Crystal Structure of Escherichia coli Agmatinase: Catalytic Mechanism and Residues Relevant for Substrate Specificity
Abstract
1. Introduction
2. Results and Discussion
2.1. Mutagenesis Studies in E. coli Agmatinase
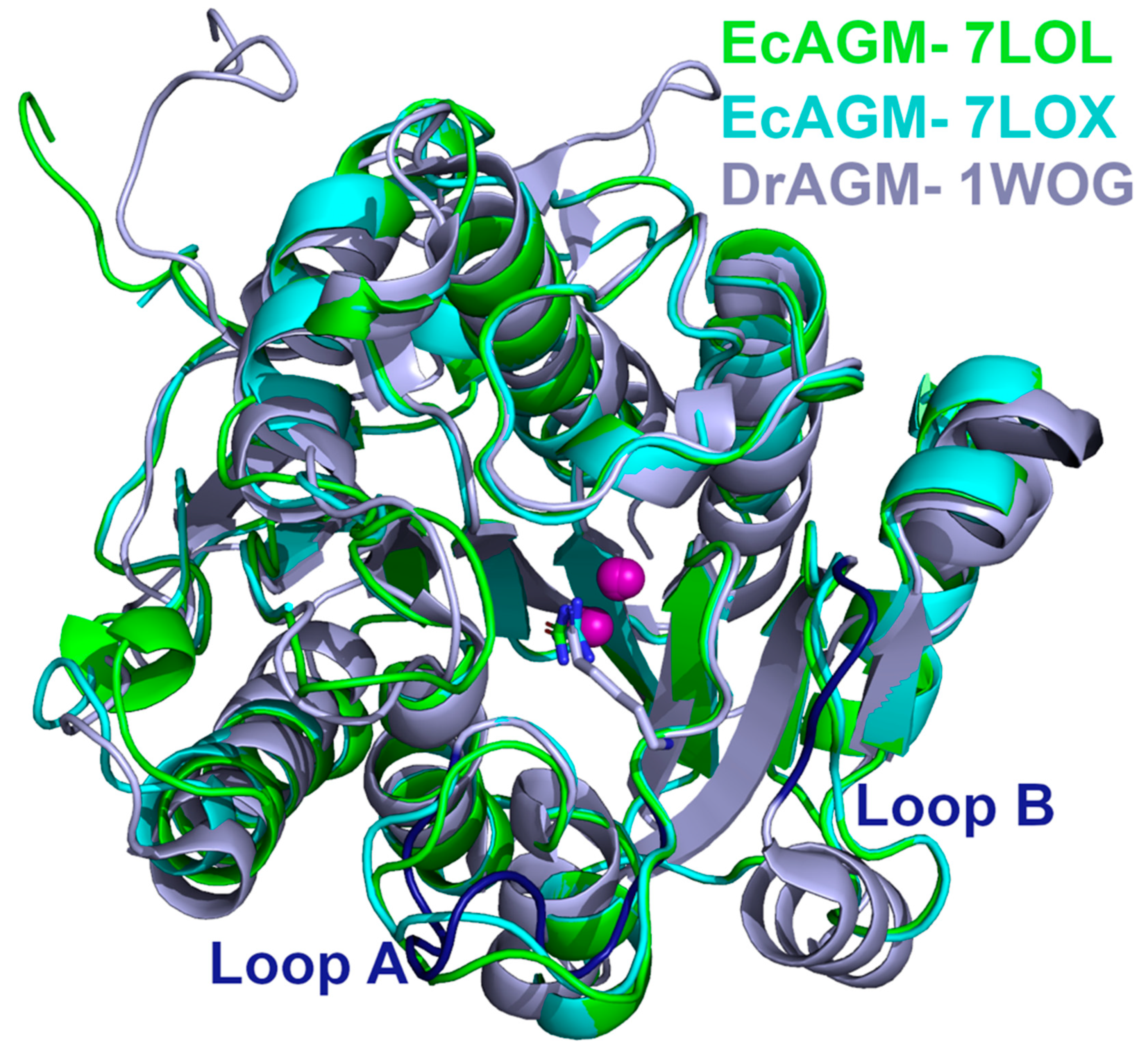
2.2. Structural Analysis of E.coli Agmatinase: Monomer Architecture and Oligomerization State
2.3. Guanidine, Urea, and Mn2+ Binding-Sites: Implications in the Catalytic Mechanism
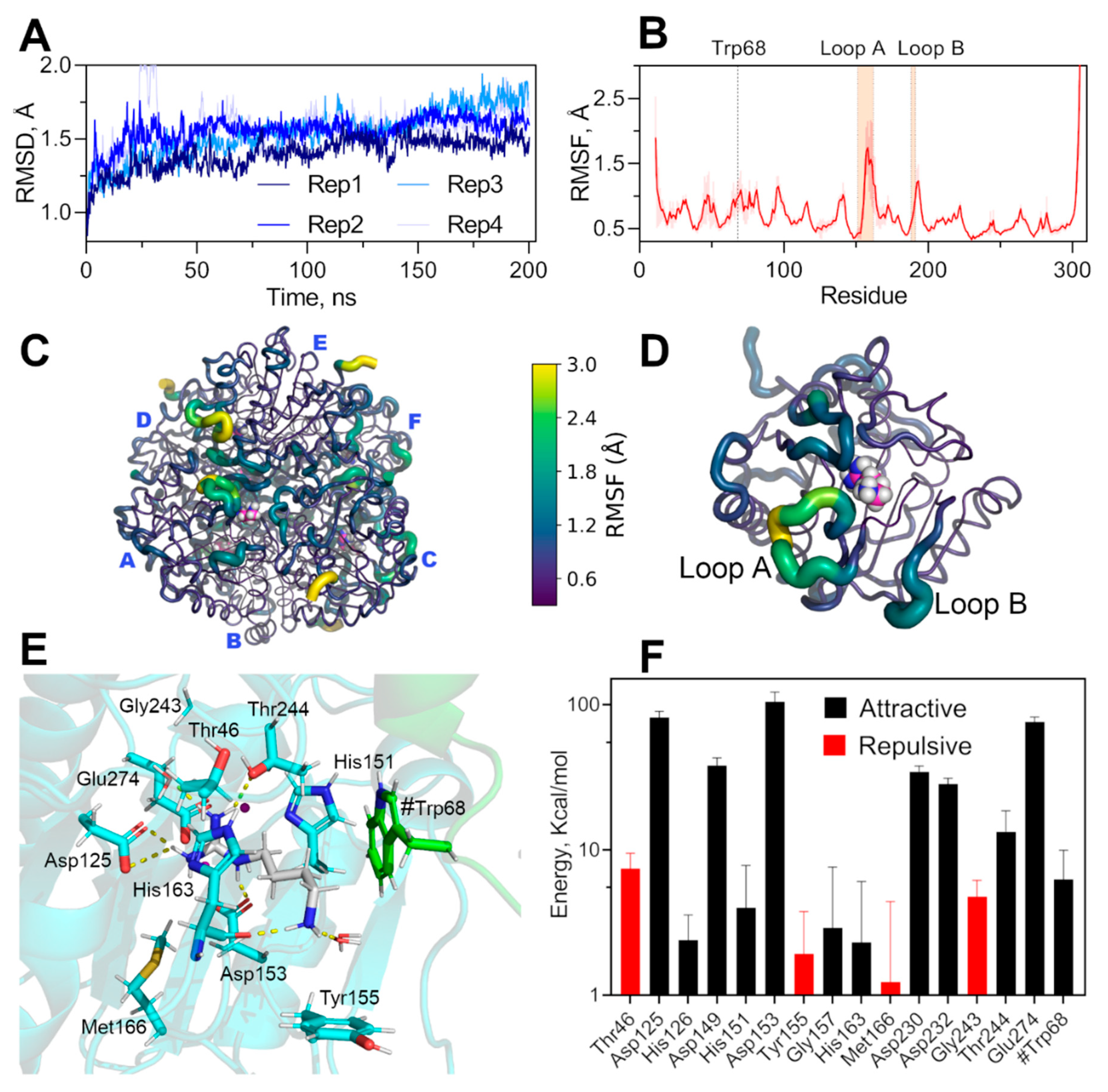
2.4. Analysis of Agmatine Binding: Molecular Dynamic Simulation and Role of Loop A and B of EcAGM
3. Conclusions
4. Materials and Methods
4.1. EcAGM Expression and Purification
4.2. EcAGM Site-Directed Mutagenesis
4.3. EcAGM Activity Determination and Kinetics Experiments
4.4. X-ray Crystallography
4.5. Sequence and Structural Analysis
4.6. Molecular Dynamic Simulation of EcAGM in Complex with Agmatine
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tabor, C.W.; Tabor, H. Polyamines. Annu. Rev. Biochem. 1984, 53, 749–790. [Google Scholar] [CrossRef]
- Li, G.; Regunathan, S.; Barrow, C.J.; Eshraghi, J.; Cooper, R.; Reis, D.J. Agmatine: An endogenous clonidine-displacing substance in the brain. Science 1994, 263, 966–969. [Google Scholar] [CrossRef]
- Demady, D.R.; Jianmongkol, S.; Vuletich, J.L.; Bender, A.T.; Osawa, Y. Agmatine enhances the NADPH oxidase activity of neuronal NO synthase and leads to oxidative inactivation of the enzyme. Mol. Pharmacol. 2001, 59, 24–29. [Google Scholar] [CrossRef]
- Satriano, J.; Schwartz, D.; Ishizuka, S.; Lortie, M.J.; Thomson, S.C.; Gabbai, F.; Kelly, C.J.; Blantz, R.C. Suppression of inducible nitric oxide generation by agmatine aldehyde: Beneficial effects in sepsis. J. Cell. Physiol. 2001, 188, 313–320. [Google Scholar] [CrossRef]
- Regunathan, S.; Piletz, J.E. Regulation of Inducible Nitric Oxide Synthase and Agmatine Synthesis in Macrophages and Astrocytes. Ann. N. Y. Acad. Sci. 2003, 1009, 20–29. [Google Scholar] [CrossRef]
- Mun, C.H.; Lee, W.T.; Park, K.A.; Lee, J.E. Regulation of endothelial nitric oxide synthase by agmatine after transient global cerebral ischemia in rat brain. Anat. Cell Biol. 2010, 43, 230. [Google Scholar] [CrossRef]
- Piletz, J.E.; Aricioglu, F.; Cheng, J.T.; Fairbanks, C.A.; Gilad, V.H.; Haenisch, B.; Halaris, A.; Hong, S.; Lee, J.E.; Li, J.; et al. Agmatine: Clinical applications after 100 years in translation. Drug Discov. Today 2013, 18, 880–893. [Google Scholar] [CrossRef]
- Loring, R.H. Agmatine acts as an antagonist of neuronal nicotinic receptors. Br. J. Pharmacol. 1990, 99, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.L.F. Clonidine-displacing substance and its putative role in control of insulin secretion: A minireview. Gen. Pharmacol. 1998, 31, 525–529. [Google Scholar] [CrossRef]
- Lee, J.H.; Park, G.H.; Lee, Y.K.; Park, J.H. Changes in the arginine methylation of organ proteins during the development of diabetes mellitus. Diabetes Res. Clin. Pract. 2011, 94, 111–118. [Google Scholar] [CrossRef]
- Su, C.H.; Liu, I.M.; Chung, H.H.; Cheng, J.T. Activation of I2-imidazoline receptors by agmatine improved insulin sensitivity through two mechanisms in type-2 diabetic rats. Neurosci. Lett. 2009, 457, 125–128. [Google Scholar] [CrossRef]
- Satriano, J.; Cunard, R.; Peterson, O.W.; Dousa, T.; Gabbai, F.B.; Blantz, R.C. Effects on kidney filtration rate by agmatine requires activation of ryanodine channels for nitric oxide generation. Am. J. Physiol. Ren. Physiol. 2008, 294, 795–800. [Google Scholar] [CrossRef]
- Schwartz, D.; Peterson, O.W.; Mendonca, M.; Satriano, J.; Lortie, M.; Blantz, R.C. Agmatine affects glomerular filtration via a nitric oxide synthase- dependent mechanism. Am. J. Physiol. Ren. Physiol. 1997, 272. [Google Scholar] [CrossRef]
- Cunha, A.S.; Matheus, F.C.; Moretti, M.; Sampaio, T.B.; Poli, A.; Santos, D.B.; Colle, D.; Cunha, M.P.; Blum-Silva, C.H.; Sandjo, L.P.; et al. Agmatine attenuates reserpine-induced oral dyskinesia in mice: Role of oxidative stress, nitric oxide and glutamate NMDA receptors. Behav. Brain Res. 2016, 312, 64–76. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.T.; Hong, S.; Yoon, S.H.; Kim, J.H.; Park, K.A.; Seong, G.J.; Lee, J.E. Neuroprotective effects of agmatine on oxygen-glucose deprived primary-cultured astrocytes and nuclear translocation of nuclear factor-kappa B. Brain Res. 2009, 1281, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Moretti, M.; Matheus, F.C.; De Oliveira, P.A.; Neis, V.B.; Ben, J.; Walz, R.; Rodrigues, A.L.S.; Prediger, R.D. Role of agmatine in neurodegenerative diseases and epilepsy. Front. Biosci. Elit. 2014, 6, 341–359. [Google Scholar] [CrossRef]
- Lee, L.M.; Tsai, T.C.; Chung, H.H.; Tong, Y.C.; Cheng, J.T. Prostatic relaxation induced by agmatine is decreased in spontaneously hypertensive rats. BJU Int. 2012, 110, 253–258. [Google Scholar] [CrossRef]
- Carvajal, N.; López, V.; Salas, M.; Uribe, E.; Herrera, P.; Cerpa, J. Manganese is essential for catalytic activity of Escherichia coli agmatinase. Biochem. Biophys. Res. Commun. 1999, 258, 808–811. [Google Scholar] [CrossRef] [PubMed]
- Carvajal, N.; Orellana, M.S.; Salas, M.; Enríquez, P.; Alarcón, R.; Uribe, E.; López, V. Kinetic studies and site-directed mutagenesis of Escherichia coli agmatinase. a role for Glu274 in binding and correct positioning of the substrate guanidinium group. Arch. Biochem. Biophys. 2004, 430, 185–190. [Google Scholar] [CrossRef]
- Uribe, E.; Reyes, M.-B.; Martínez, I.; Mella, K.; Salas, M.; Tarifeño-Saldivia, E.; López, V.; García-Robles, M.; Martínez-Oyanedel, J.; Figueroa, M.; et al. Functional analysis of the Mn2+ requirement in the catalysis of ureohydrolases arginase and agmatinase—A historical perspective. J. Inorg. Biochem. 2020, 202, 110812. [Google Scholar] [CrossRef]
- Hyung, J.A.; Kyoung, H.K.; Lee, J.; Ha, J.Y.; Hyung, H.L.; Kim, D.; Yoon, H.J.; Kwon, A.R.; Se, W.S. Crystal structure of agmatinase reveals structural conservation and inhibition mechanism of the ureohydrolase superfamily. J. Biol. Chem. 2004, 279, 50505–50513. [Google Scholar] [CrossRef]
- Baugh, L.; Gallagher, L.A.; Patrapuvich, R.; Clifton, M.C.; Gardberg, A.S.; Edwards, T.E.; Armour, B.; Begley, D.W.; Dieterich, S.H.; Dranow, D.M.; et al. Combining Functional and Structural Genomics to Sample the Essential Burkholderia Structome. PLoS ONE 2013, 8. [Google Scholar] [CrossRef]
- Iyer, R.K.; Kim, H.K.; Tsoa, R.W.; Grody, W.W.; Cederbaum, S.D. Cloning and characterization of human agmatinase. Mol. Genet. Metab. 2002, 75, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Salas, M.; López, V.; Uribe, E.; Carvajal, N. Studies on the interaction of Escherichia coli agmatinase with manganese ions: Structural and kinetic studies of the H126N and H151N variants. J. Inorg. Biochem. 2004, 98, 1032–1036. [Google Scholar] [CrossRef] [PubMed]
- Carvajal, N.; Olate, J.; Salas, M.; López, V.; Cerpa, J.; Herrera, P.; Uribe, E. Evidence that histidine-163 is critical for catalytic activity, but not for substrate binding to Escherichia coli agmatinase. Biochem. Biophys. Res. Commun. 1999, 264, 196–200. [Google Scholar] [CrossRef] [PubMed]
- Salas, M.; Rodríguez, R.; López, N.; Uribe, E.; López, V.; Carvajal, N. Insights into the reaction mechanism of Escherichia coli agmatinase by site-directed mutagenesis and molecular modelling: A critical role for aspartate 153. Eur. J. Biochem. 2002, 269, 5522–5526. [Google Scholar] [CrossRef]
- Lee, S.J.; Kim, D.J.; Kim, H.S.; Lee, B.I.; Yoon, H.J.; Yoon, J.Y.; Kim, K.H.; Jang, J.Y.; Im, H.N.; An, D.R.; et al. Crystal structures of Pseudomonas aeruginosa guanidinobutyrase and guanidinopropionase, members of the ureohydrolase superfamily. J. Struct. Biol. 2011, 175, 329–338. [Google Scholar] [CrossRef]
- Elkins, J.M.; Clifton, I.J.; Hernández, H.; Doan, L.X.; Robinson, C.V.; Schofield, C.J.; Hewitson, K.S. Oligomeric structure of proclavaminic acid amidino hydrolase: Evolution of a hydrolytic enzyme in clavulanic acid biosynthesis. Biochem. J. 2002, 366, 423–434. [Google Scholar] [CrossRef]
- Alarcón, R.; Orellana, M.S.; Neira, B.; Uribe, E.; García, J.R.; Carvajal, N. Mutational analysis of substrate recognition by human arginase type I—Agmatinase activity of the N130D variant. FEBS J. 2006, 273, 5625–5631. [Google Scholar] [CrossRef]
- Sekowska, A.; Danchin, A.; Risler, J.L. Phylogeny of related functions: The case of polyamine biosynthetic enzymes. Microbiology 2000, 146, 1815–1828. [Google Scholar] [CrossRef][Green Version]
- Sekula, B. The Neighboring Subunit Is Engaged to Stabilize the Substrate in the Active Site of Plant Arginases. Front. Plant Sci. 2020, 11, 1–12. [Google Scholar] [CrossRef]
- Bewley, M.C.; Jeffrey, P.D.; Patchett, M.L.; Kanyo, Z.F.; Baker, E.N. Crystal structures of Bacillus caldovelox arginase in complex with substrate and inhibitors reveal new insights into activation, inhibition and catalysis in the arginase superfamily. Structure 1999, 7, 435–448. [Google Scholar] [CrossRef]
- Kabsch, W. XDS. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef]
- Evans, P.R.; Murshudov, G.N. How good are my data and what is the resolution? Acta Crystallogr. Sect. D Biol. Crystallogr. 2013, 69, 1204–1214. [Google Scholar] [CrossRef]
- Long, F.; Vagin, A.A.; Young, P.; Murshudov, G.N. BALBES: A molecular-replacement pipeline. Acta Crystallogr. Sect. D Biol. Crystallogr. 2007, 64, 125–132. [Google Scholar] [CrossRef]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef]
- Afonine, P.V.; Grosse-Kunstleve, R.W.; Echols, N.; Headd, J.J.; Moriarty, N.W.; Mustyakimov, M.; Terwilliger, T.C.; Urzhumtsev, A.; Zwart, P.H.; Adams, P.D. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012, 68, 352–367. [Google Scholar] [CrossRef]
- Adams, P.D.; Afonine, P.V.; Bunkóczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef]
- Bell, J. CLVII.—The hydrolysis of guanidine. J. Chem. Soc. 1926, 129, 1213–1219. [Google Scholar] [CrossRef]
- Chen, V.B.; Arendall, W.B.; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 12–21. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Krissinel, E.; Henrick, K. Inference of Macromolecular Assemblies from Crystalline State. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef] [PubMed]
- Søndergaard, C.R.; Olsson, M.H.M.; Rostkowski, M.; Jensen, J.H. Improved treatment of ligands and coupling effects in empirical calculation and rationalization of p K a values. J. Chem. Theory Comput. 2011, 7, 2284–2295. [Google Scholar] [CrossRef] [PubMed]
- Olsson, M.H.M.; SØndergaard, C.R.; Rostkowski, M.; Jensen, J.H. PROPKA3: Consistent treatment of internal and surface residues in empirical pKa predictions. J. Chem. Theory Comput. 2011, 7, 525–537. [Google Scholar] [CrossRef] [PubMed]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef]
- Bradbrook, G.M.; Gleichmann, T.; Harrop, S.J.; Habash, J.; Raftery, J.; Kalb, J.; Yariv, J.; Hillier, I.H.; Helliwell, J.R. X-ray and molecular dynamics studies of concanavalin-A glucoside and mannoside complexes. Relating structure to thermodynamics of binding. J. Chem. Soc. Faraday Trans. 1998, 94, 1603–1611. [Google Scholar] [CrossRef]
- Wang, J.; Wang, W.; Kollman, P.A.; Case, D.A. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Model. 2006, 25, 247–260. [Google Scholar] [CrossRef]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general Amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef]
- Jorgensen, W.L. Transferable Intermolecular Potential Functions for Water, Alcohols, and Ethers. Application to Liquid Water. J. Am. Chem. Soc. 1981, 103, 335–340. [Google Scholar] [CrossRef]
- Case, D.A.; Ben-Shalon, I.Y.; Brozell, S.R.; Cerutti, T.E.; Cheatham, T.E.; Cruzeiro, V.W.D.; Darden, T.A.; Duke, R.E.; Ghoreishi, D.; Giambasu, G.; et al. AMBER 2019; University of California: San Francisco, CA, USA, 2019. [Google Scholar]
- Berendsen, H.J.C.; Postma, J.P.M.; van Gunsteren, W.F.; Hermans, J. Interaction Models for Water in Relation to Protein Hydration. In Intermolecular Forces. The Jerusalem Symposia on Quantum Chemistry and Biochemistry; Pullman, B., Ed.; Springer: Dordrecht, The Netherlands, 1981; Volume 14, pp. 331–342. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; Van Gunsteren, W.F.; Dinola, A.; Haak, J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Ryckaert, J.P.; Ciccotti, G.; Berendsen, H.J.C. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 1977, 23, 327–341. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Roe, D.R.; Cheatham, T.E. PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef]
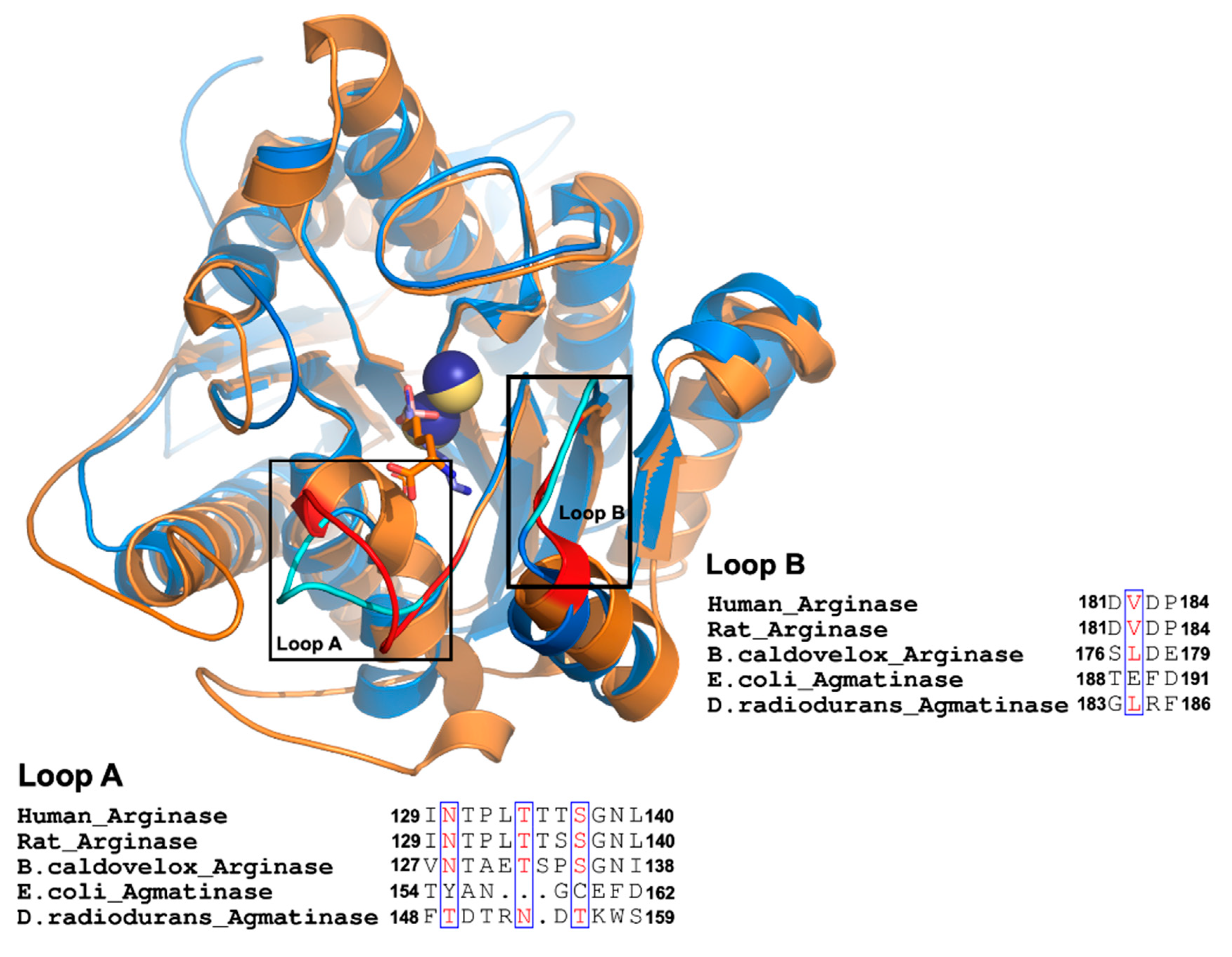
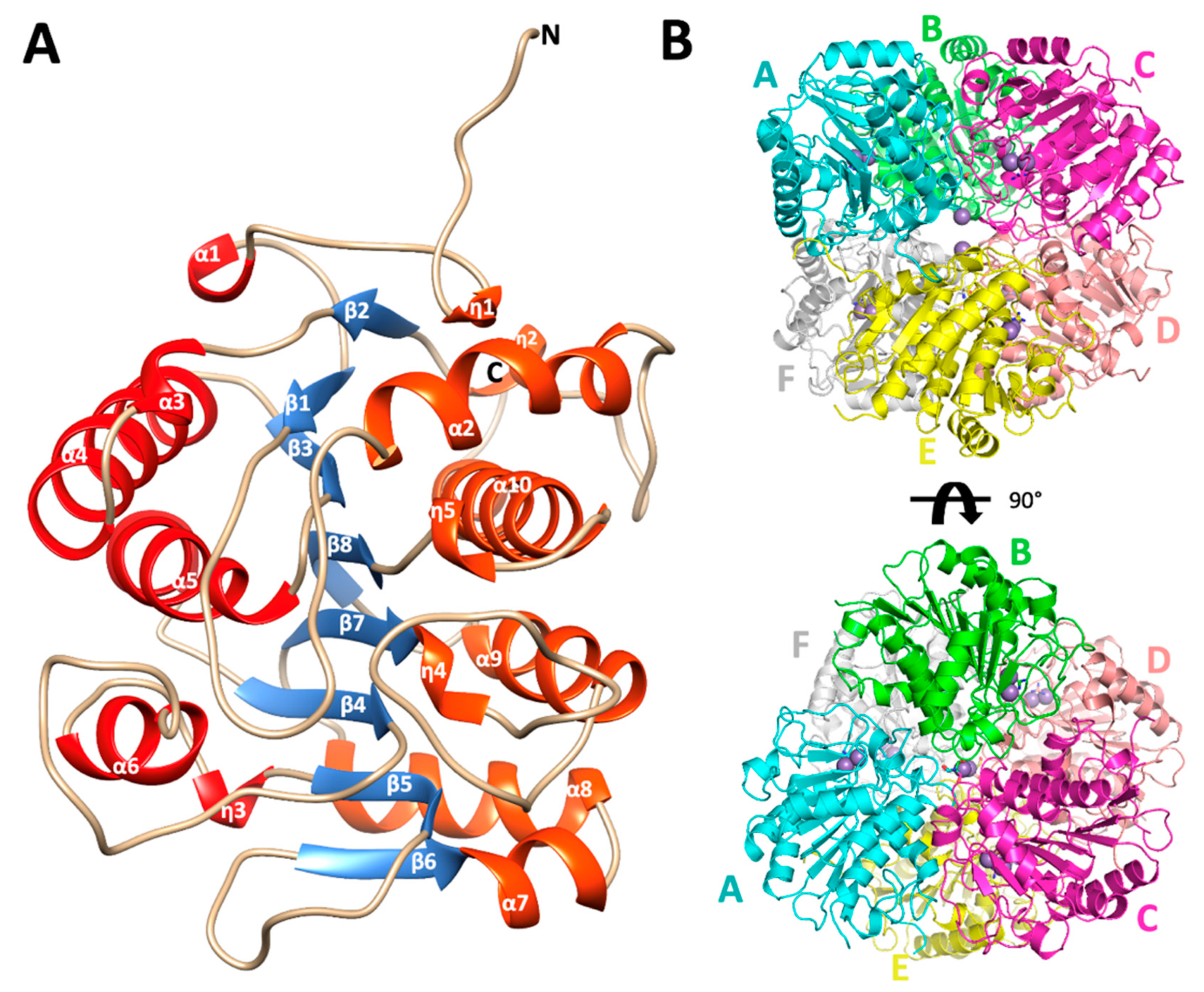
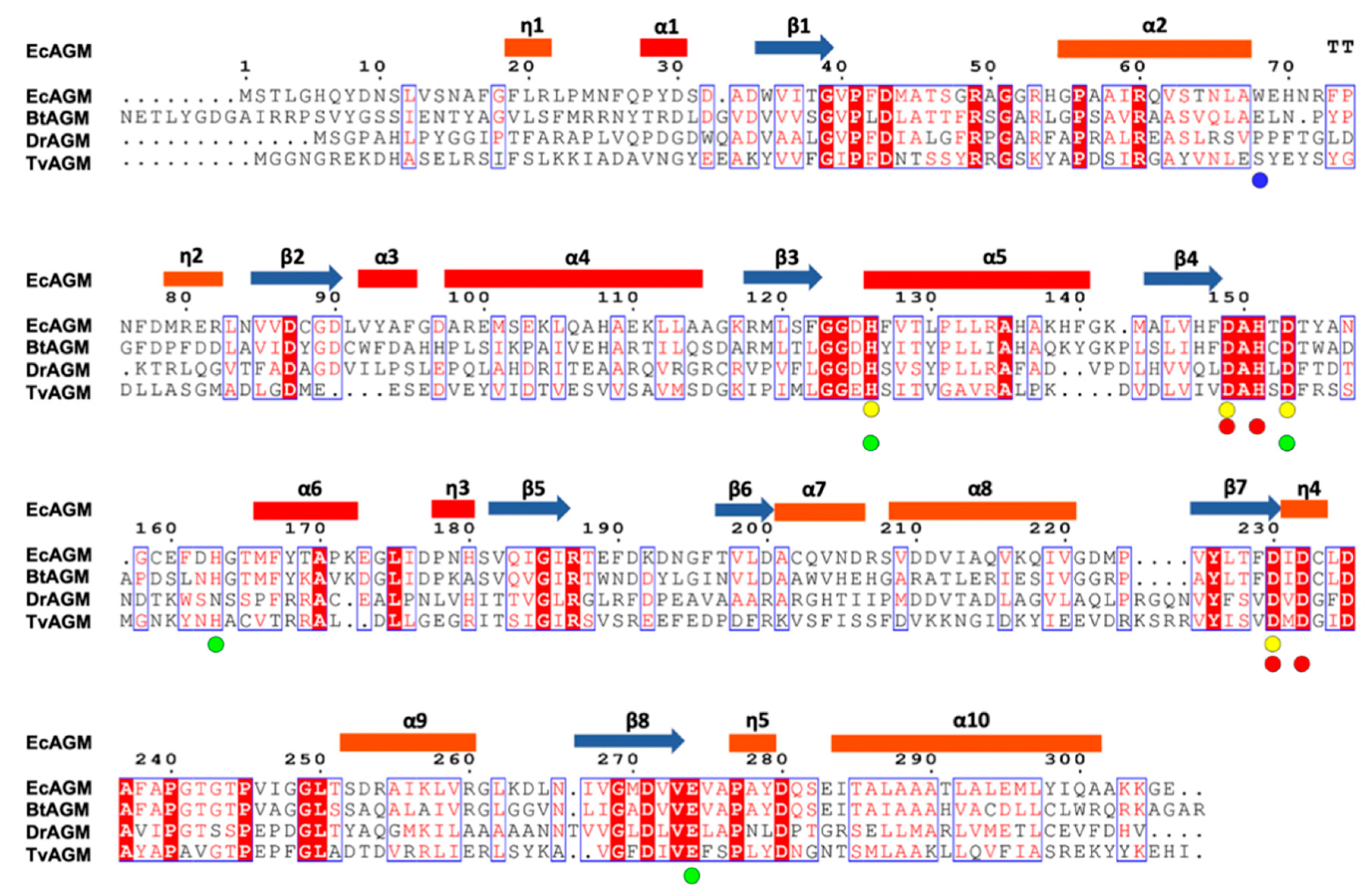
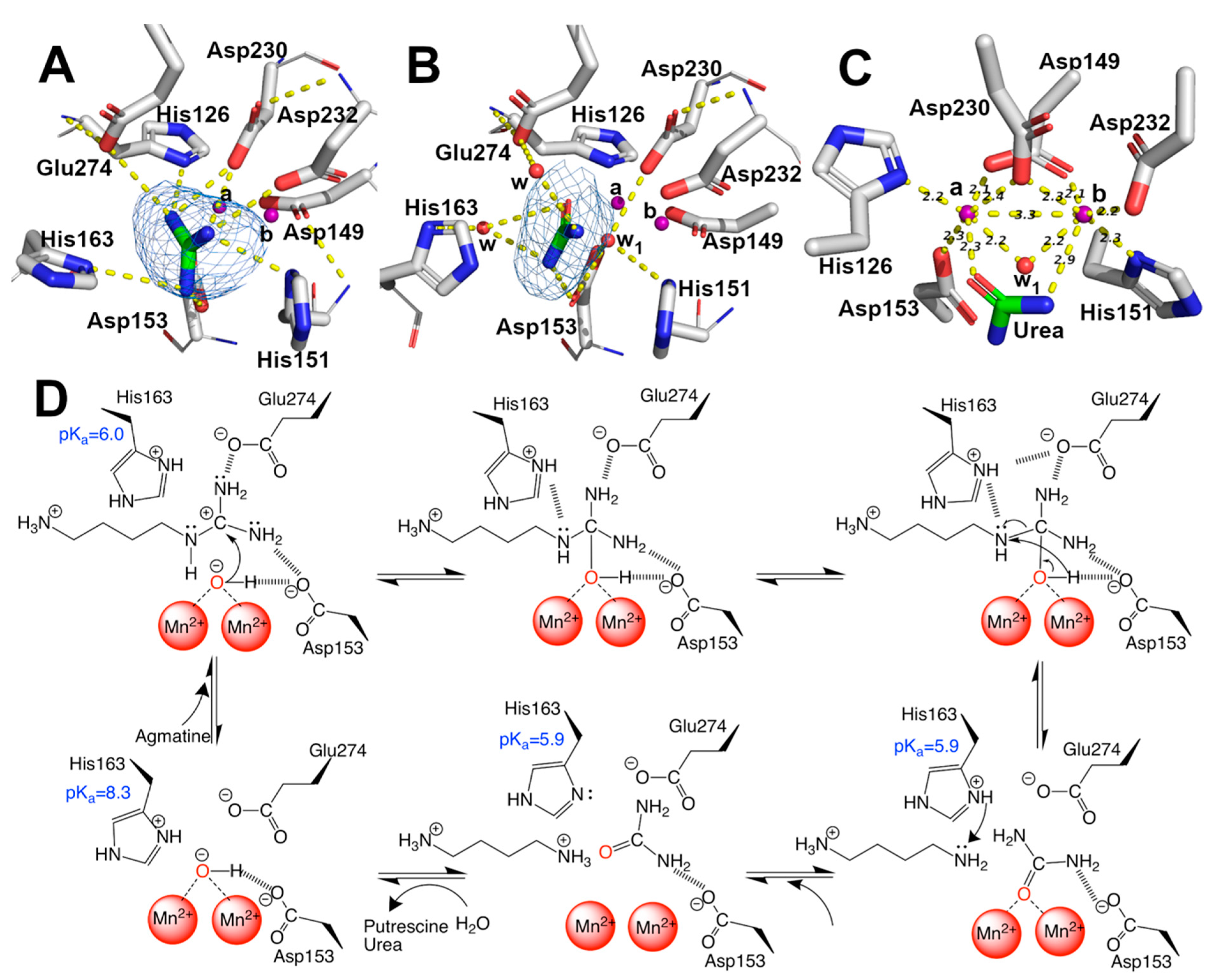
| Mutants | Arginine | Agmatine | ||||
|---|---|---|---|---|---|---|
| kcat (s−1) | KM (mM) | kcat/KM (M−1 s−1) | kcat (s−1) | KM (mM) | kcat/KM (M−1 s−1) | |
| Wild type | ND | 120 | 1.1 | 1.1 × 105 | ||
| Loop A mutants | ||||||
| (1) T154I-Y155N-A156T-N157P-/ins L-T-T | ND | 55 | 12 | 4.6 × 103 | ||
| (2) T154I-Y155N-A156T-N157P-/ins L-T-T/-G158T-C159S-E160G | ND | 48 | 15 | 3.2 × 103 | ||
| (3) T154I-Y155N-A156T-N157P-/ins L-T-T/-G158T-C159S-E160G-F161N-D162L | ND | 46 | 18 | 2.5 × 103 | ||
| Loop B mutants | ||||||
| (4) T188D-E189V | ND | 0.96 | 12 | 8 × 102 | ||
| (5) T188D-E189V-/ΔF190 | ND | 0.24 | 6.5 | 3.7 × 101 | ||
| Mutants in both loops | ||||||
| (6) T154I-Y155N-A156T-N157P-/ins L-T-T/-G158T-C159S-E160G-F161N-D162L-T188D-E189V | ND | 7.3 | 7 | 1 × 103 | ||
| (7) T154I-Y155N-A156T-N157P-/ins L-T-T/-G158T-C159S-E160G-F161N-D162L-T188D-E189V-/ΔF190 | ND | 8.4 | 8 | 1.1 × 103 | ||
| EcAGM-Urea (7LOL) | EcAGM-Gnd (7LOX) | |
|---|---|---|
| Wavelength | 1.033 | 1.771 |
| Resolution range | 35.79–1.8 (1.86–1.8) | 64.51–3.2 (3.31–3.2) |
| Space group | R 3 2:H | P 31 2 1 |
| Unit cell | 81.746 81.746 207.436 90 90 120 | 129.028 129.028 88.29 90 90 120 |
| Total reflections | 499876 (47706) | 564172 (55308) |
| Unique reflections | 25154 (2475) | 14314 (1418) |
| Multiplicity | 19.9 (19.3) | 39.4 (39.0) |
| Completeness (%) | 99.87 (99.88) | 99.87 (100.00) |
| Mean I/sigma(I) | 28.76 (3.61) | 26.70 (4.25) |
| Wilson B-factor | 27.1 | 84.6 |
| R-merge | 0.06576 (0.892) | 0.1561 (1.066) |
| R-meas | 0.06753 (0.9161) | 0.1582 (1.08) |
| R-pim | 0.01521 (0.2075) | 0.02515 (0.1718) |
| CC1/2 | 1 (0.933) | 0.999 (0.929) |
| CC* | 1 (0.983) | 1 (0.982) |
| R-work | 0.1702 (0.2355) | 0.1727 (0.2256) |
| R-free | 0.1947 (0.2617) | 0.2172 (0.2679) |
| Number of non-hydrogen atoms | 2432 | 6539 |
| macromolecules | 2259 | 6517 |
| ligands | 24 | 22 |
| solvent | 149 | 0 |
| Protein residues | 294 | 852 |
| RMS (bonds) | 0.015 | 0.012 |
| RMS (angles) | 1.35 | 1.40 |
| Ramachandran favored (%) | 99.32 | 98.33 |
| Ramachandran allowed (%) | 0.68 | 1.67 |
| Ramachandran outliers (%) | 0.00 | 0.00 |
| Rotamer outliers (%) | 1.28 | 0.00 |
| Clashscore | 3.11 | 9.98 |
| Average B-factor | 33.35 | 81.50 |
| macromolecules | 32.90 | 81.51 |
| ligands | 34.70 | 76.68 |
| solvent | 39.81 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maturana, P.; Orellana, M.S.; Herrera, S.M.; Martínez, I.; Figueroa, M.; Martínez-Oyanedel, J.; Castro-Fernandez, V.; Uribe, E. Crystal Structure of Escherichia coli Agmatinase: Catalytic Mechanism and Residues Relevant for Substrate Specificity. Int. J. Mol. Sci. 2021, 22, 4769. https://doi.org/10.3390/ijms22094769
Maturana P, Orellana MS, Herrera SM, Martínez I, Figueroa M, Martínez-Oyanedel J, Castro-Fernandez V, Uribe E. Crystal Structure of Escherichia coli Agmatinase: Catalytic Mechanism and Residues Relevant for Substrate Specificity. International Journal of Molecular Sciences. 2021; 22(9):4769. https://doi.org/10.3390/ijms22094769
Chicago/Turabian StyleMaturana, Pablo, María S. Orellana, Sixto M. Herrera, Ignacio Martínez, Maximiliano Figueroa, José Martínez-Oyanedel, Victor Castro-Fernandez, and Elena Uribe. 2021. "Crystal Structure of Escherichia coli Agmatinase: Catalytic Mechanism and Residues Relevant for Substrate Specificity" International Journal of Molecular Sciences 22, no. 9: 4769. https://doi.org/10.3390/ijms22094769
APA StyleMaturana, P., Orellana, M. S., Herrera, S. M., Martínez, I., Figueroa, M., Martínez-Oyanedel, J., Castro-Fernandez, V., & Uribe, E. (2021). Crystal Structure of Escherichia coli Agmatinase: Catalytic Mechanism and Residues Relevant for Substrate Specificity. International Journal of Molecular Sciences, 22(9), 4769. https://doi.org/10.3390/ijms22094769







