Cell Shape and Matrix Stiffness Impact Schwann Cell Plasticity via YAP/TAZ and Rho GTPases
Abstract
:1. Introduction
2. Results
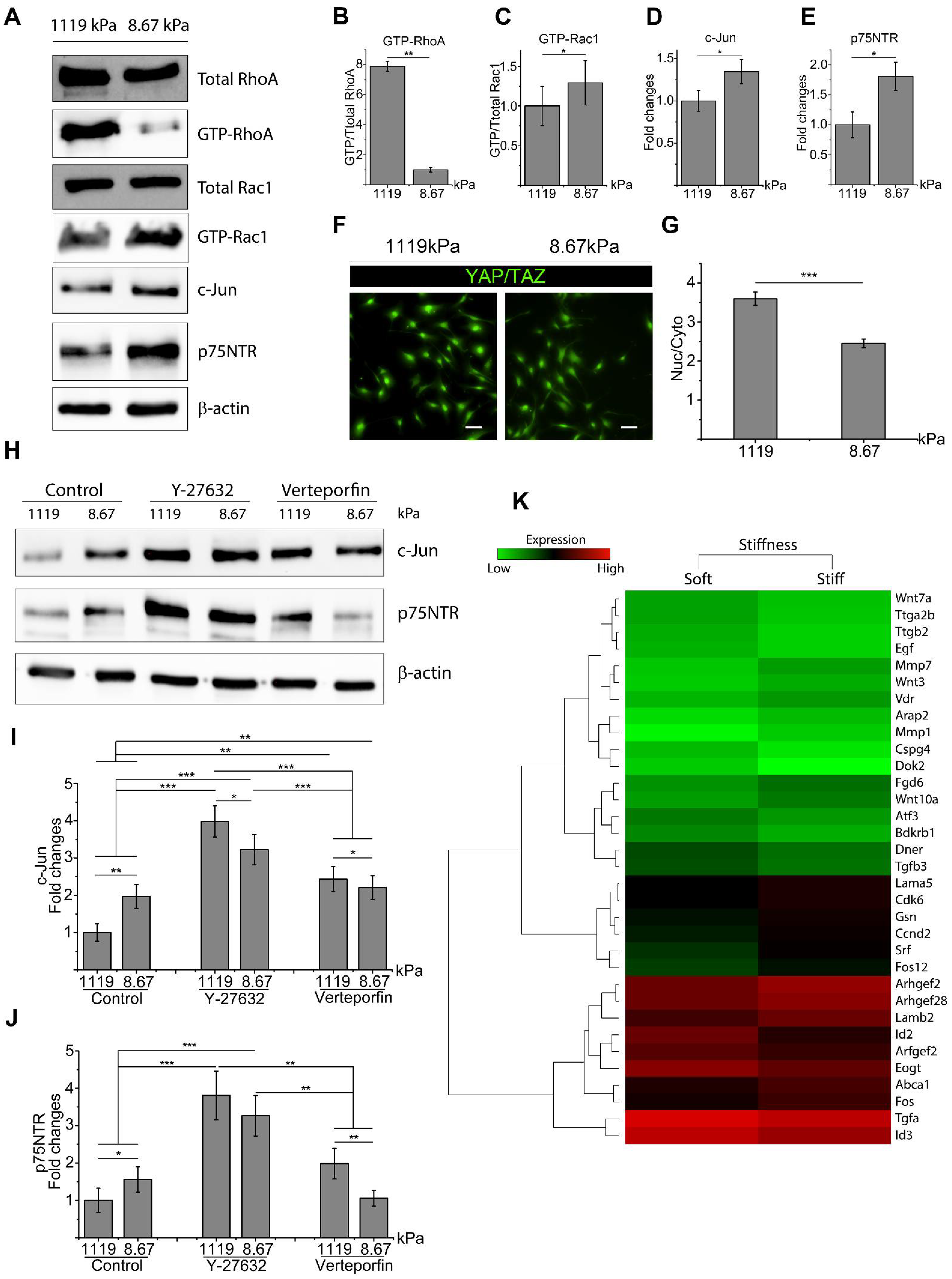
2.1. Matrix Stiffness Inhibits c-Jun and p75NTR with Activation of RhoA and YAP/TAZ
2.2. RhoA and YAP/TAZ Inhibition Promote SC Regenerative Markers
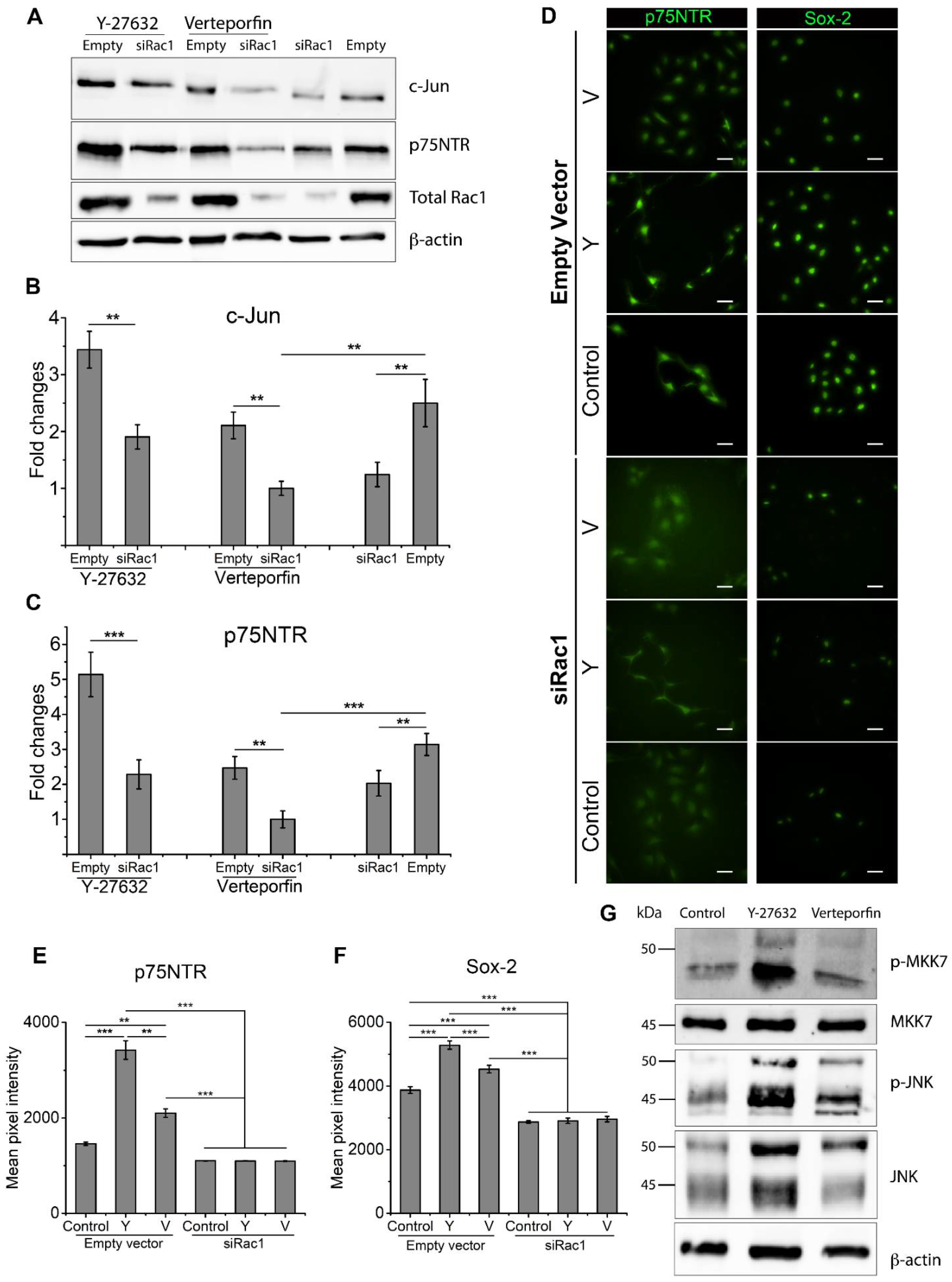
2.3. Rac1 Cooperatively Modulates SC Phenotype with RhoA and YAP/TAZ
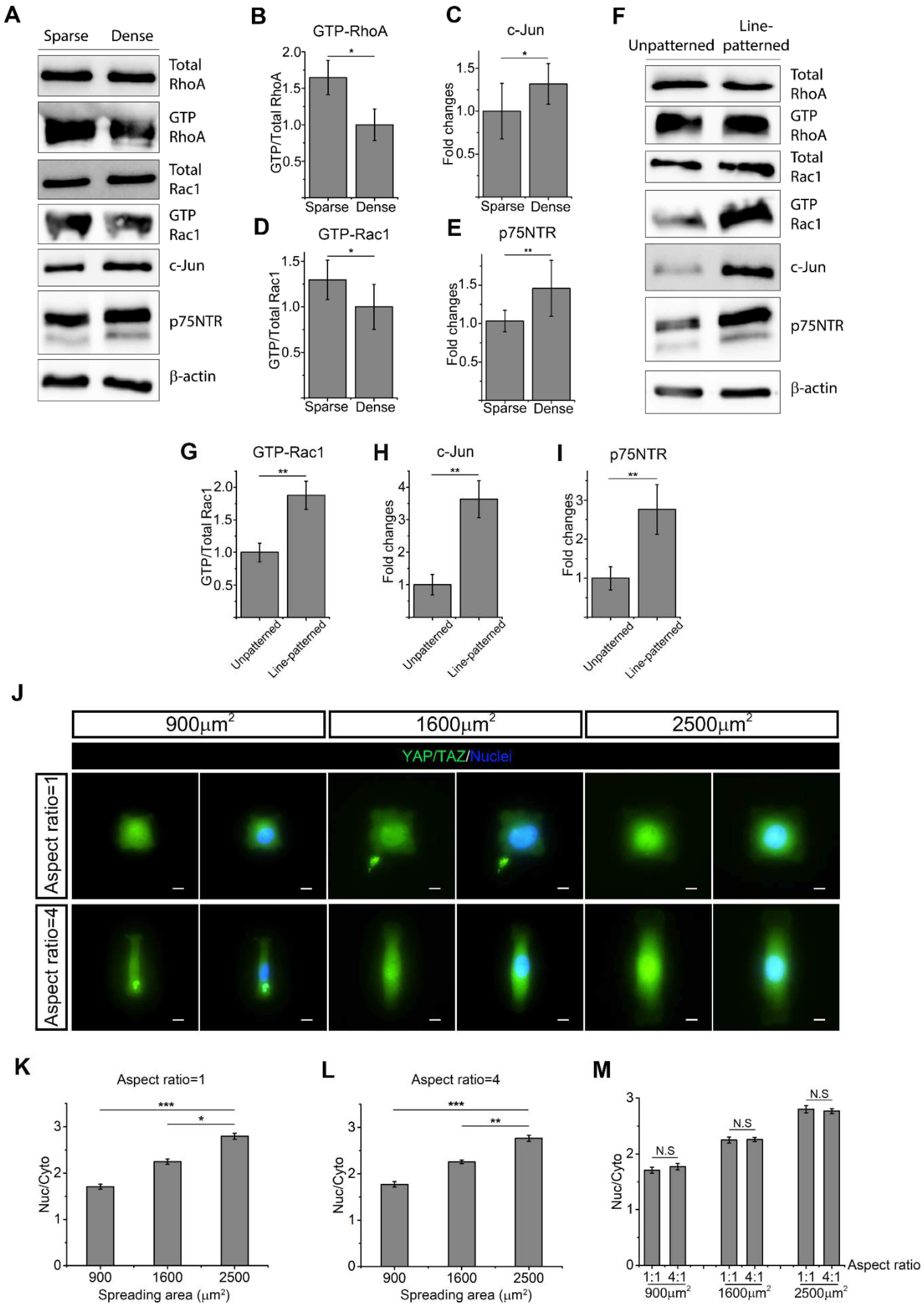
2.4. Cell Morphology Impacts RhoA, Rac1, and YAP/TAZ to Determine SC Phenotype
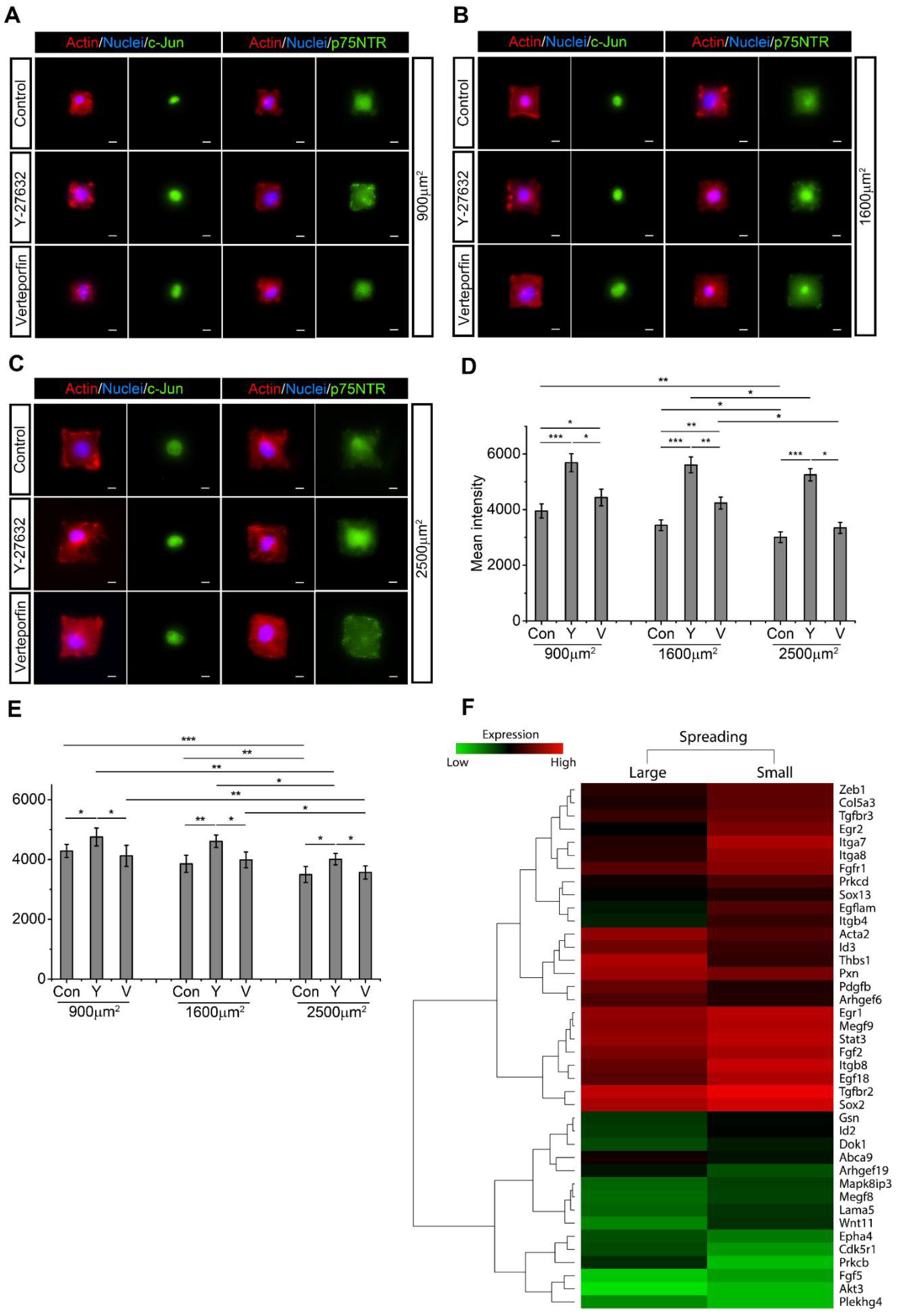
2.5. SC Spreading Promotes Active RhoA and YAP/TAZ and Inhibits c-Jun and p75NTR
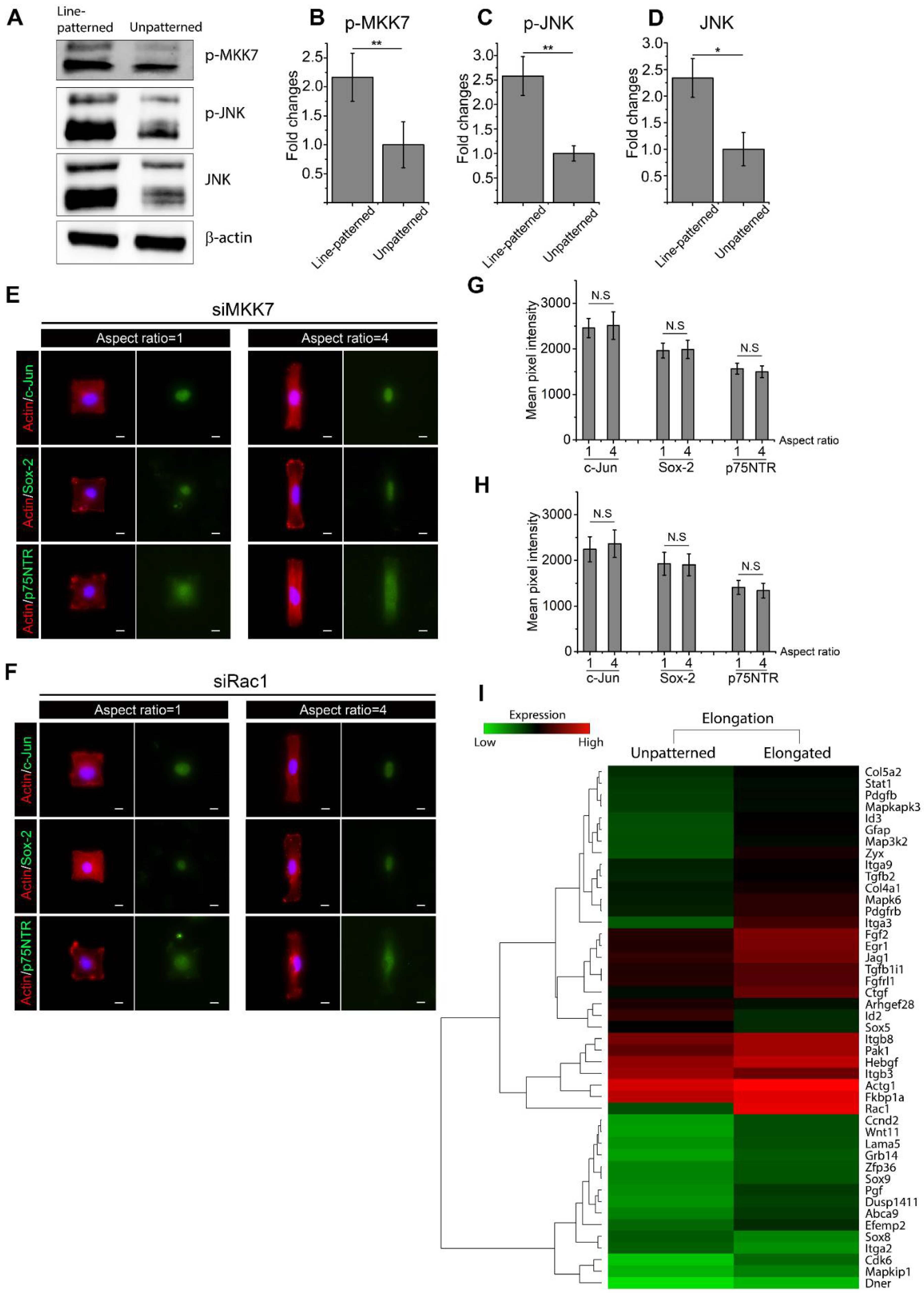
2.6. Active Rac1 Promotes SC Regenerative Markers through SC Elongation
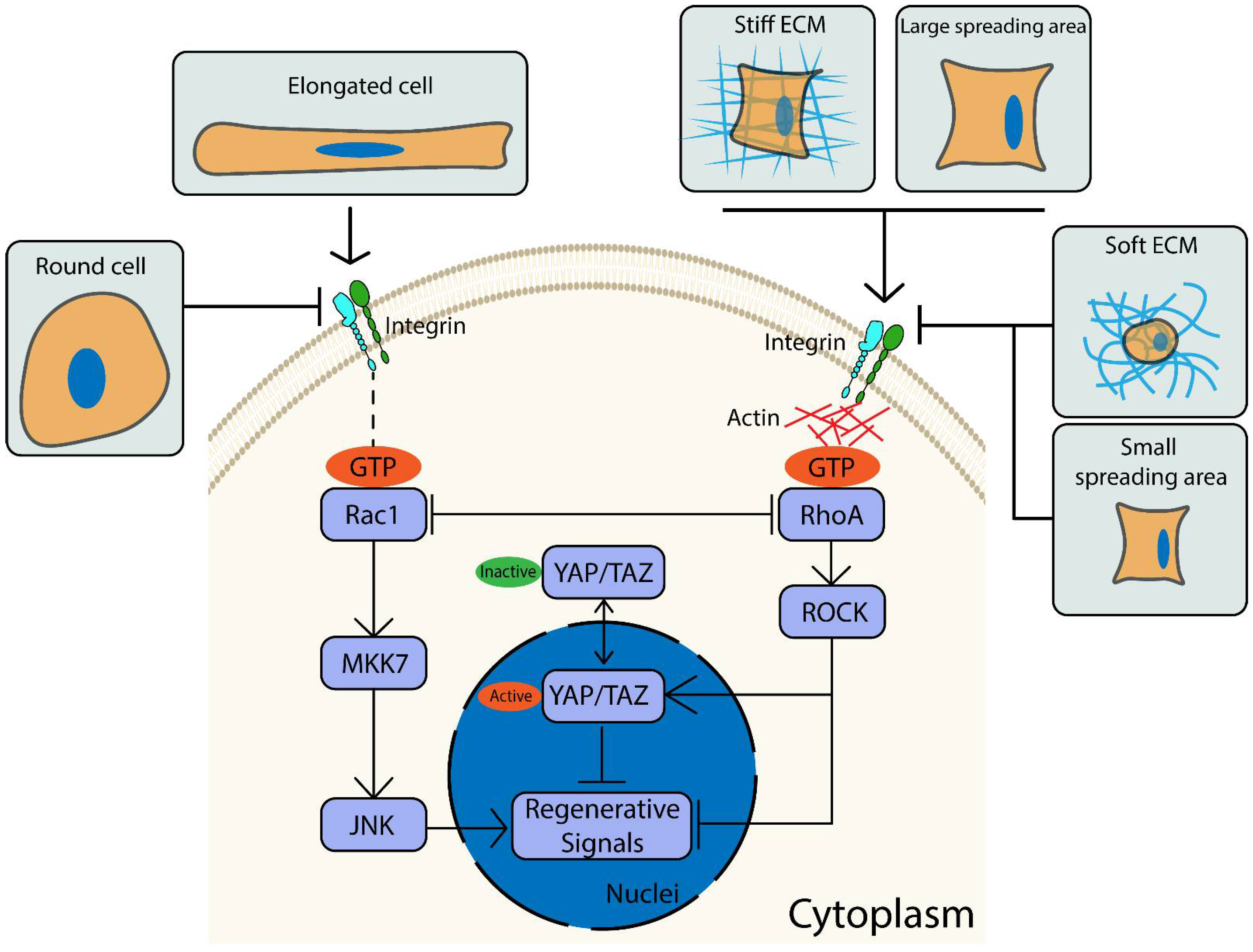
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Preparation of Engineered Substrates
4.3. Western Blot, Active Rho GTPases Assay
4.4. siRNA Transfection
4.5. Immunofluorescent Staining and Cell Analysis
4.6. Microarray Analysis
4.7. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jessen, K.R.; Mirsky, R. The Success and Failure of the Schwann Cell Response to Nerve Injury. Front. Cell. Neurosci. 2019, 13, 33. [Google Scholar] [CrossRef]
- Nocera, G.; Jacob, C. Mechanisms of Schwann cell plasticity involved in peripheral nerve repair after injury. Cell. Mol. Life Sci. 2020. [Google Scholar] [CrossRef] [Green Version]
- Ibanez, C.F.; Simi, A. p75 neurotrophin receptor signaling in nervous system injury and degeneration: Paradox and opportunity. Trends Neurosci. 2012, 35, 431–440. [Google Scholar] [CrossRef] [PubMed]
- Roberts, S.L.; Dun, X.-P.; Doddrell, R.D.S.; Mindos, T.; Drake, L.K.; Onaitis, M.W.; Florio, F.; Quattrini, A.; Lloyd, A.C.; D’Antonio, M.; et al. Sox2 expression in Schwann cells inhibits myelination in vivo and induces influx of macrophages to the nerve. Development 2017, 144, 3114–3125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deumens, R.; Bozkurt, A.; Meek, M.F.; Marcus, M.A.E.; Joosten, E.A.J.; Weis, J.; Brook, G.A. Repairing injured peripheral nerves: Bridging the gap. Prog. Neurobiol. 2010, 92, 245–276. [Google Scholar] [CrossRef]
- Parrinello, S.; Napoli, I.; Ribeiro, S.; Digby, P.W.; Fedorova, M.; Parkinson, D.B.; Doddrell, R.D.S.; Nakayama, M.; Adams, R.H.; Lloyd, A.C. EphB signaling directs peripheral nerve regeneration through Sox2-dependent Schwann cell Sorting. Cell 2010, 143, 145–155. [Google Scholar] [CrossRef] [Green Version]
- Fu, S.Y.; Gordon, T. The cellular and molecular basis of peripheral nerve regeneration. Mol. Neurobiol. 1997, 14, 67–116. [Google Scholar] [CrossRef] [PubMed]
- Gordon, T.; Wood, P.; Sulaiman, O.A.R. Long-Term Denervated Rat Schwann Cells Retain Their Capacity to Proliferate and to Myelinate Axons in vitro. Front. Cell. Neurosci. 2018, 12, 511. [Google Scholar] [CrossRef] [PubMed]
- Frantz, C.; Stewart, K.M.; Weaver, V.M. The extracellular matrix at a glance. J. Cell Sci. 2010, 123, 4195–4200. [Google Scholar] [CrossRef] [Green Version]
- Belin, S.; Zuloaga, K.L.; Poitelon, Y. Influence of Mechanical Stimuli on Schwann Cell Biology. Front. Cell. Neurosci. 2017, 11, 1–11. [Google Scholar] [CrossRef]
- Harris, G.M.; Madigan, N.N.; Lancaster, K.Z.; Enquist, L.W.; Windebank, A.J.; Schwartz, J.; Schwarzbauer, J.E. Nerve Guidance by a Decellularized Fibroblast Extracellular Matrix. Matrix Biol. 2017, 60, 176–189. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Ji, Y.; Zhao, Y.; Liu, Y.; Ding, F.; Gu, X.; Yang, Y. The influence of substrate stiffness on the behavior and functions of Schwann cells in culture. Biomaterials 2012, 33, 6672–6681. [Google Scholar] [CrossRef]
- Evans, E.B.; Brady, S.W.; Tripathi, A.; Hoffman-kim, D. Schwann cell durotaxis can be guided by physiologically relevant stiffness gradients. Biomater. Res. 2018, 22, 14. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.; Orkwis, J.A.; DeVine, B.M.; Harris, G.M. Extracellular matrix cues modulate Schwann cell morphology, proliferation, and protein expression. J. Tissue Eng. Regen. Med. 2019. [Google Scholar] [CrossRef]
- Coillard, A.; Segura, E. In vivo Differentiation of Human Monocytes. Front. Immunol. 2019, 10, 1907. [Google Scholar] [CrossRef] [PubMed]
- Chernousov, M.A.; Yu, W.-M.; Chen, Z.-L.; Carey, D.J.; Strickland, S. Regulation of Schwann cell function by the extracellular matrix. Glia 2008, 56, 1498–1507. [Google Scholar] [CrossRef]
- Xu, Z.; Orkwis, J.A.; Harris, G.M. Preparation of Tunable Extracellular Matrix Microenvironments to Evaluate Schwann Cell Phenotype Specification. J. Vis. Exp. 2020. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.S. Mechanotransduction—A field pulling together? J. Cell Sci. 2008, 121, 3285–3292. [Google Scholar] [CrossRef] [Green Version]
- Rausch, V.; Hansen, C.G. The Hippo Pathway, YAP/TAZ, and the Plasma Membrane. Trends Cell Biol. 2020, 30, 32–48. [Google Scholar] [CrossRef]
- Piccolo, S.; Dupont, S.; Cordenonsi, M. The Biology of YAP/TAZ: Hippo Signaling and Beyond. Physiol. Rev. 2014, 94, 1287–1312. [Google Scholar] [CrossRef] [PubMed]
- Urbanski, M.M.; Kingsbury, L.; Moussouros, D.; Kassim, I.; Mehjabeen, S.; Paknejad, N.; Melendez-Vasquez, C.V. Myelinating glia differentiation is regulated by extracellular matrix elasticity. Sci. Rep. 2016, 6, 33751. [Google Scholar] [CrossRef]
- Grove, M.; Kim, H.; Santerre, M.; Krupka, A.J.; Han, S.B.; Zhai, J.; Cho, J.Y.; Park, R.; Harris, M.; Kim, S.; et al. YAP/TAZ initiate and maintain schwann cell myelination. Elife 2017, 6, 1–27. [Google Scholar] [CrossRef] [Green Version]
- Sophie, B.; Jacob, H.; Jordan, V.J.S.; Yungki, P.; Laura, F.M.; Yannick, P. YAP and TAZ Regulate Cc2d1b and Purbeta in Schwann Cells. Front. Mol. Neurosci. 2019, 12, 177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grove, M.; Lee, H.; Zhao, H.; Son, Y.J. Axon-dependent expression of YAP/TAZ mediates Schwann cell remyelination but not proliferation after nerve injury. Elife 2020, 9. [Google Scholar] [CrossRef] [PubMed]
- Poitelon, Y.; Lopez-Anido, C.; Catignas, K.; Berti, C.; Palmisano, M.; Williamson, C.; Ameroso, D.; Abiko, K.; Hwang, Y.; Gregorieff, A.; et al. YAP and TAZ control peripheral myelination and the expression of laminin receptors in Schwann cells. Nat. Neurosci. 2016, 19, 879–887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopez-Anido, C.; Poitelon, Y.; Gopinath, C.; Moran, J.J.; Ma, K.H.; Law, W.D.; Antonellis, A.; Feltri, M.L.; Svaren, J. Tead1 regulates the expression of Peripheral Myelin Protein 22 during Schwann cell development. Hum. Mol. Genet. 2016, 25, 3055–3069. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mindos, T.; Dun, X.P.; North, K.; Doddrell, R.D.; Schulz, A.; Edwards, P.; Russell, J.; Gray, B.; Roberts, S.L.; Shivane, A.; et al. Merlin controls the repair capacity of Schwann cells after injury by regulating Hippo/YAP activity. J. Cell Biol. 2017, 216, 495–510. [Google Scholar] [CrossRef] [Green Version]
- Hall, A. Rho GTPases and the Actin Cytoskeleton. Science 1998, 279, 509–514. [Google Scholar] [CrossRef] [Green Version]
- Provenzano, P.P.; Keely, P.J. Mechanical signaling through the cytoskeleton regulates cell proliferation by coordinated focal adhesion and Rho GTPase signaling. J. Cell Sci. 2011, 124, 1195–1205. [Google Scholar] [CrossRef] [Green Version]
- Etienne-manneville, S.A.H. Rho GTPases in cell biology. Nature 2002, 420, 629–635. [Google Scholar] [CrossRef]
- O’Brien, L.E.; Jou, T.-S.; Pollack, A.L.; Zhang, Q.; Hansen, S.H.; Yurchenco, P.; Mostov, K.E. Rac1 orientates epithelial apical polarity through effects on basolateral laminin assembly. Nat. Cell Biol. 2001, 3, 831–838. [Google Scholar] [CrossRef]
- Chauhan, B.K.; Lou, M.; Zheng, Y.; Lang, R.A. Balanced Rac1 and RhoA activities regulate cell shape and drive invagination morphogenesis in epithelia. Proc. Natl. Acad. Sci. USA 2011, 108, 18289–18294. [Google Scholar] [CrossRef] [Green Version]
- Melendez-Vasquez, C.V.; Einheber, S.; Salzer, J.L. Rho kinase regulates schwann cell myelination and formation of associated axonal domains. J. Neurosci. 2004, 24, 3953–3963. [Google Scholar] [CrossRef]
- Marinissen, M.J.; Chiariello, M.; Tanos, T.; Bernard, O.; Narumiya, S.; Gutkind, J.S. The small GTP-binding protein RhoA regulates c-jun by a ROCK-JNK signaling axis. Mol. Cell 2004, 14, 29–41. [Google Scholar] [CrossRef] [Green Version]
- Nodari, A.; Zambroni, D.; Quattrini, A.; Court, F.A.; D’Urso, A.; Recchia, A.; Tybulewicz, V.L.; Wrabetz, L.; Feltri, M.L. Beta1 integrin activates Rac1 in Schwann cells to generate radial lamellae during axonal sorting and myelination. J. Cell Biol. 2007, 177, 1063–1075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, H.T.; Feltri, M.L. Rac1 GTPase controls myelination and demyelination. Bioarchitecture 2011, 1, 110–113. [Google Scholar] [CrossRef] [Green Version]
- Shin, Y.K.; Jang, S.Y.; Park, J.Y.; Park, S.Y.; Lee, H.J.; Suh, D.J.; Park, H.T. The Neuregulin-Rac-MKK7 pathway regulates antagonistic c-jun/Krox20 expression in Schwann cell dedifferentiation. Glia 2013, 61, 892–904. [Google Scholar] [CrossRef] [PubMed]
- Martin, E.; Ouellette, M.H.; Jenna, S. Rac1/RhoA antagonism defines cell-to-cell heterogeneity during epidermal morphogenesis in nematodes. J. Cell Biol. 2016, 215, 483–498. [Google Scholar] [CrossRef] [Green Version]
- Bros, M.; Haas, K.; Moll, L.; Grabbe, S. RhoA as a Key Regulator of Innate and Adaptive Immunity. Cells 2019, 8, 733. [Google Scholar] [CrossRef] [Green Version]
- Hong, E.-H.; Kim, J.-Y.; Kim, J.-H.; Lim, D.-S.; Kim, M.; Kim, J.-Y. BIG2-ARF1-RhoA-mDia1 Signaling Regulates Dendritic Golgi Polarization in Hippocampal Neurons. Mol. Neurobiol. 2018, 55, 7701–7716. [Google Scholar] [CrossRef]
- Liu, H.W.; Halayko, A.J.; Fernandes, D.J.; Harmon, G.S.; McCauley, J.A.; Kocieniewski, P.; McConville, J.; Fu, Y.; Forsythe, S.M.; Kogut, P.; et al. The RhoA/Rho kinase pathway regulates nuclear localization of serum response factor. Am. J. Respir. Cell Mol. Biol. 2003, 29, 39–47. [Google Scholar] [CrossRef] [Green Version]
- Koo, J.H.; Plouffe, S.W.; Meng, Z.; Lee, D.H.; Yang, D.; Lim, D.S.; Wang, C.Y.; Guan, K.L. Induction of AP-1 by YAP/TAZ contributes to cell proliferation and organ growth. Genes Dev. 2020, 34, 72–86. [Google Scholar] [CrossRef]
- Li, Z.; Razavi, P.; Li, Q.; Toy, W.; Liu, B.; Ping, C.; Hsieh, W.; Sanchez-Vega, F.; Brown, D.N.; Da Cruz Paula, A.F.; et al. Loss of the FAT1 Tumor Suppressor Promotes Resistance to CDK4/6 Inhibitors via the Hippo Pathway. Cancer Cell 2018, 34, 893–905. [Google Scholar] [CrossRef] [Green Version]
- Schulze-Topphoff, U.; Prat, A.; Prozorovski, T.; Siffrin, V.; Paterka, M.; Herz, J.; Bendix, I.; Ifergan, I.; Schadock, I.; Mori, M.A.; et al. Activation of kinin receptor B1 limits encephalitogenic T lymphocyte recruitment to the central nervous system. Nat. Med. 2009, 15, 788–793. [Google Scholar] [CrossRef] [Green Version]
- Mburu, P.; Romero, M.R.; Hilton, H.; Parker, A.; Townsend, S.; Kikkawa, Y.; Brown, S.D. Gelsolin plays a role in the actin polymerization complex of hair cell stereocilia. PLoS ONE 2010, 5, e11627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boerboom, A.; Dion, V.; Chariot, A.; Franzen, R. Molecular Mechanisms Involved in Schwann Cell Plasticity. Front. Mol. Neurosci. 2017, 10, 38. [Google Scholar] [CrossRef] [Green Version]
- Dahlin, L.B. Nerve injury-induced c-Jun activation in Schwann cells is JNK independent. BioMed Res. Int. 2014, 2014, 392971. [Google Scholar] [CrossRef] [Green Version]
- Schmid, D.; Zeis, T.; Schaeren-Wiemers, N. Transcriptional regulation induced by cAMP elevation in mouse Schwann cells. ASN Neuro 2014, 6, 137–157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Zhang, P.; Yu, J.; Zhang, F.; Dai, W.; Yi, S. Matrix metalloproteinase 7 promoted Schwann cell migration and myelination after rat sciatic nerve injury. Mol. Brain 2019, 12, 101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jessen, K.R.; Arthur-Farraj, P. Repair Schwann cell update: Adaptive reprogramming, EMT, and stemness in regenerating nerves. Glia 2019, 67, 421–437. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ye, Z.; Chen, S.; Pan, Z.; Zhou, Q.; Li, Y.Z.; Shuai, W.D.; Kuang, C.M.; Peng, Q.H.; Shi, W.; et al. ARHGEF19 interacts with BRAF to activate MAPK signaling during the tumorigenesis of non-small cell lung cancer. Int. J. Cancer 2018, 142, 1379–1391. [Google Scholar] [CrossRef] [Green Version]
- Gawlak, G.; Tian, Y.; O’Donnell, J.J., 3rd; Tian, X.; Birukova, A.A.; Birukov, K.G. Paxillin mediates stretch-induced Rho signaling and endothelial permeability via assembly of paxillin-p42/44MAPK-GEF-H1 complex. FASEB J. 2014, 28, 3249–3260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cayuso, J.; Xu, Q.; Addison, M.; Wilkinson, D.G. Actomyosin regulation by Eph receptor signaling couples boundary cell formation to border sharpness. Elife 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Mullin, B.H.; Mamotte, C.; Prince, R.L.; Wilson, S.G. Influence of ARHGEF3 and RHOA knockdown on ACTA2 and other genes in osteoblasts and osteoclasts. PLoS ONE 2014, 9, e98116. [Google Scholar] [CrossRef] [Green Version]
- Joung, I.; Yoo, M.; Woo, J.H.; Chang, C.Y.; Heo, H.; Kwon, Y.K. Secretion of EGF-like domain of heregulinbeta promotes axonal growth and functional recovery of injured sciatic nerve. Mol. Cells 2010, 30, 477–484. [Google Scholar] [CrossRef]
- Castelnovo, L.F.; Bonalume, V.; Melfi, S.; Ballabio, M.; Colleoni, D.; Magnaghi, V. Schwann cell development, maturation and regeneration: A focus on classic and emerging intracellular signaling pathways. Neural Regen. Res. 2017, 12, 1013–1023. [Google Scholar] [CrossRef]
- Sun, Z.; Huang, S.; Li, Z.; Meininger, G.A. Zyxin is involved in regulation of mechanotransduction in arteriole smooth muscle cells. Front. Physiol. 2012, 3, 472. [Google Scholar] [CrossRef] [Green Version]
- Sabra, H.; Brunner, M.; Mandati, V.; Wehrle-Haller, B.; Lallemand, D.; Ribba, A.S.; Chevalier, G.; Guardiola, P.; Block, M.R.; Bouvard, D. beta1 integrin-dependent Rac/group I PAK signaling mediates YAP activation of Yes-associated protein 1 (YAP1) via NF2/merlin. J. Biol. Chem. 2017, 292, 19179–19197. [Google Scholar] [CrossRef] [Green Version]
- Lopez-Fagundo, C.; Bar-Kochba, E.; Livi, L.L.; Hoffman-Kim, D.; Franck, C. Three-dimensional traction forces of Schwann cells on compliant substrates. J. R. Soc. Interface 2014, 11, 20140247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Yang, X.; Yue, Y.; Ye, J.; Yao, Y.; Fu, Y.; Li, G.; Yao, Q.; Lin, Y.; Gong, P. Cyclic mechanical stress modulates neurotrophic and myelinating gene expression of Schwann cells. Cell Prolif. 2015, 48, 59–66. [Google Scholar] [CrossRef]
- Zhou, W.; Stukel, J.M.; Cebull, H.L.; Willits, R.K. Tuning the Mechanical Properties of Poly(Ethylene Glycol) Microgel-Based Scaffolds to Increase 3D Schwann Cell Proliferation. Macromol. Biosci. 2016, 16, 535–544. [Google Scholar] [CrossRef]
- Rosso, G.; Guck, J. Mechanical changes of peripheral nerve tissue microenvironment and their structural basis during development. APL Bioeng. 2019, 3, 036107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wells, R.G. Tissue mechanics and fibrosis. Biochim. Biophys. Acta Mol. Basis Dis. 2013, 1832, 884–890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asano, S.; Ito, S.; Takahashi, K.; Furuya, K.; Kondo, M.; Sokabe, M.; Hasegawa, Y. Matrix stiffness regulates migration of human lung fibroblasts. Physiol. Rep. 2017, 5. [Google Scholar] [CrossRef] [PubMed]
- Ozcelikkale, A.; Dutton, J.C.; Grinnell, F.; Han, B. Effects of dynamic matrix remodelling on en masse migration of fibroblasts on collagen matrices. J. R. Soc. Interface 2017, 14. [Google Scholar] [CrossRef] [Green Version]
- El-Mohri, H.; Wu, Y.; Mohanty, S.; Ghosh, G. Impact of matrix stiffness on fibroblast function. Mater. Sci. Eng. C Mater. Biol. Appl. 2017, 74, 146–151. [Google Scholar] [CrossRef]
- Blaker, J.J.; Faroni, A.; Li, X.; Gough, J.E.; Magaz, A.; Reid, A.J. Bioactive Silk-Based Nerve Guidance Conduits for Augmenting Peripheral Nerve Repair. Adv. Healthc. Mater. 2018, 7, 1800308. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, M.; Ffrench-Constant, C. Extracellular Matrix Regulation of Stem Cell Behavior. Curr. Stem Cell Rep. 2016, 2, 197–206. [Google Scholar] [CrossRef] [Green Version]
- Chang, T.Y.; Chen, C.; Lee, M.; Chang, Y.C.; Lu, C.H.; Lu, S.T.; Wang, D.Y.; Wang, A.; Guo, C.L.; Cheng, P.L. Paxillin facilitates timely neurite initiation on soft-substrate environments by interacting with the endocytic machinery. Elife 2017, 6. [Google Scholar] [CrossRef] [Green Version]
- Kovar, H.; Bierbaumer, L.; Radic-Sarikas, B. The YAP/TAZ Pathway in Osteogenesis and Bone Sarcoma Pathogenesis. Cells 2020, 9, 972. [Google Scholar] [CrossRef] [Green Version]
- Boopathy, G.T.K.; Hong, W. Role of Hippo Pathway-YAP/TAZ Signaling in Angiogenesis. Front. Cell Dev. Biol. 2019, 7, 49. [Google Scholar] [CrossRef]
- Jessen, K.R.; Mirsky, R. The repair Schwann cell and its function in regenerating nerves. J. Physiol. 2016, 594, 3521–3531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aragona, M.; Panciera, T.; Manfrin, A.; Giulitti, S.; Michielin, F.; Elvassore, N.; Dupont, S.; Piccolo, S. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell 2013, 154, 1047–1059. [Google Scholar] [CrossRef] [Green Version]
- Deng, Y.; Wu, L.M.N.; Bai, S.; Zhao, C.; Wang, H.; Wang, J.; Xu, L.; Sakabe, M.; Zhou, W.; Xin, M.; et al. A reciprocal regulatory loop between TAZ/YAP and G-protein Galphas regulates Schwann cell proliferation and myelination. Nat. Commun. 2017, 8, 15161. [Google Scholar] [CrossRef] [Green Version]
- Tan, D.; Wen, J.; Li, L.; Wang, X.; Qian, C.; Pan, M.; Lai, M.; Deng, J.; Hu, X.; Zhang, H.; et al. Inhibition of RhoA-Subfamily GTPases Suppresses Schwann Cell Proliferation Through Regulating AKT Pathway Rather Than ROCK Pathway. Front. Cell. Neurosci. 2018, 12, 437. [Google Scholar] [CrossRef]
- Yamauchi, J.; Chan, J.R.; Shooter, E.M. Neurotrophins regulate Schwann cell migration by activating divergent signaling pathways dependent on Rho GTPases. Proc. Natl. Acad. Sci. USA 2004, 101, 8774–8779. [Google Scholar] [CrossRef] [Green Version]
- Mantuano, E.; Jo, M.; Gonias, S.L.; Campana, W.M. Low density lipoprotein receptor-related protein (LRP1) regulates Rac1 and RhoA reciprocally to control Schwann cell adhesion and migration. J. Biol. Chem. 2010, 285, 14259–14266. [Google Scholar] [CrossRef] [Green Version]
- Arthur, W.T.; Burridge, K.; Carolina, N.; Hill, C.; Carolina, N. RhoA Inactivation by p190RhoGAP Regulates Cell Spreading and Migration by Promoting Membrane Protrusion and Polarity. Mol. Biol. Cell 2001, 12, 2711–2720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colciago, A.; Melfi, S.; Giannotti, G.; Bonalume, V.; Ballabio, M.; Caffino, L.; Fumagalli, F.; Magnaghi, V. Tumor suppressor Nf2/merlin drives Schwann cell changes following electromagnetic field exposure through Hippo-dependent mechanisms. Cell Death Discov. 2015, 1, 15021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benninger, Y.; Thurnherr, T.; Pereira, J.A.; Krause, S.; Wu, X.; Chrostek-Grashoff, A.; Herzog, D.; Nave, K.A.; Franklin, R.J.; Meijer, D.; et al. Essential and distinct roles for cdc42 and rac1 in the regulation of Schwann cell biology during peripheral nervous system development. J. Cell Biol. 2007, 177, 1051–1061. [Google Scholar] [CrossRef]
- Sanz-moreno, V.; Gadea, G.; Ahn, J.; Paterson, H.; Marra, P.; Pinner, S.; Sahai, E.; Marshall, C.J. Rac Activation and Inactivation Control Plasticity of Tumor Cell Movement. Cell 2008, 510–523. [Google Scholar] [CrossRef] [Green Version]
- Dupont, S.; Morsut, L.; Aragona, M.; Enzo, E.; Giulitti, S.; Cordenonsi, M.; Zanconato, F.; Le Digabel, J.; Forcato, M.; Bicciato, S.; et al. Role of YAP/TAZ in mechanotransduction. Nature 2011, 474, 179–184. [Google Scholar] [CrossRef]
- Doddrell, R.D.; Dun, X.P.; Shivane, A.; Feltri, M.L.; Wrabetz, L.; Wegner, M.; Sock, E.; Hanemann, C.O.; Parkinson, D.B. Loss of SOX10 function contributes to the phenotype of human Merlin-null schwannoma cells. Brain 2013, 136, 549–563. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, I.; Fernando, A.; Gurgel, R.; Jason Clark, J.; Xu, L.; Hansen, M.R. Merlin status regulates p75(NTR) expression and apoptotic signaling in Schwann cells following nerve injury. Neurobiol. Dis. 2015, 82, 114–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaw, R.J.; Paez, J.G.; Curto, M.; Yaktine, A.; Pruitt, W.M.; Saotome, I.; O’Bryan, J.P.; Gupta, V.; Ratner, N.; Der, C.J.; et al. The Nf2 Tumor Suppressor, Merlin, Functions in Rac-Dependent Signaling. Dev. Cell 2001, 1, 63–72. [Google Scholar] [CrossRef] [Green Version]
- Yamauchi, J.; Miyamoto, Y.; Chan, J.R.; Tanoue, A. ErbB2 directly activates the exchange factor Dock7 to promote Schwann cell migration. J. Cell Biol. 2008, 181, 351–365. [Google Scholar] [CrossRef] [Green Version]
- Yamauchi, J.; Chan, J.R.; Shooter, E.M. Neurotrophin 3 activation of TrkC induces Schwann cell migration through the c-Jun N-terminal kinase pathway. Proc. Natl. Acad. Sci. USA 2003, 100, 14421–14426. [Google Scholar] [CrossRef] [Green Version]
- Manchanda, P.K.; Jones, G.N.; Lee, A.A.; Pringle, D.R.; Zhang, M.; Yu, L.; La Perle, K.M.; Kirschner, L.S. Rac1 is required for Prkar1a-mediated Nf2 suppression in Schwann cell tumors. Oncogene 2013, 32, 3491–3499. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Zhao, H.; Li, Y.; Xia, D.; Yang, L.; Ma, Y.; Li, H. The role of YAP/TAZ activity in cancer metabolic reprogramming. Mol. Cancer 2018, 17, 134. [Google Scholar] [CrossRef] [PubMed]
- Ju, M.S.; Lin, C.C.K.; Chang, C.T. Researches on biomechanical properties and models of peripheral nerves—A review. J. Biomech. Sci. Eng. 2017, 12. [Google Scholar] [CrossRef] [Green Version]
- Cady, E.; Orkwis, J.A.; Weaver, R.; Conlin, L.; Madigan, N.N.; Harris, G.M. Micropatterning Decellularized ECM as a Bioactive Surface to Guide Cell Alignment, Proliferation, and Migration. Bioengineering 2020, 7, 102. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Z.; Orkwis, J.A.; Harris, G.M. Cell Shape and Matrix Stiffness Impact Schwann Cell Plasticity via YAP/TAZ and Rho GTPases. Int. J. Mol. Sci. 2021, 22, 4821. https://doi.org/10.3390/ijms22094821
Xu Z, Orkwis JA, Harris GM. Cell Shape and Matrix Stiffness Impact Schwann Cell Plasticity via YAP/TAZ and Rho GTPases. International Journal of Molecular Sciences. 2021; 22(9):4821. https://doi.org/10.3390/ijms22094821
Chicago/Turabian StyleXu, Zhenyuan, Jacob A. Orkwis, and Greg M. Harris. 2021. "Cell Shape and Matrix Stiffness Impact Schwann Cell Plasticity via YAP/TAZ and Rho GTPases" International Journal of Molecular Sciences 22, no. 9: 4821. https://doi.org/10.3390/ijms22094821
APA StyleXu, Z., Orkwis, J. A., & Harris, G. M. (2021). Cell Shape and Matrix Stiffness Impact Schwann Cell Plasticity via YAP/TAZ and Rho GTPases. International Journal of Molecular Sciences, 22(9), 4821. https://doi.org/10.3390/ijms22094821






