lines [47]. Moreover, fatty acids produce NADPH and FADPH2 through β-oxidation in Schwann cells, dorsal root ganglion and axons [48]. Patients with diabetes—due to the increased accumulation of the acetyl-CoA molecules that are formed in Schwann cells from long-chain fatty acids—convert such molecules into acylcarnitines that affect Schwann cells and dorsal root ganglions, thus such a process contributes to diabetic neuropathy [48]. In a systematic review and meta-analysis of Cai et al., it was stated that the free fatty acids could bind to pyruvate in mitochondria and thus increasing the ROS production [49].
In this environment with increased oxidative stress, the oxidation of cholesterol to oxysterols occurs. The production of oxysterol might be via autoxidation or by the enzymatic oxidation of cholesterol, where those originated by autoxidation are speculated to be the most important oxidative stress biomarkers [50]. In oxysterol metabolism, three main groups of enzymes are described: oxidoreductases that are subcellularly localized in the endoplasmic reticulum, mitochondria and cytosol; hydrolases that are subcellularly localized in the endoplasmic reticulum; and transferases that are subcellularly localized in cytosol [51]. In the study of Ferderbar et al. [52], it was stated that cholesterol oxides could induce apoptosis of several cell types: vascular, endothelial, smooth muscle of the artery as well as T cells and monocyte macrophages. Moreover, they can act proinflammatory by modulating the synthesis of cytokines, adhesion molecules and growth factors [52]. Additionally, oxidized low-density lipoproteins (LDL) are binding to oxidized LDL receptor 1 (LOX1), Toll-like receptor 4 (TLR4) and RAGE contributing to the inflammation processes and increase of ROS [48].
7. Genetic Basis of Polyol Pathway
The complex role of the AR gene in the etiopathogenesis of complications in individuals with DM could be referred to the fact that AR gene expression might be induced by methylglyoxal (MGO), advance glycation end-product (AGE) and oxidative stress in a hyperglycemic state [53]. Previous investigations of DN pathology on rodent models and humans pointed that mice do not tend to have higher loss of nerve fibers even long-term in DM [53]. Thus, it might be assumed that other factors could be involved in the progression of DN, while it is suggested that AR could be involved in the early stage of DN [53].
The AR is encoded by the ALR2 gene, while SD, the second enzyme in polyol pathway, is encoded by the SDH gene [54]. In the study of Sivenius et al. for a group of patients with DM type 2, it was noticed that the-106C/T polymorphism of promoter region of ALR2 gene is associated with a decrease in the nerve conduction velocities of the motor peroneal nerve and a lower amplitudes of the tested sensory nerves versus subjects with-106C/C genotype [55]. Heesom et al., in the study of patients with insulin dependent DM, found the association between the DN and another polymorphism located at the 5‘, upstream regulatory region of ALR2 [56]. This is the 5′-(CA)n microsatellite polymorphism represented with more than 10 alleles; two major alleles are Z-2 and Z+2, whereby Z corresponds to 24 CA repeats. The group of patients with DN had significantly decreased frequency of the Z+2 allele [56], while it was noticed that the Z-2 allele is associated with the increased susceptibility of diabetic complications in both types of DM [53]. It should be pointed that these markers are located outside of the protein coded region of AR gene, indicating that they should not directly affect the enzyme structure and function [53]. Thus, the possible mechanisms of action for some of these alleles could be enhancement of AR gene expression or the transcription of the one [53].
Considering the SD role in DN, it should be noticed that the nerve sorbitol levels in SD deficient mice are not suggestive for DN susceptibility [53].
8. Other Genetic Risk Factors for Diabetic Neuropathy
In their review, Witzel et al. contributed to the creation of a genetic-metabolic model that should help to elucidate the cause of diabetic neuropathies, evaluate a patient’s risk profile and facilitate preventative and targeted treatment for the individual [1]. After evaluation of published candidate genetic markers in different forms of DN, the authors summarized common genetic risk factors. Polymorphisms in genes ACE (angiotensin-converting enzyme), APOE (apolipoprotein E), MTHFR (methylene tetrahydropholate reductase), NOS3 or ENOS (nitric oxide synthase 3 or endothelial nitric oxide synthase) and VEGF (vascular endothelial growth factor) have been shown to contribute to DPN as well as DN. In all of these genes, the minor polymorphic allele is associated with DN [1]. It was speculated that having in mind functions of these genes, it is possible that the same variants also contribute to UN (uremic neuropathy) and CAN (cardiac autonomic neuropathy). Considering candidate genes that have been implied in but not significantly associated with the diabetic neuropathies, this number of potentially genetic risk factors could be larger. Authors emphasized that all of the listed genes are involved in key molecular pathways that have been linked to diabetes and its complications, mentioned above [57,58,59].
Very recently, Zhao Y et al. performed a systematic review, meta-analysis and trial sequential analysis (TSA) of the association between genetic polymorphisms and DN risk [2]. They conducted a systematic review of more than 1350 articles and publications reporting on 136 polymorphisms in 76 genes. More than 100 meta-analyses on 36 studies were performed involving 12,221 subjects to derive pooled effect estimates for analyzed polymorphisms.
Data for the most important genes processed in the mentioned studies will be presented in the following text.
The ACE is an enzyme and vasoconstrictor playing a crucial role in the renin–angiotensin system by doing conversion of angiotensin I to angiotensin II. As a reminder, it is well documented that angiotensin II is involved in regulation of glucose and insulin levels, and among other effects that induce oxidative stress, inflammation and vascular changes [4,44]. The ACE gene insertion/deletion (I/D) polymorphism is widely studied because this variant has a regulatory role on gene expression and itself is responsible for about 46% of enzyme serum variance [1]. The ACE I allele is defined by presence of 287bp Alu sequence in intron 16, while in the D allele this sequence is deleted; D allele and DD genotype are associated with elevated ACE enzyme levels [29]. A number of original investigations and several meta-analyses confirmed the role of ACE I/D polymorphism in DN, indicating D allele as a risk factor, particularly for DPN [4,60,61,62,63]. In addition, association of D allele with renal disease, as well as with blood pressure variations in DM, indicates its role in UN and CAN, respectively. On the contrary, the ACE II genotype is assigned as protective regarding DN. The ACE I/D polymorphism has been considered as potential pharmacogenetic marker because it could affect efficiency of ACE inhibitors, angiotensin receptor blockers and statins, which are widely used in DM. However, the clinical benefit of routine ACE I/D genotyping in this purpose is still controversial [1,2,4].
APOE is a cholesterol transporter and LDL receptor ligand involved in lipid metabolism, nerve repair and regeneration. Well-known allelic variants e2, e3 and e4 in APOE gene are defined by two functional single nucleotide polymorphisms (SNPs), rs429358 C/T and rs7412 T/C, which cause amino acid changes 112Cys/Arg and 158Arg/Cys, respectively, at the protein level [1,2,44]. APOE gene variants have an impact on protein function trough the isoforms E2, E3 and E4, which differ in charge and stability. In general, the e2 allele is associated with higher circulatory APOE and lower levels of LDL cholesterol, while e4 allele shows opposite effects. The APOE genotype is associated with disorders of both the central and peripheral nervous system, and e4 allele is assigned as a risk variant for neuropathology. Regarding the role of APOE in DM and DN, studies show some population specificities. Meta-analyses confirmed e4 allele as a risk variant for DPN, and there are data about particularly severe DPN in e4 carriers also [1,2,64,65]. In addition, both clinical studies and animal models suggest APOE as candidate gene for CAN risk. Besides, pharmacogenetic evaluation of APOE is a very current topic. It has been found that the antidiabetic drug Metformin enhances APOE expression, which consecutively stimulates nerve regeneration. Some studies showed association of APOE e4 form with a worse response to statins, but a better response to life style changes in DM patients. These results are still inconsistent but could have effect on various aspects of DM and DN monitoring and control [1,2].
MTHFR is an enzyme in folate cycle and homocysteine metabolism with a role in neurotransmitter production, protein synthesis, immune response and inflammation. MTHFR plays important role in remethylation of homocysteine to methionine and its lower activity is linked to hyperhomocysteinemia, which has damaging effects on blood vessels, lipid metabolism, neuronal function, etc. [1,4,6]. In DM, high blood homocysteine is associated with neurovascular complications. Frequently studied polymorphisms MTHFR 677C/T (rs1801133) and 1298A/C (rs1801131) lead to amino acid changes 222Ala/Val and 429Glu/Ala, respectively, and affect enzyme structure and function. After conflicting results of numerous studies, meta-analyses showed significant association of DN, including DPN and CAN, and MTHFR 1298A/C variant, but no clear association with 677C/T [1,2,4,66,67,68,69]. It is important to emphasize that about 35% of variations in homocysteine levels are due to folate and vitamin B12 levels, and that diet may diminish negative genetic effects, at least to some extent. This is of particular interest in assessment of MTHFR variants as pharmacogenetic markers. It has been found that MTHFR polymorphism affect response to various drugs, including antidiabetic drug Metformin. As Metformin usage leads to vitamin B12 deficiency, DM patients at treatment who have risky MTHFR genotypes should take vitamin supplements [1].
NOS3 (eNOS) regulates NO circulating levels by production of NO from L-arginine, maintains endothelial cell function and homeostasis. In the NOS3 gene, three important polymorphisms are: promoter variant-786T/C (rs2070744), functional SNP 894G/T (rs1799983) and variation in tandem repeats in intron 4, known as 4a/b. Several studies detected association of DN and DPN occurrence and progression with all three NOS3 polymorphisms, while for UN are CAN results are negative or not consistent [1,70,71]. NOS3 is interesting therapeutic target, because of its association to neuropathic pain and inflammation, and mentioned variants are potential pharmacogenetic markers [1].
VEGF is chemokine that modulates vascular permeability, regulates angiogenesis and contributes positively to neurogenesis [1,2,44]. Related to VEGF gene there are two polymorphic sites, rs6921438 and rs10738760, which contribute to about 50% of VEGF level variation. Association of VEGF related polymorphisms and DPN are observed, but a significant link to CAN and UN is not clearly established. Further investigation about contribution and pharmacogenetic effects of these genetic variants in DNs is needed [1,2,72].
Considering anti-oxidative mechanisms in DN, variants in genes encoding ubiquitous enzymes that catalyze the removal of hydrogen peroxide, or that quench reactive molecules have been widely studied. Meta-analyses confirmed association of polymorphism 599C/T (rs1050450) in the gene GPx-1 (glutathione peroxidase 1) and-262C/T in the gene CAT (catalase) with DN, while no significant association was observed with null/present variants in GSTM1 and GSTMT1 (glutathione S-transferases M1 or T1) [2,73].
Contemporary genomic studies are also conducted in the field of DN. Meng et al. published results of a genome-wide association study, which was undertaken in Scotland. They found that chromosomal loci 1p35.1 and 8p21.3 were associated with neuropathic pain in DPN, with a more prevalent association in women [74]. However, a noticed weakness of this study is about criteria for painful-DPN. Future well-designed studies are needed to confirm these results.
In Table 2, the most relevant gene polymorphisms associated to DN were presented.

Table 2.
The most relevant gene polymorphisms associated to diabetic neuropathy.
9. Epigenomic Modifications in Diabetic Neuropathy
Major epidemiological studies revealed that an early intensive glycemic control is able to decrease the long-term risk of diabetic complications, the phenomenon termed as “metabolic memory”. Although the mechanism of “metabolic memory” is not completely understood, recent studies implicated that continuing metabolic changes are established trough oxidative stress, non-enzymatic glycation of proteins, chronic inflammation as well as dysregulation of epigenetic mechanisms [75]. Epigenetic modifications are defined as heritable changes in gene expression patterns without alterations in the DNA sequence, emerging from genome–environment interactions [76]. Mounting evidence suggests a key role of different epigenetic mechanisms such as DNA methylation, non-coding RNAs and post-translational histone modifications in “metabolic memory” and the risk of developing diabetic complication [77] (Figure 3).
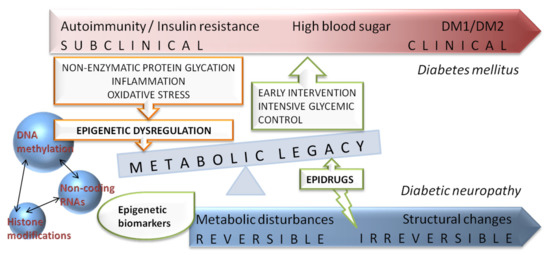
Figure 3.
Schematic representation of diabetes and diabetic neuropathy progression with integrated view of potential benefits of epigenetic related biomarkers and novel epidrugs on “metabolic memory” or “metabolic legacy” phenomenon.
DNA methylation is a stable and reversible attachment of methyl (CH3) group to a cytosine in palindromic sites in DNA, named CpG islands. Those sites are usually located near or in promoter and regulatory regions allowing the family of enzymes, DNA methyltransferases (DNMTs), to regulate gene function trough methylation process [78]. DNA methylation is regulating gene expression and has an important role in many biological functions including disease susceptibility [79]. Genome-wide methylation analysis of samples collected 16–17 years apart from the same diabetic patients revealed a persistency of DNA methylation over time at key genomic loci associated with diabetic complications [80]. Hyperglycemia is changing DNA methylation status inducing alterations in gene expression related to PDN and significantly reduced methylation of whole genomic DNA in white blood cells represents a potential biomarker for PDN [81]. Additionally, new evidence has pointed out that different DNA methylation patterns in genes related to nerve regeneration and functionality have an important role in the development of cardiovascular autonomic neuropathy (CAN) in patients with DM type 1 [82]. For instance, an increased methylation, correlated with the decreased expression, was observed in the ninjurin 2 (NINJ2) gene. NINJ2 protein in Schwann cells is promoting nerve regeneration after injury [83], therefore downregulating NINJ2 expression may contribute to neuropathies development. Decreased methylation of genes involved in neuronal polarization (BR serine/threonine kinase 2 (BRSK2)) was also observed, but not confirmed. An interesting finding from the same study was a strong hypomethylation of the 5′UTR region of the claudin 4 (CLDN4) gene. Although it is supposed to promote the attachment of the transcriptional factor, it was associated with the significant downregulation of gene expression. The authors suggested that the involvement of other epigenetic mechanisms may be involved in decreasing gene expression [82,84]. Promising findings from genome-wide profiling of DNA methylation in sural nerves of patients with differences in progression of PDN is linking specific DNA methylation profiles with PDN progression through pathways related to nervous system development and/or axon guidance (netrin-4 (NTN4) and dihydropyrimidinase like 2 (DPYSL2) genes), glycerophospholipid metabolism (phospholipase A2 and phosphatidylserine decarboxylase) and MAPK signaling [85]. Moreover, integrated analysis of the sural nerve methylome and transcriptome identified DN candidate genes and pathways (immune response, ECM regulation and PI3K-Akt signaling) [86]. Guo et al. also observed multiple differentially methylated CpGs within regulatory regions that may affect the expression of lncRNAs or miRNAs. Highest methylation difference is observed in miR3138 whose target is erb-b2 receptor tyrosine kinase 4 (ERBB4), a gene previously associated with progression of PDN [87].
MicroRNAs (MiRNAs) are group of evolutionarily conserved, regulatory non-coding RNAs comprise of less than 200 nucleotides in length (about 20) that are complementary to 3′-UTR of different messenger RNAs (mRNAs). Binding miRNA with targeted mRNA disrupts translation and leads to mRNA cleavage [88]. The role of miRNAs different expression patterns in multifactorial diseases like DN is a rapidly evolving field because of their potential use as biomarkers of disease progression and response to therapy, as well as promising targets for developing epidrugs [89,90]. Study in patients with diabetic neuropathy showed an increased expression of miR199a3p and subsequently downregulation of extracellular protein Serpin E2 in disease progression [91]. Increased expression of miR9 in PDN rat model upregulated CALHM1, a protein involved in DN pathophysiology [92]. Conversely, several studies in diabetic animal models identified decreased miR25, miR146 and miR190a5p expression [93,94,95,96] (Table 3). Besides studies focused on alternations in miRNAs expression, it has been shown that polymorphisms in miRNAs genes are affecting the maturation process or target recognition and binding, and may be involved in pathological processes in different diseases [97]. Genetic variants in MIR128A, MIR146A, MIR27A and MIR499A genes have been associated with the risk of PDN and CAN in Italian patients with DM type 2 [98,99]. Furthermore, the association between decreased number of mtDNA copies and polymorphism in MIR499A gene has been recently described in DM type 2 patients, particularly with DN, suggesting a possible role of oxidative stress in the development of DN [100].
Long non-coding RNA (LncRNAs), RNAs with more than 200 nucleotides in length, have a regulatory function in numerous metabolic processes and pathways in the nucleus and cytoplasm [101] and it is not surprising that several recent studies revealed specific lncRNAs (48+, LncRNANONRATT021972 and BC168687) as players in pathogenesis of DN and neuropathic pain [102,103,104]. Their role is further confirmed with findings that small interference RNAs (siRNA) for specific lncRNAs in diabetic rat models are attenuating PDN potentially through reduced production of inflammatory factors [105,106,107,108,109]. Genome-wide expression differences of lncRNAs between DM patients with and without PDN were identified using microarray analysis. The lncRNA/mRNA coexpression network in PDN patients indicated involvement of the neurotrophin-MAPK signaling pathway. Several lncRNAs (CCNT2-AS1, RP1-249H1.2, CTD-3239E11.2, RP11-51B23.3, STAM-AS1 and LINC00629) were indicated as potential interplayers in the abovementioned signaling pathways and additional, previously reported diabetes-associated lncRNAs (MALAT1, H19, PVT1, MEG3, MIAT and MIR143HG) followed trends observed in the microarray analysis [110] (Table 3). Results from similar animal studies also revealed enriched pathways including retrograde endocannabinoid signaling, amphetamine addiction and dopaminergic synapse [111], as well as PI3K-Akt, MAPK, PPAR and Toll-like receptor signaling pathway [112].

Table 3.
Summary of recent studies of non-coding RNAs expression in diabetic neuropathy pathogenesis.
Table 3.
Summary of recent studies of non-coding RNAs expression in diabetic neuropathy pathogenesis.
| Non-Coding RNAs | Potential Mechanism in PDN | Model System | References |
|---|---|---|---|
| MiRNA | |||
| miR199a3p | Increased expression, Serpin E2 downregulation | DM Patients (peripheral blood) | [91] |
| mir9 | Increased expression, CALHM1 upregulated | PDN rat model (spinal dorsal horn) | [92] |
| miR25 | Decreased expression, regulation of oxidative/nitrative stress | Diabetic mouse (sciatic nerves) | [93] |
| miR146 | Decreased expression, negatively correlated with the levels of IL-1β, TNF-α and NF-κB | T2DM and PDN rat | [94,95] |
| miR190a5p | Decreased expression, SLC17A6 upregulated | Diabetic mouse | [96] |
| LncRNA | |||
| 48+ | Increased expression, P2X3 Receptor upregulated | Diabetic rat (dorsal root ganglia) | [102] |
| Increased expression, P2X7 Receptor upregulated | Diabetic rat (superior cervical ganglia) | [107] | |
| NONRATT021972 | Increased expression, neuropathic pain higher scores/TNF-a upregulated | DM2 Patients (peripheral blood)/diabetic rat | [103] |
| Increased expression, P2X7 Receptor upregulated | PC12 Cells | [106] | |
| Diabetic rat (dorsal root ganglia) | [108] | ||
| BC168687 | Increased expression, TRPV1 Receptor upregulated Increased expression, P2X7 Receptor upregulated | Diabetic rat (dorsal root ganglia) | [104,109] |
| CCNT2-AS1 | Neurotrophin-MAPK signaling pathway affecting | DM/PDN patients (peripheral blood) | [110] |
| RP1-249H1.2 | |||
| CTD-3239E11.2 | |||
| RP11-51B23.3 | |||
| STAM-AS1 | |||
| LINC00629 | |||
| MALAT1 (NEAT2) | Increased expression in DM and PDN patients, affecting multiple physiological processes | ||
| MIR143HG | Decreased expression in PDN patients | ||
| PVT1 | |||
| H19 | Decreased expression in PDN patients VDAC1 downregulation in diabetic cardiomyopathy | DM/PDN patients (peripheral blood) Cultured neonatal rat cardiomyocytes | [110,113] |
DM–diabetes mellites PDN–peripheral diabetic neuropathy; T2DM–type 2 diabetes mellitus.
Post-translational histone modifications (acetylation, methylation etc.) are dynamic processes regulating DNA wrapping in nucleosome formation and chromatin organization. The most prominent mechanism, histone acetylation, is under the regulation of two groups of enzymes: histone acetyltransferases (HATs) and histone deacetylases (HDACs) [114]. Inhibitors of HDACs interfere with the epigenetic process of acetylation and have a role in neuropathic pain [115], peripheral nerve injury–induced neuropathic hypersensitivity [116], but also in diabetic retinopathy and nephropathy [117,118]. HDACs represent potential therapeutic targets in the treatment of diabetic peripheral neuropathy. Studies of the peripheral nerves of diabetic animal models and in vitro high glucose–cultured Schwann cells (RSC96) revealed that STAT3 and HDAC1 are promising targets in PDN prevention via the improvement of autophagy [119].
10. Potential Treatment Targets
It is evident that DN is a complex condition, thus potential treatment management should target numerous pathogenic mechanisms and abnormalities that are shown to be related with DN onset and progression. In the statement of the American Diabetes Association published by Boulton et al. [120], certain aldolase reductase inhibitors were shown to be effective in randomized clinical trials (RCT) in reduction of nerve sorbitol, thus affecting an increased polyol pathway. α-Lipoic acid for reduction of oxygen free radicals, as well as angiotensin converting enzyme (ACE) inhibitors and prostaglandin analogs for increase of nerve blood flow were shown to be effective in RCTs as well [120]. Previously in this paper, the potential roles of ACE inhibitors and metformin were described.
Moreover, it should be noted that in treating DM as a condition, the complications including DN might be reduced or even prevented. In the meta-analysis of Vujkovic et al. [121], numerous drug-gene relationships in DM type 2 patients were described. Furthermore, there are several drugs (sulphonylureas, metformin and thiazolidinediones) that are widely used in the treatment of patients with DM type 2. In the study of Pollastro et al., it was noted that there is an association of certain polymorphisms in several genes (ABCC8, KCNJ11, TCF7L2, CYP2C9, IRS1 and CAPN10) and sulphonylureas responsiveness [122]. The presence of polymorphisms (R61C, G401S, 420del and G465R) in the SLC22A1 gene, as well as certain SLC47A1 and ATM gene variants are associated with metformin responsiveness [122,123].
Drugs currently available for the treatment of neuropathic pain show only modest improvement in symptoms, but sometimes significant side effects. In order to improve therapeutic options for DN, recent studies have analyzed genes encoding for voltage-gated sodium channels and their role in neuropathic pain. One good example is the SCN9A gene coding Nav 1.7 sodium channel, which is involved in pain signaling. Investigations have identified SCN9A gain of function mutations in idiopathic small fiber neuropathy [124] and in painful DPN [125]. In the study of Blesneac et al., rare Nav 1.7 variants were found in 10 out of 111 patients with painful-DPN, but in none of 78 respondents with painless-DPN [126]. In addition, subjects with Nav 1.7 variants showed shorter duration of diabetes and more severe pain. These results are interesting from a clinical point of view because carriers of these genetic variants treated with the anticonvulsant lacosamide had significantly improved pain compared with a placebo [127]. In the era of precision medicine, more similar relevant data are expected.
Additionally, development of novel therapeutics for DN is one of the main focuses in recent human and animal studies. The previously mentioned chemokine, VEGF, is considered as prospective new therapeutic treatment for DN, even for a gene therapy approach. Some studies indicated that upregulation of VEGF could have neuroprotective and neurotropic function, and so could be used for the treatment of DN. However, others showed pathological angiogenesis and vascular permeability after VEFG administration, with consecutive worsening of DN and other DM complications [57]. Such contradictory effects could be explained by different roles of VEGF in the DM onset and progression, as well as by differences in DM and non-DM cases.
As a potential novel epidrug, trichostatin A (TSA) treatment is targeting histone deacetylases and reversing reduced expression of brain derived neurotrophic factor (BDNF) in diabetic mice [128]. Another innovative approach in epidrug development is the application of pharmaceutical nanocarriers with different miRNAs. MiR146a5p, was used to create nanoparticle–miRNA-146a-5p polyplexes (nano-miR-146a-5p) with protective effect of on peripheral nerves in the DPN rat model, probably through the inhibition of expression of proinflammatory cytokines [129].
11. Conclusions
Although PDN has a high prevalence and strong impact on the quality of life of DM patients, it is still underdiagnosed, and current therapies are not fully efficient in the majority of cases. Accumulating evidence of epigenetic mechanisms underlying PDN pathogenesis may lead to identification of novel drug targets and development of epidrugs in the era of precision medicine.
Author Contributions
M.J., I.N. and D.N. conceptualization, investigation, methodology, supervision, and writing original draft; J.M.M., S.B. and S.D. formal analysis, and writing initial draft; I.P., D.C., M.G. and D.B. investigation, methodology. All authors approved the final version of the manuscript.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial or not-for-profit sectors.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
Authors declare no conflict of interest.
References
- Witzel, I.-I.; Jelinek, H.F.; Khalaf, K.; Lee, S.; Khandoker, A.H.; Alsafar, H. Identifying Common Genetic Risk Factors of Diabetic Neuropathies. Front. Endocrinol. (Lausanne) 2015, 6, 88. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhu, R.; Wang, D.; Liu, X. Genetics of Diabetic Neuropathy: Systematic Review, Meta-Analysis and Trial Sequential Analysis. Ann. Clin. Transl. Neurol. 2019, 6, 1996–2013. [Google Scholar] [CrossRef]
- Kallinikou, D.; Soldatou, A.; Tsentidis, C.; Louraki, M.; Kanaka-Gantenbein, C.; Kanavakis, E.; Karavanaki, K. Diabetic Neuropathy in Children and Adolescents with Type 1 Diabetes Mellitus: Diagnosis, Pathogenesis, and Associated Genetic Markers. Diabetes. Metab. Res. Rev. 2019, 35, e3178. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Han, Y.; Hu, Q.; Zhang, X.; Cui, G.; Li, Z.; Guan, Y. Effects of Common Polymorphisms in the MTHFR and ACE Genes on Diabetic Peripheral Neuropathy Progression: A Meta-Analysis. Mol. Neurobiol. 2017, 54, 2435–2444. [Google Scholar] [CrossRef] [PubMed]
- Verrotti, A.; Giuva, P.T.; Morgese, G.; Chiarelli, F. New trends in the etiopathogenesis of diabetic peripheral neuropathy. J. Child. Neurol. 2001, 16, 389–394. [Google Scholar] [CrossRef] [PubMed]
- Novakovic, I.; Maksimovic, N.; Pavlovic, A.; Zarkovic, M.; Rovcanin, B.; Mirkovic, D.; Pekmezovic, T.; Cvetkovic, D. Introduction to Molecular Genetic Diagnostics. J. Med. Biochem. 2014, 33, 3–7. [Google Scholar] [CrossRef][Green Version]
- Wilson, N.M.; Wright, D.E. Inflammatory Mediators in Diabetic Neuropathy. J. Diabetes Metab. 2011, S5, 4. [Google Scholar] [CrossRef]
- Vinik, A.I.; Nevoret, M.L.; Casellini, C.; Parson, H. Diabetic neuropathy. Endocrinol. Metab. Clin. N. Am. 2013, 42, 747–787. [Google Scholar] [CrossRef]
- Groener, J.B.; Jende, J.M.E.; Kurz, F.T.; Kender, Z.; Treede, R.D.; Schuh-Hofer, S.; Nawroth, P.P.; Bendszus, M.; Kopf, S. Understanding Diabetic Neuropathy-From Subclinical Nerve Lesions to Severe Nerve Fiber Deficits: A Cross-Sectional Study in Patients With Type 2 Diabetes and Healthy Control Subjects. Diabetes 2020, 69, 436–447. [Google Scholar] [CrossRef]
- Galosi, E.; Di Pietro, G.; La Cesa, S.; Di Stefano, G.; Leone, C.; Fasolino, A.; Di Lionardo, A.; Leonetti, F.; Buzzetti, R.; Mollica, C.; et al. Differential involvement of myelinated and unmyelinated nerve fibers in painful diabetic polyneuropathy. Muscle Nerve 2021, 63, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L. Small Fiber Neuropathy. Semin. Neurol. 2019, 39, 570–577. [Google Scholar] [CrossRef]
- Vinik, A.I.; Park, T.S.; Stansberry, K.B.; Pittenger, G.L. Diabetic neuropathies. Diabetologia 2000, 43, 957–973. [Google Scholar] [CrossRef] [PubMed]
- Freeman, R. Diabetic autonomic neuropathy. Handb. Clin. Neurol. 2014, 126, 63–79. [Google Scholar]
- Azmi, S.; Petropoulos, I.N.; Ferdousi, M.; Ponirakis, G.; Alam, U.; Malik, R.A. An update on the diagnosis and treatment of diabetic somatic and autonomic neuropathy. F1000Research 2019, 8, 186. [Google Scholar] [CrossRef]
- Gatopoulou, A.; Papanas, N.; Maltezos, E. Diabetic gastrointestinal autonomic neuropathy: Current status and new achievements for everyday clinical practice. Eur. J. Intern. Med. 2012, 23, 499–505. [Google Scholar] [CrossRef] [PubMed]
- Bissinger, A. Cardiac Autonomic Neuropathy: Why Should Cardiologists Care about That? J. Diabetes Res. 2017, 2017, 5374176. [Google Scholar] [CrossRef] [PubMed]
- Hecht, M.J.; Neundörfer, B.; Kiesewetter, F.; Hilz, M.J. Neuropathy is a major contributing factor to diabetic erectile dysfunction. Neurol. Res. 2001, 23, 651–654. [Google Scholar] [CrossRef] [PubMed]
- Agochukwu-Mmonu, N.; Pop-Busui, R.; Wessells, H.; Sarma, A.V. Autonomic neuropathy and urologic complications in diabetes. Auton. Neurosci. 2020, 229, 102736. [Google Scholar] [CrossRef]
- Nielsen, F.S.; Hansen, H.P.; Jacobsen, P.; Rossing, P.; Smidt, U.M.; Christensen, N.J.; Pevet, P.; Vivien-Roels, B.; Parving, H.H. Increased sympathetic activity during sleep and nocturnal hypertension in Type 2 diabetic patients with diabetic nephropathy. Diabet. Med. 1999, 16, 555–562. [Google Scholar] [CrossRef]
- Gandhi, P.G.; Gundu, H.R. Detection of neuropathy using a sudomotor test in type 2 diabetes. Degener. Neurol. Neuromuscul. Dis. 2015, 5, 1–7. [Google Scholar] [PubMed]
- Schmidt, R.E. Neuropathology and pathogenesis of diabetic autonomic neuropathy. Int. Rev. Neurobiol. 2002, 50, 257–292. [Google Scholar]
- Naruse, K. Schwann Cells as Crucial Players in Diabetic Neuropathy. Adv. Exp. Med. Biol. 2019, 1190, 345–356. [Google Scholar]
- De Gregorio, C.; Contador, D.; Campero, M.; Ezquer, M.; Ezquer, F. Characterization of diabetic neuropathy progression in a mouse model of type 2 diabetes mellitus. Biol. Open. 2018, 7, bio036830. [Google Scholar] [CrossRef] [PubMed]
- Younger, D.S. Diabetic neuropathy: A clinical and neuropathological study of 107 patients. Neurol. Res. Int. 2010, 2010, 140379. [Google Scholar] [CrossRef] [PubMed]
- Zenker, J.; Ziegler, D.; Chrast, R. Novel pathogenic pathways in diabetic neuropathy. Trends Neurosci. 2013, 36, 439–449. [Google Scholar] [CrossRef] [PubMed]
- Mizisin, A.P. Mechanisms of diabetic neuropathy: Schwann cells. Handb. Clin. Neurol. 2014, 126, 401–428. [Google Scholar] [PubMed]
- Robertson, D.M.; Sima, A.A. Diabetic Neuropathy in the Mutant Mouse [C57BL/Ks(Db/Db)]: A Morphometric Study. Diabetes 1980, 29, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, N.P.; Vægter, C.B.; Andersen, H.; Østergaard, L.; Calcutt, N.A.; Jensen, T.S. Schwann cell interactions with axons and microvessels in diabetic neuropathy. Nat. Rev. Neurol. 2017, 13, 135–147. [Google Scholar] [CrossRef] [PubMed]
- Akude, E.; Zherebitskaya, E.; Chowdhury, S.K.R.; Smith, D.R.; Dobrowsky, R.T.; Fernyhough, P. Diminished Superoxide Generation Is Associated with Respiratory Chain Dysfunction and Changes in the Mitochondrial Proteome of Sensory Neurons from Diabetic Rats. Diabetes 2011, 60, 288–297. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.-J.; Price, S.A.; Chilton, L.; Calcutt, N.A.; Tomlinson, D.R.; Verkhratsky, A.; Fernyhough, P. Insulin Prevents Depolarization of the Mitochondrial Inner Membrane in Sensory Neurons of Type 1 Diabetic Rats in the Presence of Sustained Hyperglycemia. Diabetes 2003, 52, 2129–2136. [Google Scholar] [CrossRef] [PubMed]
- Roy Chowdhury, S.K.; Smith, D.R.; Saleh, A.; Schapansky, J.; Marquez, A.; Gomes, S.; Akude, E.; Morrow, D.; Calcutt, N.A.; Fernyhough, P. Impaired adenosine monophosphate-activated protein kinase signalling in dorsal root ganglia neurons is linked to mitochondrial dysfunction and peripheral neuropathy in diabetes. Brain 2012, 135 Pt 6, 1751–1766. [Google Scholar] [CrossRef]
- Sifuentes-Franco, S.; Pacheco-Moisés, F.P.; Rodríguez-Carrizalez, A.D.; Miranda-Díaz, A.G. The Role of Oxidative Stress, Mitochondrial Function, and Autophagy in Diabetic Polyneuropathy. J. Diabetes Res. 2017, 2017, 1673081. [Google Scholar] [CrossRef]
- Román-Pintos, L.M.; Villegas-Rivera, G.; Rodríguez-Carrizalez, A.D.; Miranda-Díaz, A.G.; Cardona-Muñoz, E.G. Diabetic Polyneuropathy in Type 2 Diabetes Mellitus: Inflammation, Oxidative Stress, and Mitochondrial Function. J. Diabetes. Res. 2016, 2016, 3425617. [Google Scholar] [CrossRef] [PubMed]
- Verge, V.M.K.; Andreassen, C.S.; Arnason, T.G.; Andersen, H. Mechanisms of Disease: Role of Neurotrophins in Diabetes and Diabetic Neuropathy. Handb. Clin. Neurol. 2014, 126, 443–460. [Google Scholar]
- Singh, R.; Kishore, L.; Kaur, N. Diabetic Peripheral Neuropathy: Current Perspective and Future Directions. Pharmacol. Res. 2014, 80, 21–35. [Google Scholar] [CrossRef] [PubMed]
- Dewanjee, S.; Das, S.; Das, A.K.; Bhattacharjee, N.; Dihingia, A.; Dua, T.K.; Kalita, J.; Manna, P. Molecular Mechanism of Diabetic Neuropathy and Its Pharmacotherapeutic Targets. Eur. J. Pharmacol. 2018, 833, 472–523. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, K.; Kato, N.; Makino, M.; Suzuki, T.; Shindo, M. Continuous Inhibition of Excessive Polyol Pathway Flux in Peripheral Nerves by Aldose Reductase Inhibitor Fidarestat Leads to Improvement of Diabetic Neuropathy. J. Diabetes Complicat. 1999, 13, 141–150. [Google Scholar] [CrossRef]
- Vincent, A.M.; Russell, J.W.; Low, P.; Feldman, E.L. Oxidative Stress in the Pathogenesis of Diabetic Neuropathy. Endocr. Rev. 2004, 25, 612–628. [Google Scholar] [CrossRef]
- Buyukaydin, B.; Guler, E.M.; Karaaslan, T.; Olgac, A.; Zorlu, M.; Kiskac, M.; Kocyigit, A. Relationship between Diabetic Polyneuropathy, Serum Visfatin, and Oxidative Stress Biomarkers. World J. Diabetes 2020, 11, 309–321. [Google Scholar] [CrossRef]
- Pop-Busui, R.; Sima, A.; Stevens, M. Diabetic Neuropathy and Oxidative Stress. Diabetes. Metab. Res. Rev. 2006, 22, 257–273. [Google Scholar] [CrossRef]
- Jin, H.Y.; Park, T.S. Role of Inflammatory Biomarkers in Diabetic Peripheral Neuropathy. J. Diabetes Investig. 2018, 9, 1016–1018. [Google Scholar] [CrossRef] [PubMed]
- Doupis, J.; Lyons, T.E.; Wu, S.; Gnardellis, C.; Dinh, T.; Veves, A. Microvascular Reactivity and Inflammatory Cytokines in Painful and Painless Peripheral Diabetic Neuropathy. J. Clin. Endocrinol. Metab. 2009, 94, 2157–2163. [Google Scholar] [CrossRef]
- Bönhof, G.J.; Herder, C.; Strom, A.; Papanas, N.; Roden, M.; Ziegler, D. Emerging Biomarkers, Tools, and Treatments for Diabetic Polyneuropathy. Endocr. Rev. 2019, 40, 153–192. [Google Scholar] [CrossRef]
- Nikolic, D.; Jankovic, M.; Petrovic, B.; Novakovic, I. Genetic Aspects of Inflammation and Immune Response in Stroke. Int. J. Mol. Sci. 2020, 21, 7409. [Google Scholar] [CrossRef] [PubMed]
- Bonini, S.; Rasi, G.; Bracci-Laudiero, M.L.; Procoli, A.; Aloe, L. Nerve Growth Factor: Neurotrophin or Cytokine? Int. Arch. Allergy Immunol. 2003, 131, 80–84. [Google Scholar] [CrossRef] [PubMed]
- Stino, A.M.; Rumora, A.E.; Kim, B.; Feldman, E.L. Evolving concepts on the role of dyslipidemia, bioenergetics, and inflammation in the pathogenesis and treatment of diabetic peripheral neuropathy. J. Peripher. Nerv. Syst. 2020, 25, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Vincent, A.M.; Callaghan, B.C.; Smith, A.L.; Feldman, E.L. Diabetic neuropathy: Cellular mechanisms as therapeutic targets. Nat. Rev. Neurol. 2011, 7, 573–583. [Google Scholar] [CrossRef] [PubMed]
- Feldman, E.L.; Callaghan, B.C.; Pop-Busui, R.; Zochodne, D.W.; Wright, D.E.; Bennett, D.L.; Bril, V.; Russel, J.W.; Viswanathan, V. Diabetic neuropathy. Nat. Rev. Dis. Primers 2019, 5, 41. [Google Scholar] [CrossRef]
- Cai, Z.; Yang, Y.; Zhang, J. A systematic review and meta-analysis of the serum lipid profile in prediction of diabetic neuropathy. Sci. Rep. 2021, 11, 499. [Google Scholar] [CrossRef]
- Samadi, A.; Gurlek, A.; Sendur, S.N.; Karahan, S.; Akbiyik, F.; Lay, I. Oxysterol species: Reliable markers of oxidative stress in diabetes mellitus. J. Endocrinol. Investig. 2019, 42, 7–17. [Google Scholar] [CrossRef]
- Mutemberezi, V.; Guillemot-Legris, O.; Muccioli, G.G. Oxysterols: From cholesterol metabolites to key mediators. Prog. Lipid. Res. 2016, 64, 152–169. [Google Scholar] [CrossRef] [PubMed]
- Ferderbar, S.; Pereira, E.C.; Apolinário, E.; Bertolami, M.C.; Faludi, A.; Monte, O.; Calliari, L.E.; Sales, J.E.; Gagliardi, A.R.; Xavier, H.T.; et al. Cholesterol oxides as biomarkers of oxidative stress in type 1 and type 2 diabetes mellitus. Diabetes Metab. Res. Rev. 2007, 23, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.S.M.; Chung, S.K. Genetic Analysis of Aldose Reductase in Diabetic Complications. Curr. Med. Chem. 2003, 10, 1375–1387. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Chen, S.; Mei, Z.; Zhao, F.; Xiang, Y. Polymorphisms in Sorbitol-Aldose Reductase (Polyol) Pathway Genes and Their Influence on Risk of Diabetic Retinopathy among Han Chinese. Med. Sci. Monit. 2019, 25, 7073–7078. [Google Scholar] [CrossRef] [PubMed]
- Sivenius, K.; Pihlajamäki, J.; Partanen, J.; Niskanen, L.; Laakso, M.; Uusitupa, M. Aldose Reductase Gene Polymorphisms and Peripheral Nerve Function in Patients with Type 2 Diabetes. Diabetes Care 2004, 27, 2021–2026. [Google Scholar] [CrossRef]
- Heesom, A.E.; Millward, A.; Demaine, A.G. Susceptibility to Diabetic Neuropathy in Patients with Insulin Dependent Diabetes Mellitus Is Associated with a Polymorphism at the 5′ End of the Aldose Reductase Gene. J. Neurol. Neurosurg. Psychiatry 1998, 64, 213–216. [Google Scholar] [CrossRef]
- Boulton, A.J.M.; Malik, R.A.; Arezzo, J.C.; Sosenko, J.M. Diabetic Somatic Neuropathies. Diabetes Care 2004, 27, 1458–1486. [Google Scholar] [CrossRef]
- Vinik, A.; Maser, R.; Nakave, A. Diabetic cardiovascular autonomic nerve dysfunction. US Endocr. Dis. 2007, 2, 66–74. [Google Scholar] [CrossRef]
- Forbes, J.M.; Cooper, M.E. Mechanisms of Diabetic Complications. Physiol. Rev. 2013, 93, 137–188. [Google Scholar] [CrossRef]
- Li, Y.; Tong, N. Angiotensin-converting enzyme I/D polymorphism and diabetic peripheral neuropathy in type 2 diabetes mellitus: A meta-analysis. J. Renin Angiotensin. Aldosterone Syst. 2015, 16, 787–792. [Google Scholar] [CrossRef]
- Rahimi, Z. ACE insertion/deletion (I/D) polymorphism and diabetic nephropathy. J. Nephropathol. 2012, 1, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Fan, D.; Zhang, Y. Angiogenin gene polymorphism: A risk factor for diabetic peripheral neuropathy in the northern Chinese Han population. Neural. Regen. Res. 2013, 8, 3434–3440. [Google Scholar]
- Mooyaart, A.L.; Valk, E.J.; van Es, L.A.; Bruijn, J.A.; de Heer, E.; Freedman, B.I.; Dekkers, O.M.; Baelde, H.J. Genetic associations in diabetic nephropathy: A meta-analysis. Diabetologia 2011, 54, 544–553. [Google Scholar] [CrossRef]
- Bedlack, R.S.; Edelman, D.; Gibbs, J.W., 3rd; Kelling, D.; Strittmatter, W.; Saunders, A.M.; Morgenlander, J. APOE genotype is a risk factor for neuropathy severity in diabetic patients. Neurology 2003, 60, 1022–1024. [Google Scholar] [CrossRef] [PubMed]
- Monastiriotis, C.; Papanas, N.; Veletza, S.; Maltezos, E. APOE gene polymorphisms and diabetic peripheral neuropathy. Arch. Med. Sci. 2012, 8, 583–588. [Google Scholar] [CrossRef]
- Yigit, S.; Karakus, N.; Inanir, A. Association of MTHFR gene C677T mutation with diabetic peripheral neuropathy and diabetic retinopathy. Mol. Vis. 2013, 19, 1626–1630. [Google Scholar] [PubMed]
- Tutuncu, N.B.; Erbas, T.; Alikasifoglu, M.; Tuncbilek, E. Thermolabile methylenetet-rahydrofolate reductase enzyme genotype is frequent in type 2 diabetic patients with normal fasting homocysteine levels. J. Intern. Med. 2005, 257, 446–453. [Google Scholar] [CrossRef]
- Wang, H.; Fan, D.; Hong, T. Is the C677T polymorphism in methylenetetrahydrofo-late reductase gene or plasma homocysteine a risk factor for diabetic peripheral neuropathy in Chinese individuals? Neural Regen. Res. 2012, 7, 2384–2391. [Google Scholar] [PubMed]
- Rahimi, M.; Hasanvand, A.; Rahimi, Z.; Vaisi-Raygani, A.; Mozafari, H.; Rezaei, M.; Zargooshi, J.; Najafi, F.; Shakiba, E. Synergistic effects of the MTHFR C677T and A1298C polymorphisms on the increased risk of micro-and macro-albuminuria and progression of diabetic nephropathy among Iranians with type 2 diabetes mellitus. Clin. Biochem. 2010, 43, 1333–1339. [Google Scholar] [CrossRef]
- Ezzidi, I.; Mtiraoui, N.; Mohamed, M.; Mahjoub, T.; Kacem, M.; Almawi, W. Association of endothelial nitric oxide synthase Glu298Asp, 4b/a, and-786T>C gene vari-ants with diabetic nephropathy. J. Diabetes Complicat. 2008, 22, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Dellamea, B.S.; Pinto, L.C.; Leitão, C.B.; Santos, K.G.; Canani, L.H. Endothelial nitric oxide synthase gene polymorphisms and risk of diabetic nephropa-thy: A systematic review and meta-analysis. BMC Med. Genet. 2014, 15, 9. [Google Scholar] [CrossRef] [PubMed]
- Bonnefond, A.; Saulnier, P.J.; Stathopoulou, M.G.; Grarup, N.; Ndiaye, N.C.; Roussel, R.; Nezhad, M.A.; Dechaume, A.; Lantieri, O.; Hercberg, S.; et al. What is the contribution of two genetic variants regulating VEGF levels to type 2 diabetes risk and to microvascular complications? PLoS ONE 2013, 8, e55921. [Google Scholar]
- Babizhayev, M.A.; Strokov, I.A.; Nosikov, V.V.; Savel’yeva, E.L.; Sitnikov, V.F.; Yegorov, Y.E.; Lankin, V.Z. The Role of Oxidative Stress in Diabetic Neuropathy: Generation of Free Radical Species in the Glycation Reaction and Gene Polymorphisms Encoding Antioxidant Enzymes to Genetic Susceptibility to Diabetic Neuropathy in Population of Type I Diabetic Patients. Cell Biochem. Biophys. 2015, 71, 1425–1443. [Google Scholar] [CrossRef]
- Meng, W.; Deshmukh, H.A.; Donnelly, L.A.; Torrance, N.; Colhoun, H.M.; Palmer, C.N.; Smith, B.H.; Wellcome Trust Case Control Consortium 2 (WTCCC2); Surrogate Markers for Micro-and Macro-Vascular Hard Endpoints for Innovative Diabetes Tools (SUMMIT) Study Group. A Genome-wide Association Study Provides Evidence of Sex-specific Involvement of Chr1p35.1 (ZSCAN20-TLR12P) and Chr8p23.1 (HMGB1P46) With Diabetic Neuropathic Pain. EBioMedicine 2015, 2, 1386–1393. [Google Scholar] [CrossRef] [PubMed]
- Testa, R.; Bonfigli, A.R.; Prattichizzo, F.; La Sala, L.; De Nigris, V.; Ceriello, A. The “Metabolic Memory” Theory and the Early Treatment of Hyperglycemia in Prevention of Diabetic Complications. Nutrients 2017, 9, 437. [Google Scholar] [CrossRef]
- Berger, S.L.; Kouzarides, T.; Shiekhattar, R.; Shilatifard, A. An Operational Definition of Epigenetics. Genes Dev. 2009, 23, 781–783. [Google Scholar] [CrossRef] [PubMed]
- Reddy, M.A.; Zhang, E.; Natarajan, R. Epigenetic Mechanisms in Diabetic Complications and Metabolic Memory. Diabetologia 2015, 58, 443–455. [Google Scholar] [CrossRef] [PubMed]
- Moore, L.D.; Le, T.; Fan, G. DNA methylation and its basic function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef]
- Luo, C.; Hajkova, P.; Ecker, J.R. Dynamic DNA methylation: In the right place at the right time. Science 2018, 361, 1336–1340. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Miao, F.; Paterson, A.D.; Lachin, J.M.; Zhang, L.; Schones, D.E.; Wu, X.; Wang, J.; Tompkins, J.D.; Genuth, S.; et al. DCCT/EDIC Research Group. Epigenomic Profiling Reveals an Association between Persistence of DNA Methylation and Metabolic Memory in the DCCT/EDIC Type 1 Diabetes Cohort. Proc. Natl. Acad. Sci. USA 2016, 113, E3002–E3011. [Google Scholar] [CrossRef]
- Zhang, H.-H.; Han, X.; Wang, M.; Hu, Q.; Li, S.; Wang, M.; Hu, J. The Association between Genomic DNA Methylation and Diabetic Peripheral Neuropathy in Patients with Type 2 Diabetes Mellitus. J. Diabetes Res. 2019, 2019, 2494057. [Google Scholar] [CrossRef] [PubMed]
- Gastoł, J.; Kapusta, P.; Polus, A.; Pitera, E.; Biela, M.; Wołkow, P.; Pawliński, Ł.; Kieć-Wilk, B. Epigenetic Mechanism in Search for the Pathomechanism of Diabetic Neuropathy Development in Diabetes Mellitus Type 1 (T1DM). Endocrine 2020, 68, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Araki, T.; Milbrandt, J. Ninjurin2, a Novel Homophilic Adhesion Molecule, Is Expressed in Mature Sensory and Enteric Neurons and Promotes Neurite Outgrowth. J. Neurosci. 2000, 20, 187–195. [Google Scholar] [CrossRef] [PubMed]
- Blattler, A.; Farnham, P.J. Cross-talk between site-specific transcription factors and DNA methylation states. J. Biol. Chem. 2013, 288, 34287–34294. [Google Scholar] [CrossRef] [PubMed]
- Guo, K.; Elzinga, S.; Eid, S.; Figueroa-Romero, C.; Hinder, L.M.; Pacut, C.; Feldman, E.L.; Hur, J. Genome-Wide DNA Methylation Profiling of Human Diabetic Peripheral Neuropathy in Subjects with Type 2 Diabetes Mellitus. Epigenetics 2019, 14, 766–779. [Google Scholar] [CrossRef]
- Guo, K.; Eid, S.A.; Elzinga, S.E.; Pacut, C.; Feldman, E.L.; Hur, J. Genome-Wide Profiling of DNA Methylation and Gene Expression Identifies Candidate Genes for Human Diabetic Neuropathy [Published Correction Appears in Clin Epigenetics. Clin. Epigenet. 2020, 12, 123. [Google Scholar] [CrossRef]
- Hur, J.; Sullivan, K.A.; Pande, M.; Hong, Y.; Sima, A.A.F.; Jagadish, H.V.; Kretzler, M.; Feldman, E.L. The Identification of Gene Expression Profiles Associated with Progression of Human Diabetic Neuropathy. Brain 2011, 134, 3222–3235. [Google Scholar] [CrossRef]
- Liu, J. Control of protein synthesis and mRNA degradation by microRNAs. Curr. Opin. Cell Biol. 2008, 20, 214–221. [Google Scholar] [CrossRef] [PubMed]
- Xourgia, E.; Papazafiropoulou, A.; Melidonis, A. Circulating MicroRNAs as Biomarkers for Diabetic Neuropathy: A Novel Approach. World, J. Exp. Med. 2018, 8, 18–23. [Google Scholar] [CrossRef]
- Simeoli, R.; Fierabracci, A. Insights into the Role of MicroRNAs in the Onset and Development of Diabetic Neuropathy. Int. J. Mol. Sci. 2019, 20, 4627. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.B.; Wu, Q.; Liu, J.; Fan, Y.Z.; Yu, K.F.; Cai, Y. miR-199a-3p is involved in the pathogenesis and progression of diabetic neuropathy through downregulation of SerpinE2. Mol. Med. Rep. 2017, 16, 2417–2424. [Google Scholar] [CrossRef]
- Liu, W.; Ao, Q.; Guo, Q.; He, W.; Peng, L.; Jiang, J.; Hu, X. miR-9 Mediates CALHM1-Activated ATP-P2X7R Signal in Painful Diabetic Neuropathy Rats. Mol. Neurobiol. 2017, 54, 922–929. [Google Scholar] [CrossRef]
- Zhang, Y.; Song, C.; Liu, J.; Bi, Y.; Li, H. Inhibition of miR-25 aggravates diabetic peripheral neuropathy. Neuroreport 2018, 29, 945–953. [Google Scholar] [CrossRef]
- Feng, Y.; Chen, L.; Luo, Q.; Wu, M.; Chen, Y.; Shi, X. Involvement of microRNA-146a in diabetic peripheral neuropathy through the regulation of inflammation. Drug Des. Devel. Ther. 2018, 12, 171–177. [Google Scholar] [CrossRef]
- Wang, L.; Chopp, M.; Lu, X.; Szalad, A.; Jia, L.; Liu, X.S.; Wu, K.H.; Lu, M.; Zhang, Z.G. miR-146a mediates thymosin β4 induced neurovascular remodeling of diabetic peripheral neuropathy in type-II diabetic mice. Brain Res. 2019, 1707, 198–207. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Yang, Q.; Wei, X.; Liu, Y.; Ma, D.; Li, J.; Wan, Y.; Luo, Y. The role of miR-190a-5p contributes to diabetic neuropathic pain via targeting SLC17A6. J. Pain Res. 2017, 10, 2395–2403. [Google Scholar] [CrossRef] [PubMed]
- Saunders, M.A.; Liang, H.; Li, W.-H. Human Polymorphism at MicroRNAs and MicroRNA Target Sites. Proc. Natl. Acad. Sci. USA 2007, 104, 3300–3305. [Google Scholar] [CrossRef]
- Ciccacci, C.; Morganti, R.; Di Fusco, D.; D’Amato, C.; Cacciotti, L.; Greco, C.; Rufini, S.; Novelli, G.; Sangiuolo, F.; Marfia, G.A.; et al. Common Polymorphisms in MIR146a, MIR128a and MIR27a Genes Contribute to Neuropathy Susceptibility in Type 2 Diabetes. Acta Diabetol. 2014, 51, 663–671. [Google Scholar] [CrossRef] [PubMed]
- Ciccacci, C.; Latini, A.; Greco, C.; Politi, C.; D’Amato, C.; Lauro, D.; Novelli, G.; Borgiani, P.; Spallone, V. Association between a MIR499A Polymorphism and Diabetic Neuropathy in Type 2 Diabetes. J. Diabetes Complicat. 2018, 32, 11–17. [Google Scholar] [CrossRef]
- Latini, A.; Borgiani, P.; De Benedittis, G.; D’Amato, C.; Greco, C.; Lauro, D.; Novelli, G.; Spallone, V.; Ciccacci, C. Mitochondrial DNA Copy Number in Peripheral Blood Is Reduced in Type 2 Diabetes Patients with Polyneuropathy and Associated with a MIR499A Gene Polymorphism. DNA Cell Biol. 2020, 39, 1467–1472. [Google Scholar] [CrossRef]
- Batista, P.J.; Chang, H.Y. Long noncoding RNAs: Cellular address codes in development and disease. Cell 2013, 152, 1298–1307. [Google Scholar] [CrossRef]
- Wang, S.; Xu, H.; Zou, L.; Xie, J.; Wu, H.; Wu, B.; Yi, Z.; Lv, Q.; Zhang, X.; Ying, M.; et al. 48+ Is Involved in Diabetic Neuropathic Pain Mediated by the P2X3 Receptor in the Dorsal Root Ganglia. Purinergic Signal. 2016, 12, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Zhao, G.Q.; Cao, R.J.; Zhu, Z.H.; Li, K. LncRNANONRATT021972 Was Associated with Neuropathic Pain Scoring in Patients with Type 2 Diabetes. Behav. Neurol. 2017, 2017, 2941297. [Google Scholar] [CrossRef]
- Liu, C.; Li, C.; Deng, Z.; Du, E.; Xu, C. Long non-coding RNA BC168687 is involved in TRPV1-mediated diabetic neuropathic pain in rats. Neuroscience 2018, 15, 214–222. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, X.; Chen, X.; Li, S.; Ho, I.H.T.; Liu, X.; Chan, M.T.V.; Wu, W.K.K. Emerging Roles of Long Non-Coding RNAs in Neuropathic Pain. Cell Prolif 2019, 52, e12528. [Google Scholar] [CrossRef]
- Xu, H.; He, L.; Liu, C.; Tang, L.; Xu, Y.; Xiong, M.; Yang, M.; Fan, Y.; Hu, F.; Liu, X.; et al. LncRNA NONRATT021972 SiRNA Attenuates P2X7 Receptor Expression and Inflammatory Cytokine Production Induced by Combined High Glucose and Free Fatty Acids in PC12 Cells. Purinergic Signal. 2016, 12, 259–268. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Zhang, C.; Zou, L.; Ma, Y.; Huang, K.; Lv, Q.; Zhang, X.; Wang, S.; Xue, Y.; Yi, Z.; et al. 48+ SiRNA Improved Diabetic Sympathetic Neuropathy in Type 2 Diabetic Rats Mediated By P2X7 Receptor in SCG. Auton. Neurosci. 2016, 197, 14–18. [Google Scholar] [CrossRef]
- Peng, H.; Zou, L.; Xie, J.; Wu, H.; Wu, B.; Zhu, G.; Lv, Q.; Zhang, X.; Liu, S.; Li, G.; et al. LncRNA NONRATT021972 SiRNA Decreases Diabetic Neuropathic Pain Mediated by the P2x(3) Receptor in Dorsal Root Ganglia. Mol. Neurobiol. 2017, 54, 511–523. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Tao, J.; Wu, H.; Yang, Y.; Chen, Q.; Deng, Z.; Liu, J.; Xu, C. Effects of LncRNA BC168687 siRNA on diabetic neuropathic pain mediated by P2X(7) Receptor on SGCs in DRG of rats. Biomed. Res. Int. 2017, 2017, 7831251. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Ji, L.-D.; Cai, J.-J.; Feng, M.; Zhou, M.; Hu, S.-P.; Xu, J.; Zhou, W.-H. Microarray Analysis of Long Noncoding RNAs in Female Diabetic Peripheral Neuropathy Patients. Cell. Physiol. Biochem. 2018, 46, 1209–1217. [Google Scholar] [CrossRef]
- Du, H.; Liu, Z.; Tan, X.; Ma, Y.; Gong, Q. Identification of the Genome-Wide Expression Patterns of Long Non-Coding RNAs and mRNAs in Mice with Streptozotocin-Induced Diabetic Neuropathic Pain. Neuroscience 2019, 402, 90–103. [Google Scholar] [CrossRef]
- Guo, G.; Ren, S.; Kang, Y.; Liu, Y.; Duscher, D.; Machens, H.-G.; Chen, Z. Microarray Analyses of LncRNAs and mRNAs Expression Profiling Associated with Diabetic Peripheral Neuropathy in Rats. J. Cell. Biochem. 2019, 120, 15347–15359. [Google Scholar] [CrossRef]
- Li, X.; Wang, H.; Yao, B.; Xu, W.; Chen, J.; Zhou, X. lncRNA H19/miR-675 axis regulates cardiomyocyte apoptosis by targeting VDAC1 in diabetic cardiomyopathy. Sci. Rep. 2016, 6, 36340. [Google Scholar] [CrossRef] [PubMed]
- Margueron, R.; Reinberg, D. Chromatin structure and the inheritance of epigenetic information. Nat. Rev. Genet. 2010, 11, 285–296. [Google Scholar] [CrossRef]
- Denk, F.; Huang, W.; Sidders, B.; Bithell, A.; Crow, M.; Grist, J.; Sharma, S.; Ziemek, D.; Rice, A.S.C.; Buckley, N.J.; et al. HDAC Inhibitors Attenuate the Development of Hypersensitivity in Models of Neuropathic Pain. Pain 2013, 154, 1668–1679. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Shen, X.; Xu, Y.; Xu, S.; Xia, F.; Zhu, B.; Liu, Y.; Wang, W.; Wu, H.; Wang, F. The Etiological Changes of Acetylation in Peripheral Nerve Injury-Induced Neuropathic Hypersensitivity. Mol. Pain 2018, 14, 1744806918798408. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, C.; Sun, G. Histone Acetylation and Its Modifiers in the Pathogenesis of Diabetic Nephropathy. J. Diabetes Res. 2016, 2016, 4065382. [Google Scholar] [CrossRef] [PubMed]
- Abouhish, H.; Thounaojam, M.C.; Jadeja, R.N.; Gutsaeva, D.R.; Powell, F.L.; Khriza, M.; Martin, P.M.; Bartoli, M. Inhibition of HDAC6 Attenuates Diabetes-Induced Retinal Redox Imbalance and Microangiopathy. Antioxidants (Basel) 2020, 9, 599. [Google Scholar] [CrossRef]
- Du, W.; Wang, N.; Li, F.; Jia, K.; An, J.; Liu, Y.; Wang, Y.; Zhu, L.; Zhao, S.; Hao, J. STAT3 Phosphorylation Mediates High Glucose-Impaired Cell Autophagy in an HDAC1-Dependent and-Independent Manner in Schwann Cells of Diabetic Peripheral Neuropathy. FASEB J. 2019, 33, 8008–8021. [Google Scholar] [CrossRef]
- Boulton, A.J.; Vinik, A.I.; Arezzo, J.C.; Bril, V.; Feldman, E.L.; Freeman, R.; Malik, R.A.; Maser, R.E.; Sosenko, J.M.; American Diabetes Association; et al. Diabetic neuropathies: A statement by the American Diabetes Association. Diabetes Care 2005, 28, 956–962. [Google Scholar] [CrossRef] [PubMed]
- Vujkovic, M.; Keaton, J.M.; Lynch, J.A.; Miller, D.R.; Zhou, J.; Tcheandjieu, C.; Huffman, J.E.; Assimes, T.L.; Lorenz, K.; Zhu, X.; et al. Discovery of 318 new risk loci for type 2 diabetes and related vascular outcomes among 1.4 million participants in a multi-ancestry meta-analysis. Nat. Genet. 2020, 52, 680–691. [Google Scholar] [CrossRef]
- Pollastro, C.; Ziviello, C.; Costa, V.; Ciccodicola, A. Pharmacogenomics of Drug Response in Type 2 Diabetes: Toward the Definition of Tailored Therapies? PPAR Res. 2015, 2015, 415149. [Google Scholar] [CrossRef]
- Becker, M.L.; Pearson, E.R.; Tkáč, I. Pharmacogenetics of oral antidiabetic drugs. Int. J. Endocrinol. 2013, 2013, 686315. [Google Scholar] [CrossRef]
- Faber, C.G.; Hoeijmakers, J.G.; Ahn, H.S.; Cheng, X.; Han, C.; Choi, J.S.; Estacion, M.; Lauria, G.; Vanhoutte, E.K.; Gerrits, M.M.; et al. Gain of function Naν1.7 mutations in idiopathic small fiber neuropathy. Ann. Neurol. 2012, 71, 26–39. [Google Scholar] [CrossRef]
- Li, Q.S.; Cheng, P.; Favis, R.; Wickenden, A.; Romano, G.; Wang, H. SCN9Avariants may be implicated in neuropathic pain associated with diabetic peripheral neuropathy and pain severity. Clin. J. Pain. 2015, 31, 976–982. [Google Scholar] [CrossRef] [PubMed]
- Blesneac, I.; Themistocleous, A.C.; Fratter, C.; Conrad, L.J.; Ramirez, J.D.; Cox, J.J.; Tesfaye, S.; Shillo, P.R.; Rice, A.S.C.; Tucker, S.J.; et al. Rare NaV1.7 variants associated with painful diabetic peripheral neuropathy. Pain 2018, 159, 469–480. [Google Scholar] [CrossRef] [PubMed]
- de Greef, B.T.A.; Hoeijmakers, J.G.J.; Geerts, M.; Oakes, M.; Church, T.J.E.; Waxman, S.G.; Dib-Hajj, S.D.; Faber, C.G.; Merkies, I.S.J. Lacosamide in patients with Nav1.7 mutations-related small fibre neuropathy: A randomized controlled trial. Brain 2019, 142, 263–275. [Google Scholar] [CrossRef] [PubMed]
- An, J.; Zhang, X.; Jia, K.; Zhang, C.; Zhu, L.; Cheng, M.; Li, F.; Zhao, S.; Hao, J. Trichostatin A Increases BDNF Protein Expression by Improving XBP-1s/ATF6/GRP78 Axis in Schwann Cells of Diabetic Peripheral Neuropathy. Biomed. Pharmacother. 2021, 133, 111062. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q.; Feng, Y.; Xie, Y.; Shao, Y.; Wu, M.; Deng, X.; Yuan, W.E.; Chen, Y.; Shi, X. Nanoparticle-microRNA-146a-5p polyplexes ameliorate diabetic peripheral neuropathy by modulating inflammation and apoptosis. Nanomedicine 2019, 17, 188–197. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).