Circadian Rhythms in Legumes: What Do We Know and What Else Should We Explore?
Abstract
1. Introduction
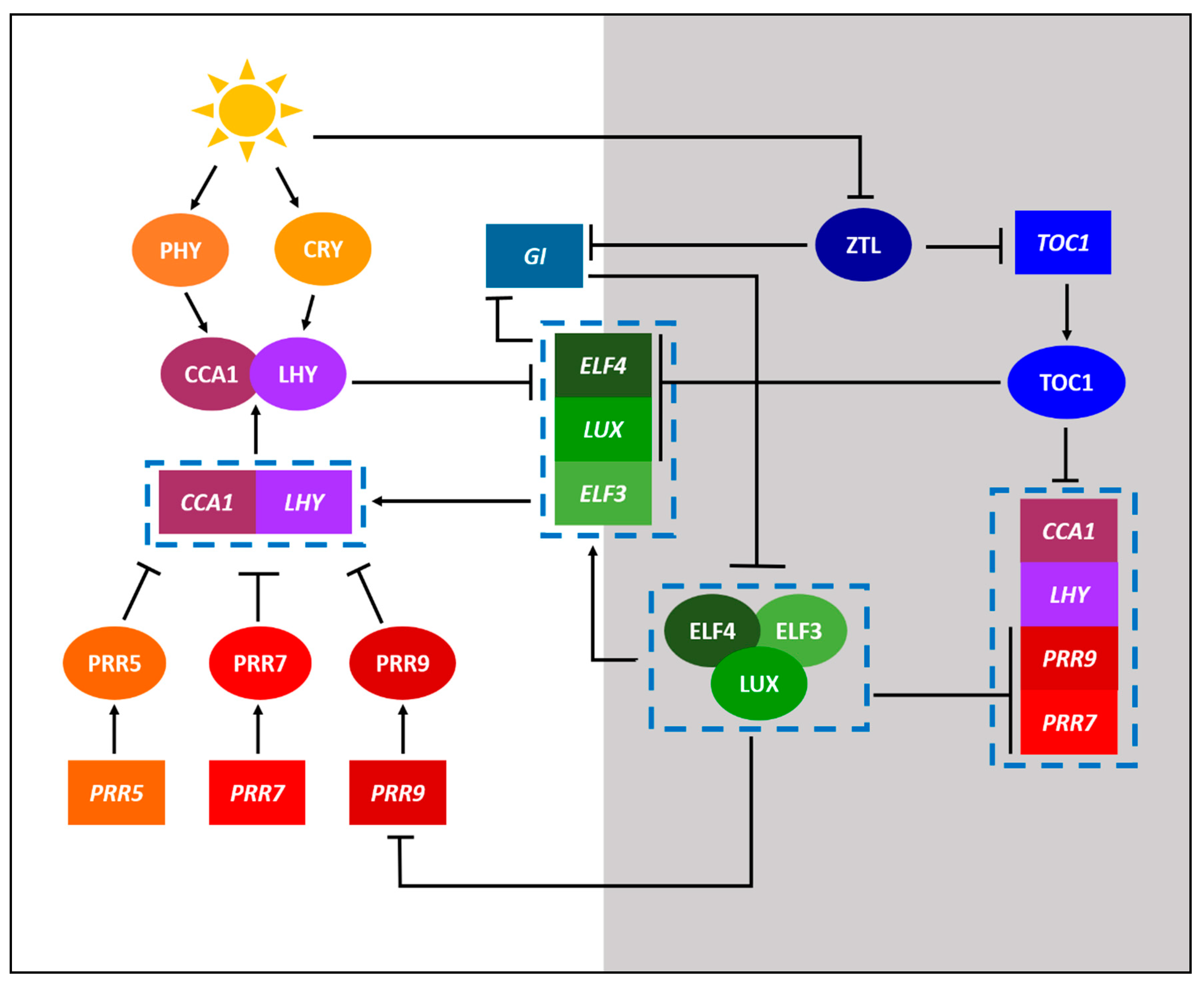
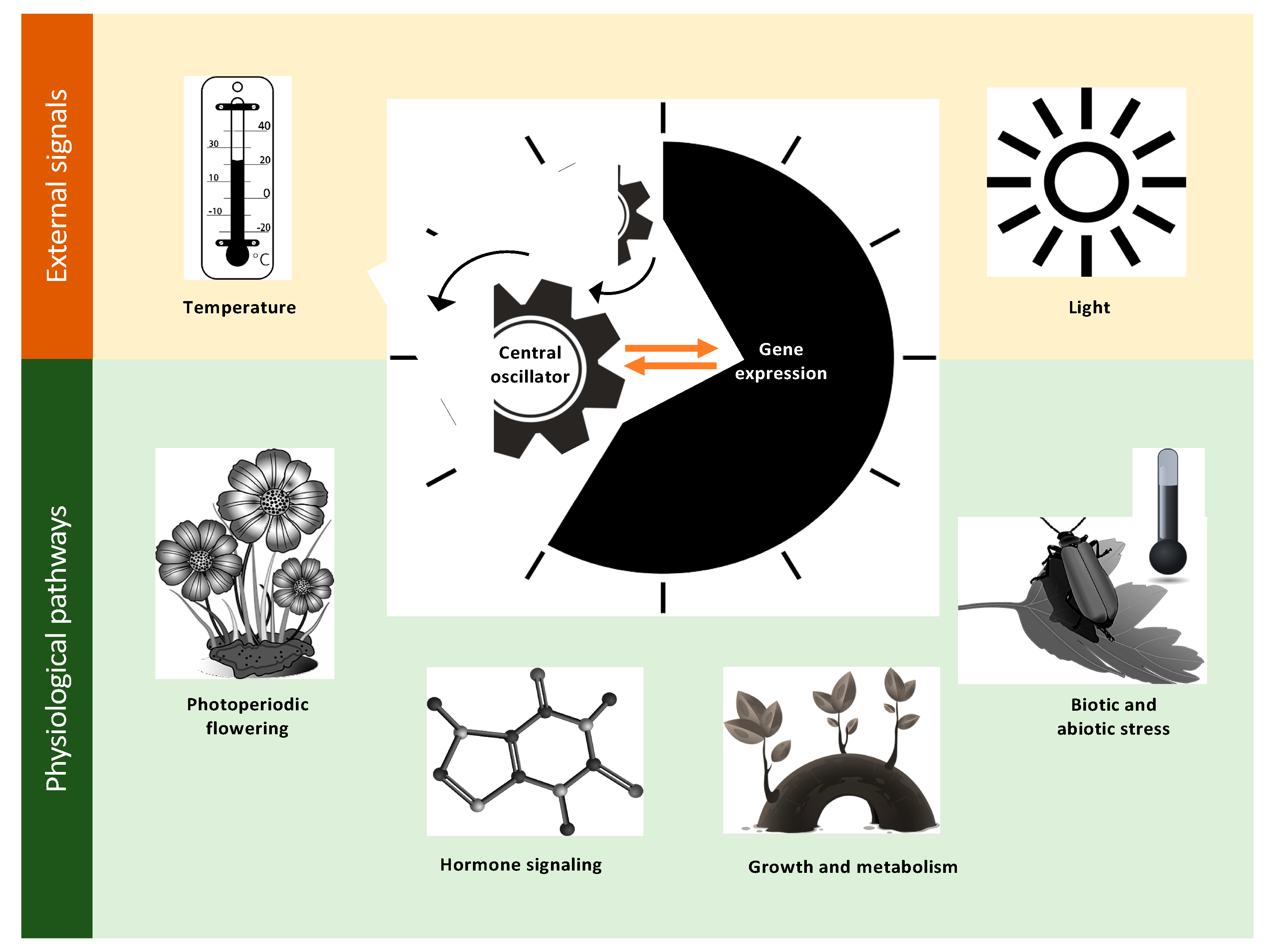
2. Defining the Important Components of Clock Research
2.1. Photoperiodic Flowering
2.2. Growth and Metabolism
2.3. Hormone Signaling
2.4. Biotic and Abiotic Stress
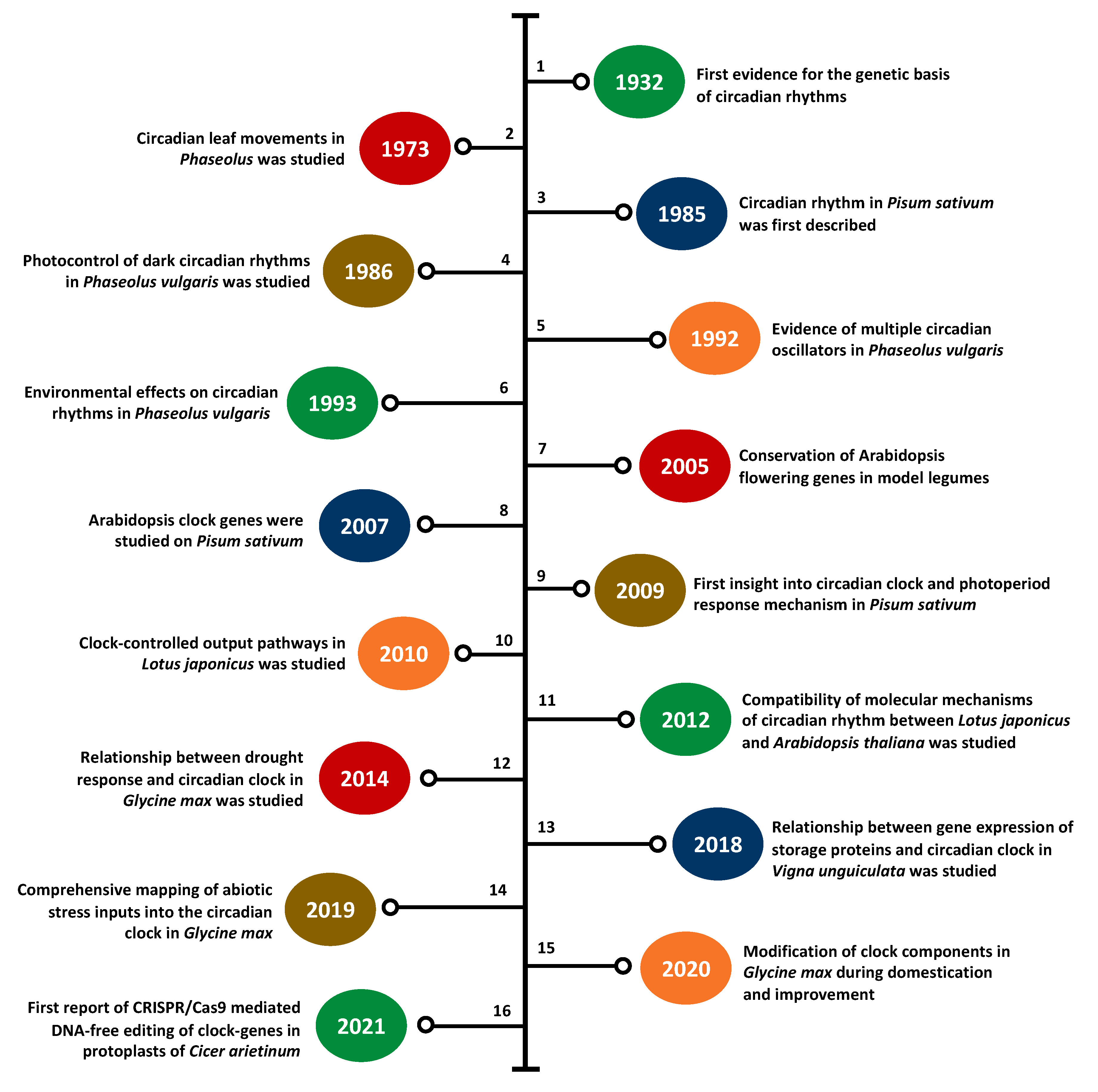
3. Legume Clock Research at a Glance
3.1. Clock Research in Model Legumes
3.1.1. Barrel Medic
3.1.2. Birds-Foot Trefoil
3.1.3. Soybean
3.1.4. Common Pea
3.2. Clock Research in Underutilized Legumes
4. Concluding Remarks and Perspective
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- McClung, C.R. The plant circadian oscillator. Biology 2019, 8, 14. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, D.; Shamim, M.; Kumar, M.; Mishra, A.; Maurya, R.; Sharma, D.; Pandey, P.; Singh, K. Role of circadian rhythm in plant system: An update from development to stress response. Environ. Exp. Bot. 2019, 162, 256–271. [Google Scholar] [CrossRef]
- McClung, C.R. Circadian clock components offer targets for crop domestication and improvement. Genes 2021, 12, 374. [Google Scholar] [CrossRef]
- Salmela, M.J.; Weinig, C. The fitness benefits of genetic variation in circadian clock regulation. Curr. Opin. Plant Biol. 2019, 49, 86–93. [Google Scholar] [CrossRef]
- Kim, J.A.; Kim, H.S.; Choi, S.H.; Jang, J.Y.; Jeong, M.J.; Lee, S.I. The importance of the circadian clock in regulating plant metabolism. Int. J. Mol. Sci. 2017, 18, 2680. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Nusinow, D.A. Into the evening: Complex interactions in the Arabidopsis circadian clock. Trends Genet. 2016, 32, 674–686. [Google Scholar] [CrossRef]
- Sahruzaini, N.A.; Rejab, N.A.; Harikrishna, J.A.; Ikram, N.K.K.; Ismail, I.; Kugan, H.M.; Cheng, A. Pulse crop genetics for a sustainable future: Where we are now and where we should be heading. Front. Plant Sci. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Cheng, A.; Raai, M.N.; Zain, N.A.M.; Massawe, F.; Singh, A.; Wan, W.A.A.Q.I. In search of alternative proteins: Unlocking the potential of underutilized tropical legumes. Food Secur. 2019, 11, 1205–1215. [Google Scholar] [CrossRef]
- Kumar, S.; Pandey, G. Biofortification of pulses and legumes to enhance nutrition. Heliyon 2020, 6, e03682. [Google Scholar] [CrossRef] [PubMed]
- Smýkal, P.; Aubert, G.; Burstin, J.; Coyne, C.J.; Ellis, N.T.; Flavell, A.J.; Ford, R.; Hýbl, M.; Macas, J.; Neumann, P. Pea (Pisum sativum L.) in the genomic era. Agronomy 2012, 2, 74–115. [Google Scholar] [CrossRef]
- Inoue, K.; Araki, T.; Endo, M. Integration of input signals into the gene network in the plant circadian clock. Plant Cell Physiol. 2017, 58, 977–982. [Google Scholar] [CrossRef]
- Gil, K.E.; Park, C.M. Thermal adaptation and plasticity of the plant circadian clock. New Phytol. 2018, 221, 1215–1229. [Google Scholar] [CrossRef]
- Oakenfull, R.J.; Davis, S.J. Shining a light on the Arabidopsis circadian clock. Plant Cell Environ. 2017, 40, 2571–2585. [Google Scholar] [CrossRef]
- Sanchez, S.E.; Kay, S.A. The plant circadian clock: From a simple timekeeper to a complex developmental manager. Cold Spring Harb. Perspect. Biol. 2016, 8, a027748. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.A. Retrograde signalling as an informant of circadian timing. New Phytol. 2019, 221, 1749–1753. [Google Scholar] [CrossRef]
- Ionescu, I.A.; Møller, B.L.; Sánchez-Pérez, R. Chemical control of flowering time. J. Exp. Bot. 2016, 68, 369–382. [Google Scholar] [CrossRef] [PubMed]
- Hecht, V.; Foucher, F.; Ferrándiz, C.; Macknight, R.; Navarro, C.; Morin, J.; Vardy, M.E.; Ellis, N.; Beltrán, J.P.; Rameau, C.; et al. Conservation of Arabidopsis flowering genes in model legumes. Plant Physiol. 2005, 137, 1420–1434. [Google Scholar] [CrossRef]
- Cheng, X.; Krom, N.; Zhang, S.; Mysore, K.S.; Udvardi, M.; Wen, J. Enabling Reverse Genetics in Medicago truncatula using high-throughput sequencing for Tnt1 flanking sequence recovery. Methods Mol. Biol. 2017, 1610, 25–37. [Google Scholar]
- Magne, K.; George, J.; Tornero, A.B.; Broquet, B.; Madueño, F.; Andersen, S.U.; Ratet, P. Lotus japonicus NOOT-BOP-COCH-LIKE1is essential for nodule, nectary, leaf and flower development. Plant J. 2018, 94, 880–894. [Google Scholar] [CrossRef]
- Sun, L.; Gill, U.S.; Nandety, R.S.; Kwon, S.; Mehta, P.; Dickstein, R.; Udvardi, M.K.; Mysore, K.S.; Wen, J. Genome-wide analysis of flanking sequences reveals that Tnt1 insertion is positively correlated with gene methylation in Medicago truncatula. Plant J. 2019, 98, 1106–1119. [Google Scholar] [CrossRef]
- Cronk, Q.; Ojeda, I.; Pennington, R.T. Legume comparative genomics: Progress in phylogenetics and phylogenomics. Curr. Opin. Plant Biol. 2006, 9, 99–103. [Google Scholar] [CrossRef] [PubMed]
- Hecht, V.; Knowles, C.L.; Vander Schoor, J.K.; Liew, L.C.; Jones, S.E.; Lambert, M.J.; Weller, J.L. Pea LATE BLOOMER1 is a GIGANTEA ortholog with roles in photoperiodic flowering, deetiolation, and transcriptional regulation of circadian clock gene homologs. Plant Physiol. 2007, 144, 648–661. [Google Scholar] [CrossRef] [PubMed]
- Weller, J.L.; Ortega, R. Genetic control of flowering time in legumes. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef]
- Wong, A.; Hecht, V.F.; Picard, K.; Diwadkar, P.; Laurie, R.E.; Wen, J.; Mysore, K.; Macknight, R.C.; Weller, J.L. Isolation and functional analysis of CONSTANS-LIKE genes suggests that a central role for CONSTANS in flowering time control is not evolutionarily conserved in Medicago truncatula. Front. Plant Sci. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Hecht, V.; Laurie, R.E.; Schoor, J.K.V.; Ridge, S.; Knowles, C.L.; Liew, L.C.; Sussmilch, F.C.; Murfet, I.C.; Macknight, R.C.; Weller, J.L. The pea GIGAS Gene is a FLOWERING LOCUS T homolog necessary for graft-transmissible specification of flowering but not for responsiveness to photoperiod. Plant Cell 2011, 23, 147–161. [Google Scholar] [CrossRef]
- Dodd, A.N.; Belbin, F.E.; Frank, A.; Webb, A.A.R. Interactions between circadian clocks and photosynthesis for the temporal and spatial coordination of metabolism. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Foo, M.; Somers, D.E.; Kim, P.-J. Kernel architecture of the genetic circuitry of the Arabidopsis circadian system. PLoS Comput. Biol. 2016, 12, e1004748. [Google Scholar] [CrossRef]
- Nagel, D.H.; Kay, S.A. Complexity in the wiring and regulation of plant circadian networks. Curr. Biol. 2012, 22, R648–R657. [Google Scholar] [CrossRef]
- Pan, W.-J.; Wang, X.; Deng, Y.-R.; Li, J.-H.; Chen, W.; Chiang, J.Y.; Yang, J.-B.; Zheng, L. Nondestructive and intuitive determination of circadian chlorophyll rhythms in soybean leaves using multispectral imaging. Sci. Rep. 2015, 5, 11108. [Google Scholar] [CrossRef]
- Singh, V.K.; Rajkumar, M.S.; Garg, R.; Jain, M. Genome-wide identification and co-expression network analysis provide insights into the roles of auxin response factor gene family in chickpea. Sci. Rep. 2017, 7, 10895. [Google Scholar] [CrossRef]
- Zhuo, C.; Liang, L.; Zhao, Y.; Guo, Z.; Lu, S. A cold responsive ethylene responsive factor from Medicago falcata confers cold tolerance by up-regulation of polyamine turnover, antioxidant protection, and proline accumulation. Plant Cell Environ. 2018, 41, 2021–2032. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.; Wang, H.; Hong, L.; Xu, Y.; Zhao, Y.; Zhou, C. MtBZR1 plays an important role in nodule development in Medicago truncatula. Int. J. Mol. Sci. 2019, 20, 2941. [Google Scholar] [CrossRef]
- Tan, S.; Debellé, F.; Gamas, P.; Frugier, F.; Brault, M. Diversification of cytokinin phosphotransfer signaling genes in Medicago truncatula and other legume genomes. BMC Genom. 2019, 20, 373. [Google Scholar] [CrossRef]
- Kong, Y.; Meng, Z.; Wang, H.; Wang, Y.; Zhang, Y.; Hong, L.; Liu, R.; Wang, M.; Zhang, J.; Han, L.; et al. Brassinosteroid homeostasis is critical for the functionality of the Medicago truncatula pulvinus. Plant Physiol. 2021. [Google Scholar] [CrossRef]
- Lyu, X.; Cheng, Q.; Qin, C.; Li, Y.; Xu, X.; Ji, R.; Mu, R.; Li, H.; Zhao, T.; Liu, J.; et al. GmCRY1s modulate gibberellin metabolism to regulate soybean shade avoidance in response to reduced blue light. Mol. Plant 2021, 14, 298–314. [Google Scholar] [CrossRef]
- Goodspeed, D.; Chehab, E.W.; Min-Venditti, A.; Braam, J.; Covington, M.F. Arabidopsis synchronizes jasmonate-mediated defense with insect circadian behavior. Proc. Natl. Acad. Sci. USA 2012, 109, 4674–4677. [Google Scholar] [CrossRef] [PubMed]
- García-Martínez, J.L.; Gil, J. Light regulation of gibberellin biosynthesis and mode of action. J. Plant Growth Regul. 2001, 20, 354–368. [Google Scholar] [CrossRef] [PubMed]
- Hisamatsu, T.; King, R.W.; Helliwell, C.A.; Koshioka, M. The involvement of gibberellin 20-oxidase genes in phytochrome-regulated petiole elongation of Arabidopsis. Plant Physiol. 2005, 138, 1106–1116. [Google Scholar] [CrossRef]
- Arana, M.V.; Marín-de la Rosa, N.; Maloof, J.N.; Blázquez, M.A.; Alabadí, D. Circadian oscillation of gibberellin signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 9292–9297. [Google Scholar] [CrossRef]
- Pokhilko, A.; Bou-Torrent, J.; Pulido, P.; Rodríguez-Concepción, M.; Ebenhöh, O. Mathematical modelling of the diurnal regulation of the MEP pathway in Arabidopsis. New Phytol. 2015, 206, 1075–1085. [Google Scholar] [CrossRef]
- Rawat, R.; Schwartz, J.; Jones, M.A.; Sairanen, I.; Cheng, Y.; Andersson, C.R.; Zhao, Y.; Ljung, K.; Harmer, S.L. REVEILLE1, a Myb-like transcription factor, integrates the circadian clock and auxin pathways. Proc. Natl. Acad. Sci. USA 2009, 106, 16883–16888. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Mas, P. A functional connection between the circadian clock and hormonal timing in Arabidopsis. Genes 2018, 9, 567. [Google Scholar] [CrossRef]
- Li, M.; Cao, L.; Mwimba, M.; Zhou, Y.; Li, L.; Zhou, M.; Schnable, P.S.; O’Rourke, J.A.; Dong, X.; Wang, W. Comprehensive mapping of abiotic stress inputs into the soybean circadian clock. Proc. Natl. Acad. Sci. USA 2019, 116, 23840–23849. [Google Scholar] [CrossRef]
- Vezza, M.E.; Alderete, L.G.S.; Agostini, E.; Talano, M.A. Expression of circadian clock genes and diurnal oscillations of key physiological events in response to AsV and AsIII in soybean plants. Environ. Exp. Bot. 2020, 174. [Google Scholar] [CrossRef]
- Preuss, S.B.; Meister, R.; Xu, Q.; Urwin, C.P.; Tripodi, F.A.; Screen, S.E.; Anil, V.S.; Zhu, S.; Morrell, J.A.; Liu, G. Expression of the Arabidopsis thaliana BBX32 gene in soybean increases grain yield. PLoS ONE 2012, 7, e30717. [Google Scholar] [CrossRef]
- Johansson, M.; Köster, T. On the move through time–a historical review of plant clock research. Plant Biol. 2019, 21, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Paltiel, J.; Amin, R.; Gover, A.; Ori, N.; Samach, A. Novel roles for GIGANTEA revealed under environmental conditions that modify its expression in Arabidopsis and Medicago truncatula. Planta 2006, 224, 1255–1268. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Han, L.; Liu, X.; Wang, H.; Wen, L.; Yu, X.; Xu, X.; Kong, F.; Fu, C.; Mysore, K.S. The nodulation and nyctinastic leaf movement is orchestrated by clock gene LHY in Medicago truncatula. J. Integr. Plant Biol. 2020, 62, 1880–1895. [Google Scholar] [CrossRef]
- Ishida, K.; Niwa, Y.; Yamashino, T.; Mizuno, T. A genome-wide compilation of the two-component systems in Lotus japonicus. DNA Res. 2009, 16, 237–247. [Google Scholar] [CrossRef]
- Ueoka-Nakanishi, H.; Yamashino, T.; Ishida, K.; Kamioka, M.; Nakamichi, N.; Mizuno, T. Molecular mechanisms of circadian rhythm in Lotus japonicus and Arabidopsis thaliana are sufficiently compatible to regulate heterologous core clock genes robustly. Biosci. Biotechnol. Biochem. 2012. [Google Scholar] [CrossRef]
- Wakabayashi, T.; Oh, H.; Kawaguchi, M.; Harada, K.; Sato, S.; Ikeda, H.; Hiroaki, S. Polymorphisms of E1 and GIGANTEA in wild populations of Lotus japonicus. J. Plant Res. 2014, 127, 651–660. [Google Scholar] [CrossRef]
- Liu, H.; Wang, H.; Gao, P.; Xü, J.; Xü, T.; Wang, J.; Wang, B.; Lin, C.; Fu, Y.-F. Analysis of clock gene homologs using unifoliolates as target organs in soybean (Glycine max). J. Plant Physiol. 2009, 166, 278–289. [Google Scholar] [CrossRef]
- Marcolino-Gomes, J.; Nakayama, T.J.; Molinari, H.B.C.; Basso, M.F.; Henning, L.M.M.; Fuganti-Pagliarini, R.; Harmon, F.G.; Nepomuceno, A.L. Functional characterization of a putative Glycine max ELF4 in transgenic Arabidopsis and its role during flowering control. Front. Plant Sci. 2017, 8, 618. [Google Scholar] [CrossRef]
- Watanabe, S.; Xia, Z.; Hideshima, R.; Tsubokura, Y.; Sato, S.; Yamanaka, N.; Takahashi, R.; Anai, T.; Tabata, S.; Kitamura, K. A map-based cloning strategy employing a residual heterozygous line reveals that the GIGANTEA gene is involved in soybean maturity and flowering. Genetics 2011, 188, 395–407. [Google Scholar] [CrossRef]
- Cheng, Q.; Dong, L.; Su, T.; Li, T.; Gan, Z.; Nan, H.; Lu, S.; Fang, C.; Kong, L.; Li, H. CRISPR/Cas9-mediated targeted mutagenesis of GmLHY genes alters plant height and internode length in soybean. BMC Plant Biol. 2019, 19, 562. [Google Scholar] [CrossRef] [PubMed]
- Liew, L.C.; Singh, M.B.; Bhalla, P.L. A novel role of the soybean clock gene LUX ARRHYTHMO in male reproductive development. Sci. Rep. 2017, 7, 10605. [Google Scholar] [CrossRef]
- Xue, Z.-G.; Zhang, X.-M.; Lei, C.-F.; Chen, X.-J.; Fu, Y.-F. Molecular cloning and functional analysis of one ZEITLUPE homolog GmZTL3 in soybean. Mol. Biol. Rep. 2012, 39, 1411–1418. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Sun, S.; Wu, T.; Liu, L.; Sun, X.; Cai, Y.; Li, J.; Jia, H.; Yuan, S.; Chen, L. Natural variation and CRISPR/Cas9-mediated mutation in GmPRR37 affect photoperiodic flowering and contribute to regional adaptation of soybean. Plant Biotechnol. J. 2020, 18, 1869–1881. [Google Scholar] [CrossRef] [PubMed]
- Liew, L.; Hecht, V.; Weeden, N.; Weller, J. Isolation of pseudo response regulator genes and evaluation as candidate genes for photoperiod response loci. Pisum. Genet. 2009, 41, 21–25. [Google Scholar]
- Liew, L.C.; Hecht, V.; Laurie, R.E.; Knowles, C.L.; Vander Schoor, J.K.; Macknight, R.C.; Weller, J.L. DIE NEUTRALIS and LATE BLOOMER 1 contribute to regulation of the pea circadian clock. Plant Cell 2009, 21, 3198–3211. [Google Scholar] [CrossRef]
- Weller, J.L.; Liew, L.C.; Hecht, V.F.; Rajandran, V.; Laurie, R.E.; Ridge, S.; Wenden, B.; Vander Schoor, J.K.; Jaminon, O.; Blassiau, C. A conserved molecular basis for photoperiod adaptation in two temperate legumes. Proc. Natl. Acad. Sci. USA 2012, 109, 21158–21163. [Google Scholar] [CrossRef]
- Liew, L.C.; Hecht, V.; Sussmilch, F.C.; Weller, J.L. The pea photoperiod response gene STERILE NODES is an ortholog of LUX ARRHYTHMO. Plant. Physiol. 2014, 165, 648–657. [Google Scholar] [CrossRef] [PubMed]
- Ridge, S.; Deokar, A.; Lee, R.; Daba, K.; Macknight, R.C.; Weller, J.L.; Tar’an, B. The chickpea Early Flowering 1 (Efl1) locus is an ortholog of Arabidopsis ELF3. Plant Physiol. 2017, 175, 802–815. [Google Scholar] [CrossRef]
- Rychel, S.; Książkiewicz, M.; Tomaszewska, M.; Bielski, W.; Wolko, B. FLOWERING LOCUS T, GIGANTEA, SEPALLATA, and FRIGIDA homologs are candidate genes involved in white lupin (Lupinus albus L.) early flowering. Mol. Breed. 2019, 39, 43. [Google Scholar] [CrossRef]
- Kaldis, A.-D.; Kousidis, P.; Kesanopoulos, K.; Prombona, A. Light and circadian regulation in the expression of LHY and Lhcb genes in Phaseolus vulgaris. Plant Mol. Biol. 2003, 52, 981–997. [Google Scholar] [CrossRef]
- Kaldis, A.-D.; Prombona, A. Synergy between the light-induced acute response and the circadian cycle: A new mechanism for the synchronization of the Phaseolus vulgaris clock to light. Plant Mol. Biol. 2006, 61, 883–895. [Google Scholar] [CrossRef]
- Galeou, A.; Prombona, A. Light at night resynchronizes the evening-phased rhythms of TOC1 and ELF4 in Phaseolus vulgaris. Plant Sci. 2012, 184, 141–147. [Google Scholar] [CrossRef]
- Galeou, A.; Roussis, A.; Prombona, A. Investigation of the Phaseolus vulgaris circadian clock and the repressive role of the PvTOC1 factor by a newly established in vitro system. J. Plant Physiol. 2018, 222, 79–85. [Google Scholar] [CrossRef]
- Kwak, M.; Velasco, D.; Gepts, P. Mapping homologous sequences for determinacy and photoperiod sensitivity in common bean (Phaseolus vulgaris). J. Hered. 2008, 99, 283–291. [Google Scholar] [CrossRef] [PubMed]
- Weiss, J.; Terry, M.I.; Martos-Fuentes, M.; Letourneux, L.; Ruiz-Hernández, V.; Fernández, J.A.; Egea-Cortines, M. Diel pattern of circadian clock and storage protein gene expression in leaves and during seed filling in cowpea (Vigna unguiculata). BMC Plant Biol. 2018, 18, 33. [Google Scholar] [CrossRef]
- Bünning, E. Über die Erblichkeit der Tagesperiodizität bei den Phaseolus Blättern. Jahrb Bot. 1932, 81, 411–418. [Google Scholar]
- Bünning, E.; Moser, I. Light-induced phase shifts of circadian leaf movements of Phaseolus: Comparison with the effects of potassium and of ethyl alcohol. Proc. Natl. Acad. Sci. USA 1973, 70, 3387–3389. [Google Scholar] [CrossRef] [PubMed]
- Kloppstech, K. Diurnal and circadian rhythmicity in the expression of light-induced plant nuclear messenger RNAs. Planta 1985, 165, 502–506. [Google Scholar] [CrossRef] [PubMed]
- Holmes, M.G.; Klein, W.H. Photocontrol of dark circadian rhythms in stomata of Phaseolus vulgaris L. Plant Physiol. 1986, 82, 28–33. [Google Scholar] [CrossRef]
- Hennessey, T.L.; Field, C.B. Evidence of multiple circadian oscillators in bean plants. J. Biol. Rhythms 1992, 7, 105–113. [Google Scholar] [CrossRef]
- Hennessey, T.L.; Freeden, A.L.; Field, C.B. Environmental effects on circadian rhythms in photosynthesis and stomatal opening. Planta 1993, 189, 369–376. [Google Scholar] [CrossRef]
- Weller, J.L.; Hecht, V.; Liew, L.C.; Sussmilch, F.C.; Wenden, B.; Knowles, C.L.; Vander Schoor, J.K. Update on the genetic control of flowering in garden pea. J. Exp. Bot. 2009, 60, 2493–2499. [Google Scholar] [CrossRef] [PubMed]
- Ono, N.; Ishida, K.; Yamashino, T.; Nakanishi, H.; Sato, S.; Tabata, S.; Mizuno, T. Genomewide characterization of the light-responsive and clock-controlled output pathways in Lotus japonicus with special emphasis of its uniqueness. Plant Cell Physiol. 2010, 51, 1800–1814. [Google Scholar] [CrossRef]
- Marcolino-Gomes, J.; Rodrigues, F.A.; Fuganti-Pagliarini, R.; Bendix, C.; Nakayama, T.J.; Celaya, B.; Molinari, H.B.C.; de Oliveira, M.C.N.; Harmon, F.G.; Nepomuceno, A. Diurnal oscillations of soybean circadian clock and drought responsive genes. PLoS ONE 2014, 9, e86402. [Google Scholar] [CrossRef] [PubMed]
- Li, M.-W.; Lam, H.-M. The modification of circadian clock components in soybean during domestication and improvement. Front. Genet. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Badhan, S.; Ball, A.S.; Mantri, N. First report of CRISPR/Cas9 mediated DNA-free editing of 4CL and RVE7 genes in chickpea protoplasts. Int. J. Mol. Sci. 2021, 22, 396. [Google Scholar] [CrossRef] [PubMed]
- Young, N.D.; Debellé, F.; Oldroyd, G.E.; Geurts, R.; Cannon, S.B.; Udvardi, M.K.; Benedito, V.A.; Mayer, K.F.; Gouzy, J.; Schoof, H. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 2011, 480, 520–524. [Google Scholar] [CrossRef]
- Crane, C.; Dixon, R.A.; Wang, Z.-Y. Medicago truncatula transformation using root explants. Methods Mol. Biol. 2006, 343, 137–142. [Google Scholar]
- Cerri, M.R.; Frances, L.; Kelner, A.; Fournier, J.; Middleton, P.H.; Auriac, M.-C.; Mysore, K.S.; Wen, J.; Erard, M.; Barker, D.G. The symbiosis-related ERN transcription factors act in concert to coordinate rhizobial host root infection. Plant Physiol. 2016, 171, 1037–1054. [Google Scholar] [CrossRef]
- Schiessl, K.; Lilley, J.L.S.; Lee, T.; Tamvakis, I.; Kohlen, W.; Bailey, P.C.; Thomas, A.; Luptak, J.; Ramakrishnan, K.; Carpenter, M.D.; et al. NODULE INCEPTION recruits the lateral root developmental program for symbiotic nodule organogenesis in Medicago truncatula. Curr. Biol. 2019, 29, 3657–3668. [Google Scholar] [CrossRef]
- Ma, L.; Yi, D.; Yang, J.; Liu, X.; Pang, Y. Genome-wide identification, expression analysis and functional study of CCT Gene family in Medicago truncatula. Plants 2020, 9, 513. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xu, M. CCT family genes in cereal crops: A current overview. Crop J. 2017, 5, 449–458. [Google Scholar] [CrossRef]
- Duangkhet, M.; Thepsukhon, A.; Widyastuti, R.; Santosa, D.A.; Tajima, S.; Nomura, M. A MYB-related transcription factor affects nodule formation in Lotus japonicus. Plant Biotechnol. 2016, 33, 187–194. [Google Scholar]
- Stracke, R.; Werber, M.; Weisshaar, B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001, 4, 447–456. [Google Scholar] [CrossRef]
- Fujiwara, S.; Oda, A.; Yoshida, R.; Niinuma, K.; Miyata, K.; Tomozoe, Y.; Tajima, T.; Nakagawa, M.; Hayashi, K.; Coupland, G. Circadian clock proteins LHY and CCA1 regulate SVP protein accumulation to control flowering in Arabidopsis. Plant Cell 2008, 20, 2960–2971. [Google Scholar] [CrossRef]
- Boycheva, I.; Vassileva, V.; Revalska, M.; Zehirov, G.; Iantcheva, A. Cyclin-like F-box protein plays a role in growth and development of the three model species Medicago truncatula, Lotus japonicus, and Arabidopsis thaliana. Res. Rep. Biol. 2015, 6, 117–130. [Google Scholar]
- Shah, N.; Wakabayashi, T.; Kawamura, Y.; Skovbjerg, C.K.; Wang, M.-Z.; Mustamin, Y.; Isomura, Y.; Gupta, V.; Jin, H.; Mun, T. Extreme genetic signatures of local adaptation during Lotus japonicus colonization of Japan. Nat. Commun. 2020, 11, 253. [Google Scholar] [CrossRef]
- Homrich, M.S.; Wiebke-Strohm, B.; Weber, R.L.M.; Bodanese-Zanettini, M.H. Soybean genetic transformation: A valuable tool for the functional study of genes and the production of agronomically improved plants. Genet. Mol. Biol. 2012, 35 (Suppl. 4), 998–1010. [Google Scholar] [CrossRef]
- Christou, P.; McCabe, D.E.; Martinell, B.J.; Swain, W.F. Soybean genetic engineering-commercial production of transgenic plants. Trends Biotechnol. 1990, 8, 145–151. [Google Scholar] [CrossRef]
- Bahrman, N.; Hascoët, E.; Jaminon, O.; Dépta, F.; Hû, J.-F.; Bouchez, O.; Lejeune-Hénaut, I.; Delbreil, B.; Legrand, S. Identification of genes differentially expressed in response to cold in Pisum sativum using RNA sequencing analyses. Plants 2019, 8, 288. [Google Scholar] [CrossRef] [PubMed]
- Mohapatra, C.; Chand, R.; Tiwari, J.K.; Singh, A.K. Effect of heat stress during flowering and pod formation in pea (Pisum sativum L.). Physiol. Mol. Biol. Plants 2020, 26, 1119–1125. [Google Scholar] [CrossRef] [PubMed]
- García-García, M.C.; Celestino, M.R.; Gil-Izquierdo, A.; Egea-Gilabert, C.; Galano, J.M.; Durand, T.; Oger, C.; Fernández, J.A.; Ferreres, F.; Domínguez-Perles. The value of legume foods as a dietary source of phytoprostanes and phytofurans is dependent on species, variety, and growing conditions. Eur. J. Lipid Sci. Technol. 2019, 121. [Google Scholar] [CrossRef]
- Kerr, S.C.; Gaiti, F.; Beveridge, C.A.; Tanurdzic, M. De novo transcriptome assembly reveals high transcriptional complexity in Pisum sativum axillary buds and shows rapid changes in expression of diurnally regulated genes. BMC Genom. 2017, 18, 221. [Google Scholar] [CrossRef]
- Creux, N.; Harmer, S. Circadian rhythms in plants. Cold Spring Harb. Perspect. Biol. 2019, 11. [Google Scholar] [CrossRef]
| Species | Gene | Arabidopsis Homologue | Function(s) | Reference(s) |
|---|---|---|---|---|
| Barrel medic (Medicago truncatula) | MtGI | GI | Circadian clock component, photoperiod response | [47] |
| MtLHY | LHY | Regulation of circadian rhythm in nodules and nyctinastic leaf movement | [48] | |
| Birds-foot trefoil (Lotus japonicus) | LjaPRR5 | PRR5 | Component of the circadian rhythm | [49] |
| LjaPRR7 | PRR7 | |||
| LjaPRR9 | PRR9 | |||
| LjaLUX | LUX | |||
| LjaTOC1 | TOC1 | Central component of the circadian rhythm | ||
| LjaLHY | LHY | |||
| LjCCA1 | CCA1 | Central component of the circadian rhythm | [50] | |
| LjGI | GI | Possible regulation of flowering time | [51] | |
| Soybean (Glycine max) | GmTOC1 | TOC1 | Central component of the soybean circadian clock (expressed as an evening gene) | [52] |
| GmELF4 | ELF4 | Circadian clock function. | [53] | |
| GmGIa | GI | Photoperiod response, flowering time regulation | [54] | |
| GmLCL1 | LHY/CCA1 | Central component of the soybean circadian clock (expressed as a morning gene) | [52] | |
| GmLCL2 | ||||
| GmLHY | LHY | Regulate plant height | [55] | |
| GmLUXa | LUX | Control flowering time | [56] | |
| GmLUXb | ||||
| GmLUXc | ||||
| GmZTL3 | ZTL | Control of flowering time (inhibitor of flowering induction) and photoreceptor. | [57] | |
| GmPRR37 | PRR3 & 7 | Control of soybean photoperiodic flowering | [58] | |
| Common pea (Pisum sativum) | PsTOC1 | TOC1 | Circadian clock component | [17,59] |
| DNE | ELF4 | Circadian clock component, flowering time regulation | [60,61] | |
| HR | ELF3 | Circadian clock component, flowering time regulation, light response | [61] | |
| LATE1 | GI | Photoperiod response | [22,60] | |
| MYB1/LHY | CCA1/LHY | Circadian clock component | [60] | |
| SN | LUX | Circadian clock component | [62] | |
| PsPRR37 | PRR | Component of phospho-relay signal transduction system | [59] | |
| PsPRR59 | ||||
| Chickpea (Cicer arietinum) | Efl1 | ELF3 | Flowering regulation (light input to the circadian clock) | [63] |
| GI | GI | Flowering time regulation | [64] | |
| Common bean (Phaseolus vulgaris) | PvLHY | LHY | Circadian mechanism regulation | [65,66] |
| PvTOC1 | TOC1 | Mediating light responsiveness to circadian clock mechanism | [67,68] | |
| PvELF4 | ELF4 | Evening-expressed gene | [67] | |
| PvGI | GI | Circadian clock component | [69] | |
| PvZTL | ZTL | |||
| Pigeon pea (Cajanus cajan) | CcGI | GI | Determinacy and flower patterning | [69] |
| Cowpea (Vigna unguiculata) | VunTOC1 | TOC1 | Circadian clock function in seed filling and leaves | [70] |
| VunLHY | LHY | |||
| VunELF3 | ELF3 | |||
| VunGI | GI | |||
| Lentil (Lens culinaris) | HR | ELF3 | Flowering time regulation | [61] |
| White lupin (Lupinis albus) | GI | GI | Flowering regulation; anthracnose resistance | [64] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kugan, H.M.; Rejab, N.A.; Sahruzaini, N.A.; Harikrishna, J.A.; Baisakh, N.; Cheng, A. Circadian Rhythms in Legumes: What Do We Know and What Else Should We Explore? Int. J. Mol. Sci. 2021, 22, 4588. https://doi.org/10.3390/ijms22094588
Kugan HM, Rejab NA, Sahruzaini NA, Harikrishna JA, Baisakh N, Cheng A. Circadian Rhythms in Legumes: What Do We Know and What Else Should We Explore? International Journal of Molecular Sciences. 2021; 22(9):4588. https://doi.org/10.3390/ijms22094588
Chicago/Turabian StyleKugan, Hazel Marie, Nur Ardiyana Rejab, Nurul Amylia Sahruzaini, Jennifer Ann Harikrishna, Niranjan Baisakh, and Acga Cheng. 2021. "Circadian Rhythms in Legumes: What Do We Know and What Else Should We Explore?" International Journal of Molecular Sciences 22, no. 9: 4588. https://doi.org/10.3390/ijms22094588
APA StyleKugan, H. M., Rejab, N. A., Sahruzaini, N. A., Harikrishna, J. A., Baisakh, N., & Cheng, A. (2021). Circadian Rhythms in Legumes: What Do We Know and What Else Should We Explore? International Journal of Molecular Sciences, 22(9), 4588. https://doi.org/10.3390/ijms22094588







