Inflammasomes in Teleosts: Structures and Mechanisms That Induce Pyroptosis during Bacterial Infection
Abstract
:1. Introduction
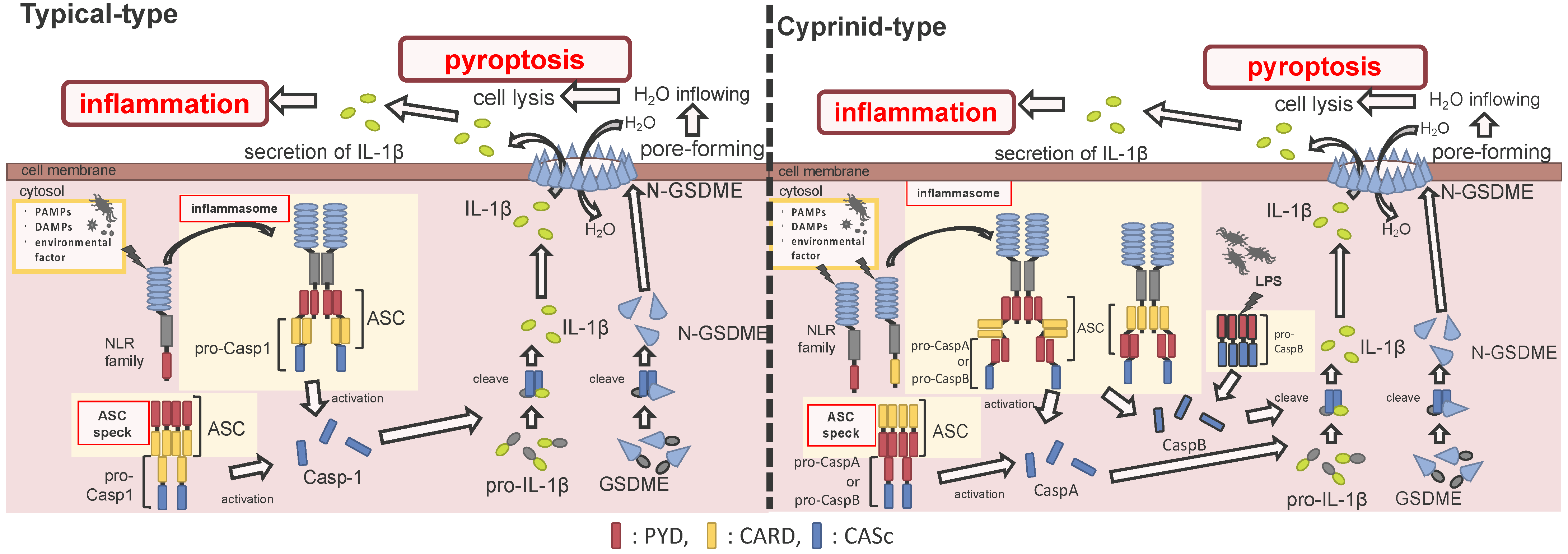
2. Inflammasome Activation to Induce Pyroptosis
2.1. Generic Structure of Inflammasome and Mechanisms of Pyroptosis
2.2. The Composition for Inflammasome and Apoptosis-Associated Speck-Like Protein Containing a Caspase Recruitment Domain Formation
2.2.1. Inflammasome Formation
2.2.2. Apoptosis-Associated Speck-Like Protein Containing a Caspase Recruitment Domain Formation
2.3. The Mechanism of Pyroptosis via Caspase 1
2.3.1. Induction of Pyroptosis through Caspase 1
2.3.2. Induction of Pyroptosis in Fish
2.3.3. Induction of Pyroptosis via Non-Canonical Inflammasome Activation in Cyprinids
3. Inflammasome-Related Gene Expression and Its Activation by Stimuli
4. Inflammasome Activation during Pathogenic Infection
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kawai, T.; Akira, S. The roles of TLRs, RLRs and NLRs in pathogen recognition. Int. Immunol. 2009, 21, 317–337. [Google Scholar] [CrossRef] [Green Version]
- Takeuchi, O.; Akira, S. Pattern recognition receptors and inflammation. Cell 2010, 140, 805–820. [Google Scholar] [CrossRef] [Green Version]
- Franchi, L.; Eigenbrod, T.; Núñez, G. Cutting Edge: TNF-α mediates sensitization to ATP and silica via the NLRP3 inflammasome in the absence of microbial stimulation. J. Immunol. 2009, 183, 792–796. [Google Scholar] [CrossRef]
- Franchi, L.; Núñez, G. The Nlrp3 inflammasome is critical for aluminium hydroxide-mediated IL-1beta secretion but dispensable for adjuvant activity. Eur. J. Immunol. 2008, 38, 2085–2089. [Google Scholar] [CrossRef]
- Brubaker, S.W.; Bonham, K.S.; Zanoni, I.; Kagan, J.C. Innate immune pattern recognition: A cell biological perspective. Annu. Rev. Immunol. 2015, 33, 257–290. [Google Scholar] [CrossRef] [Green Version]
- Sahoo, B.R. Structure of fish toll-like receptors (TLR) and NOD-like receptors (NLR). Int. J. Biol. Macromol. 2020, 161, 1602–1617. [Google Scholar] [CrossRef] [PubMed]
- Becker, C.E.; O’Neill, L.A.J. Inflammasomes in inflammatory disorders: The role of TLRs and their interactions with NLRs. Semin. Immunopathol. 2007, 29, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Davis, B.K.; Wen, H.; Ting, J.P.-Y. The inflammasome NLRs in immunity, inflammation, and associated diseases. Annu. Rev. Immunol. 2011, 29, 707–835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Netea, M.G.; Van de Veerdonk, F.L.; Kullberg, B.J.; Van der Meer, J.W.M.; Joosten, L.A.B. The role of NLRs and TLRs in the activation of the inflammasome. Expert Opin. Biol. Ther. 2008, 8, 1867–1872. [Google Scholar] [CrossRef] [PubMed]
- Bergsbaken, T.; Fink, S.L.; Cookson, B.T. Pyroptosis: Host cell death and inflammation. Nat. Rev. Microbiol. 2009, 7, 99–109. [Google Scholar] [CrossRef] [Green Version]
- Vande Walle, L.; Lamkanfi, M. Pyroptosis. Curr. Biol. 2016, 26, R568–R572. [Google Scholar] [CrossRef] [Green Version]
- Fink, S.L.; Cookson, B.T. Apoptosis, pyroptosis, and necrosis: Mechanistic description of dead and dying eukaryotic cells. Infect. Immun. 2005, 73, 1907–1916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franchi, L.; Eigenbrod, T.; Muñoz-Planillo, R.; Nuñez, G. The inflammasome: A caspase-1-activation platform that regulates immune responses and disease pathogenesis. Nat. Immunol. 2009, 10, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, M.; Ceballos-Olvera, I.; Del Barrio, L.; Re, F. Role of the inflammasome, IL-1β, and IL-18 in bacterial infections. Sci. World J. 2011, 11, 2037–2050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Netea, M.G.; Nold-Petry, C.A.; Nold, M.F.; Joosten, L.A.B.; Opitz, B.; Van Der Meer, J.H.M.; Van De Veerdonk, F.L.; Ferwerda, G.; Heinhuis, B.; Devesa, I.; et al. Differential requirement for the activation of the inflammasome for processing and release of IL-1β in monocytes and macrophages. Blood 2009, 113, 2324–2335. [Google Scholar] [CrossRef] [Green Version]
- Mariathasan, S.; Monack, D.M. Inflammasome adaptors and sensors: Intracellular regulators of infection and inflammation. Nat. Rev. Immunol. 2007, 7, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Lu, A.; Wu, H. Structural mechanisms of inflammasome assembly. FEBS J. 2015, 282, 435–444. [Google Scholar] [CrossRef]
- Guo, H.; Callaway, J.B.; Ting, J.P. Inflammasomes: Mechanism of action, role in disease, and therapeutics. Nat. Med. 2015, 21, 677–687. [Google Scholar] [CrossRef] [Green Version]
- Ta, A.; Vanaja, S.K. Inflammasome activation and evasion by bacterial pathogens. Curr. Opin. Immunol. 2021, 68, 125–133. [Google Scholar] [CrossRef]
- Miller, L.S.; Pietras, E.M.; Uricchio, L.H.; Hirano, K.; Rao, S.; Lin, H.; O’Connell, R.M.; Iwakura, Y.; Cheung, A.L.; Cheng, G.; et al. Inflammasome-mediated production of IL-1beta is required for neutrophil recruitment against Staphylococcus aureus in vivo. J. Immunol. 2007, 179, 6933–6942. [Google Scholar] [CrossRef] [Green Version]
- Varela, M.; Romero, A.; Dios, S.; van der Vaart, M.; Figueras, A.; Meijer, A.H.; Novoa, B. Cellular visualization of macrophage pyroptosis and interleukin-1β release in a viral hemorrhagic infection in zebrafish larvae. J. Virol. 2014, 88, 12026–12040. [Google Scholar] [CrossRef] [Green Version]
- Shi, J.; Zhao, Y.; Wang, K.; Shi, X.; Wang, Y.; Huang, H.; Zhuang, Y.; Cai, T.; Wang, F.; Shao, F. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 2015, 526, 660–665. [Google Scholar] [CrossRef]
- Lopez-Castejon, G.; Brough, D. Understanding the mechanism of IL-1β secretion. Cytokine Growth Factor Rev. 2011, 22, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Ceballos-Olvera, I.; Sahoo, M.; Miller, M.A.; del Barrio, L.; Re, F. Inflammasome-dependent pyroptosis and IL-18 protect against Burkholderia pseudomallei lung infection while IL-1β is deleterious. PLoS Pathog. 2011, 7. [Google Scholar] [CrossRef] [PubMed]
- Willingham, S.B.; Allen, I.C.; Bergstralh, D.T.; Brickey, W.J.; Huang, M.T.H.; Taxman, D.J.; Duncan, J.A.; Ting, J.P.Y. NLRP3 (NALP3, cryopyrin) facilitates in vivo caspase-1 activation, necrosis, and HMGB1 release via inflammasome-dependent and -independent pathways. J. Immunol. 2009, 183, 2008–2015. [Google Scholar] [CrossRef]
- Vince, J.E.; Silke, J. The intersection of cell death and inflammasome activation. Cell. Mol. Life Sci. 2016, 73, 2349–2367. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Y.; Wang, Y.Y.; Shao, T.; Fan, D.D.; Lin, A.F.; Xiang, L.X.; Shao, J.Z. The zebrafish NLRP3 inflammasome has functional roles in ASC-dependent interleukin-1β maturation and gasdermin E-mediated pyroptosis. J. Biol. Chem. 2020, 295, 1120–1141. [Google Scholar] [CrossRef]
- Jiang, S.; Gu, H.; Zhao, Y.; Sun, L. Teleost gasdermin E is cleaved by caspase 1, 3, and 7 and induces pyroptosis. J. Immunol. 2019, 203, 1369–1382. [Google Scholar] [CrossRef]
- Wang, Z.; Gu, Z.; Hou, Q.; Chen, W.; Mu, D.; Zhang, Y.; Liu, Q.; Liu, Z.; Yang, D. Zebrafish GSDMEb cleavage-gated pyroptosis drives septic acute kidney injury in vivo. J. Immunol. 2020, 204, 1929–1942. [Google Scholar] [CrossRef]
- Masumoto, J.; Zhou, W.; Chen, F.F.; Su, F.; Kuwada, J.Y.; Hidaka, E.; Katsuyama, T.; Sagara, J.; Taniguchi, S.; Ngo-Hazelett, P.; et al. Caspy, a zebrafish caspase, activated by ASC oligomerization is required for pharyngeal arch development. J. Biol. Chem. 2003, 278, 4268–4276. [Google Scholar] [CrossRef] [Green Version]
- Chavarría-Smith, J.; Vance, R.E. The NLRP1 inflammasomes. Immunol. Rev. 2015, 265, 22–34. [Google Scholar] [CrossRef]
- Schroder, K.; Tschopp, J. The Inflammasomes. Cell 2010, 140, 821–832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cassel, S.L.; Joly, S.; Sutterwala, F.S. The NLRP3 inflammasome: A sensor of immune danger signals. Semin. Immunol. 2009, 21, 194–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tuladhar, S.; Kanneganti, T.D. NLRP12 in innate immunity and inflammation. Mol. Asp. Med. 2020, 76, 100887. [Google Scholar] [CrossRef]
- Li, J.; Gao, K.; Shao, T.; Fan, D.; Hu, C.; Sun, C.; Dong, W.; Lin, A.; Xiang, L.; Shao, J. Characterization of an NLRP1 inflammasome from zebrafish reveals a unique sequential activation mechanism underlying inflammatory caspases in ancient vertebrates. J. Immunol. 2018, 201, 1946–1966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Ding, S.; Tan, J.; Yang, D.; Zhang, Y.; Liu, Q. Characterization of the Japanese flounder NLRP3 inflammasome in restricting Edwardsiella piscicida colonization in vivo. Fish Shellfish Immunol. 2020, 103, 169–180. [Google Scholar] [CrossRef]
- Chang, M.X.; Xiong, F.; Wu, X.M.; Hu, Y.W. The expanding and function of NLRC3 or NLRC3-like in teleost fish: Recent advances and novel insights. Dev. Comp. Immunol. 2021, 114, 103859. [Google Scholar] [CrossRef]
- Schneider, M.; Zimmermann, A.G.; Roberts, R.A.; Zhang, L.; Swanson, K.V.; Wen, H.; Davis, B.K.; Allen, I.C.; Holl, E.K.; Ye, Z.; et al. The innate immune sensor NLRC3 attenuates toll-like receptor signaling via modification of the signaling adaptor TRAF6 and transcription factor NF-κB. Nat. Immunol. 2012, 13, 823–831. [Google Scholar] [CrossRef]
- Li, S.; Chen, X.; Hao, G.; Geng, X.; Zhan, W.; Sun, J. Identification and characterization of a novel NOD-like receptor family CARD domain containing 3 gene in response to extracellular ATP stimulation and its role in regulating LPS-induced innate immune response in Japanese flounder (Paralichthys olivaceus). Fish Shellfish Immunol. 2016, 50, 79–90. [Google Scholar] [CrossRef]
- Gao, F.-Y.; Pang, J.-C.; Lu, M.-X.; Yang, X.-L.; Zhu, H.-P.; Ke, X.-L.; Liu, Z.-G.; Cao, J.-M.; Wang, M. Molecular characterization, expression and functional analysis of NOD1, NOD2 and NLRC3 in Nile tilapia (Oreochromis niloticus). Fish Shellfish Immunol. 2018, 73, 207–219. [Google Scholar] [CrossRef]
- Li, Y.; Huang, Y.; Cao, X.; Yin, X.; Jin, X.; Liu, S.; Jiang, J.; Jiang, W.; Xiao, T.S.; Zhou, R.; et al. Functional and structural characterization of zebrafish ASC. FEBS J. 2018, 285, 2691–2707. [Google Scholar] [CrossRef]
- Morimoto, N.; Okamura, Y.; Kono, T.; Sakai, M.; Hikima, J. Characterization and expression analysis of tandemly-replicated asc genes in the Japanese medaka, Oryzias latipes. Dev. Comp. Immunol. 2020, 115, 103894. [Google Scholar] [CrossRef]
- Xie, J.; Belosevic, M. Functional characterization of apoptosis-associated speck-like protein (ASC) of the goldfish (Carassius auratus L.). Dev. Comp. Immunol. 2016, 65, 201–210. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, Z.; Li, C.; Zhang, Y.; Wang, L.; Wei, J.; Qin, Q. Characterization of orange-spotted grouper (Epinephelus coioides) ASC and caspase-1 involved in extracellular ATP-mediated immune signaling in fish. Fish Shellfish Immunol. 2020, 97, 58–71. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, J.; Lao, H.; Yin, Z.; He, W.; Weng, S.; Yu, X.; Chan, S.M.; He, J. Molecular cloning and expression analysis of the ASC gene from mandarin fish and its regulation of NF-κB activation. Dev. Comp. Immunol. 2008, 32, 391–399. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Chen, X.; Peng, W.; Hao, G.; Geng, X.; Zhan, W.; Sun, J. Cloning and characterization of apoptosis-associated speck-like protein containing a CARD domain (ASC) gene from Japanese flounder Paralichthys olivaceus. Fish Shellfish Immunol. 2016, 54, 294–301. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Tan, J.; Wang, Z.; Zhang, Y.; Liu, Q.; Yang, D. Characterization of the inflammasome component SmASC in turbot (Scophthalmus maximus). Fish Shellfish Immunol. 2020, 100, 324–333. [Google Scholar] [CrossRef] [PubMed]
- Moriya, M.; Taniguchi, S.; Wu, P.; Liepinsh, E.; Otting, G.; Sagara, J. Role of charged and hydrophobic residues in the oligomerization of the pyrin domain of ASC. Biochemistry 2005, 44, 575–583. [Google Scholar] [CrossRef]
- Kersse, K.; Lamkanfi, M.; Bertrand, M.J.M.; Berghe, T.V.; Vandenabeele, P. Interaction patches of procaspase-1 caspase recruitment domains (CARDs) are differently involved in procaspase-1 activation and receptor-interacting protein 2 (RIP2)-dependent nuclear factor κB signaling. J. Biol. Chem. 2011, 286, 35874–35882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srinivasula, S.M.; Poyet, J.L.; Razmara, M.; Datta, P.; Zhang, Z.; Alnemri, E.S. The pyrin-CARD protein ASC is an activating adaptor for caspase-1. J. Biol. Chem. 2002, 277, 21119–21122. [Google Scholar] [CrossRef] [Green Version]
- Hoss, F.; Rodriguez-Alcazar, J.F.; Latz, E. Assembly and regulation of ASC specks. Cell. Mol. Life Sci. 2017, 74, 1211–1229. [Google Scholar] [CrossRef] [PubMed]
- Hara, H.; Tsuchiya, K.; Kawamura, I.; Fang, R.; Hernandez-Cuellar, E.; Shen, Y.; Mizuguchi, J.; Schweighoffer, E.; Tybulewicz, V.; Mitsuyama, M. Phosphorylation of the adaptor ASC acts as a molecular switch that controls the formation of speck-like aggregates and inflammasome activity. Nat. Immunol. 2013, 14, 1247–1255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Alba, E. Structure, interactions and self-assembly of ASC-dependent inflammasomes. Arch. Biochem. Biophys. 2019, 670, 15–31. [Google Scholar] [CrossRef]
- Broz, P.; Dixit, V.M. Inflammasomes: Mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 2016, 16, 407–420. [Google Scholar] [CrossRef]
- Kuri, P.; Schieber, N.L.; Thumberger, T.; Wittbrodt, J.; Schwab, Y.; Leptin, M. Dynamics of ASC speck formation during skin inflammatory responses in vivo. bioRxiv 2017, 216. [Google Scholar] [CrossRef] [Green Version]
- Broz, P.; von Moltke, J.; Jones, J.W.; Vance, R.E.; Monack, D.M. Differential requirement for caspase-1 autoproteolysis in pathogen-induced cell death and cytokine processing. Cell Host Microbe 2010, 8, 471–483. [Google Scholar] [CrossRef] [Green Version]
- Keller, M.; Rüegg, A.; Werner, S.; Beer, H.D. Active caspase-1 is a regulator of unconventional protein secretion. Cell 2008, 132, 818–831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reis, M.I.R.; do Vale, A.; Pereira, P.J.B.; Azevedo, J.E.; dos Santos, N.M.S. Caspase-1 and IL-1β processing in a teleost fish. PLoS ONE 2012, 7, e50450. [Google Scholar] [CrossRef] [Green Version]
- Rojas, V.; Camus-Guerra, H.; Guzmán, F.; Mercado, L. Pro-inflammatory caspase-1 activation during the immune response in cells from rainbow trout Oncorhynchus mykiss (Walbaum 1792) challenged with pathogen-associated molecular patterns. J. Fish Dis. 2015, 38, 993–1003. [Google Scholar] [CrossRef]
- Tang, L.; Lu, C.; Zheng, G.; Burgering, B.M.T. Emerging insights on the role of gasdermins in infection and inflammatory diseases. Clin. Transl. Immunol. 2020, 9, e1186. [Google Scholar] [CrossRef]
- Winsor, N.; Krustev, C.; Bruce, J.; Philpott, D.J.; Girardin, S.E. Canonical and non-canonical inflammasomes in intestinal epithelial cells. Cell. Microbiol. 2019, 21, e13079. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Zheng, X.; Chen, S.; Wang, Z.; Xu, W.; Tan, J.; Hu, T.; Hou, M.; Wang, W.; Gu, Z.; et al. Sensing of cytosolic LPS through caspy2 pyrin domain mediates non-canonical inflammasome activation in zebrafish. Nat. Commun. 2018, 9, 3052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, N.; Liu, Z.S.; Xue, W.; Bai, Z.F.; Wang, Q.Y.; Dai, J.; Liu, X.; Huang, Y.J.; Cai, H.; Zhan, X.Y.; et al. NLRP3 phosphorylation is an essential priming event for inflammasome activation. Mol. Cell 2017, 68, 185–197.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Unajak, S.; Santos, M.D.; Hikima, J.; Jung, T.S.S.; Kondo, H.; Hirono, I.; Aoki, T. Molecular characterization, expression and functional analysis of a nuclear oligomerization domain proteins subfamily C (NLRC) in Japanese flounder (Paralichthys olivaceus). Fish Shellfish Immunol. 2011, 31, 202–211. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Peng, W.; Li, J.; Hao, G.; Geng, X.; Sun, J. Characterization of Japanese flounder (Paralichthys olivaceus) caspase1 involved in extracellular ATP-mediated immune signaling in fish. Fish Shellfish Immunol. 2017, 67, 536–545. [Google Scholar] [CrossRef]
- Álvarez, C.A.; Ramírez-Cepeda, F.; Santana, P.; Torres, E.; Cortés, J.; Guzmán, F.; Schmitt, P.; Mercado, L. Insights into the diversity of NOD-like receptors: Identification and expression analysis of NLRC3, NLRC5 and NLRX1 in rainbow trout. Mol. Immunol. 2017, 87, 102–113. [Google Scholar] [CrossRef]
- Paria, A.; Deepika, A.; Sreedharan, K.; Makesh, M.; Chaudhari, A.; Purushothaman, C.S.; Thirunavukkarasu, A.R.; Rajendran, K.V. Identification of Nod like receptor C3 (NLRC3) in Asian seabass, Lates calcarifer: Characterisation, ontogeny and expression analysis after experimental infection and ligand stimulation. Fish Shellfish Immunol. 2016, 55, 602–612. [Google Scholar] [CrossRef]
- Biswas, G.; Bilen, S.; Kono, T.; Sakai, M.; Hikima, J. Inflammatory immune response by lipopolysaccharide-responsive nucleotide binding oligomerization domain (NOD)-like receptors in the Japanese pufferfish (Takifugu rubripes). Dev. Comp. Immunol. 2016, 55, 21–31. [Google Scholar] [CrossRef]
- Meijer, A.H.; Gabby Krens, S.F.; Medina Rodriguez, I.A.; He, S.; Bitter, W.; Snaar-Jagalska, B.E.; Spaink, H.P. Expression analysis of the toll-like receptor and TIR domain adaptor families of zebrafish. Mol. Immunol. 2004, 40, 773–783. [Google Scholar] [CrossRef]
- Sepulcre, M.P.; Alcaraz-Pérez, F.; López-Muñoz, A.; Roca, F.J.; Meseguer, J.; Cayuela, M.L.; Mulero, V. Evolution of lipopolysaccharide (LPS) recognition and signaling: Fish TLR4 does not recognize LPS and negatively regulates NF-kappaB activation. J. Immunol. 2009, 182, 1836–1845. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.; Marín-Juez, R.; Meijer, A.H.; Spaink, H.P. Common and specific downstream signaling targets controlled by Tlr2 and Tlr5 innate immune signaling in zebrafish. BMC Genom. 2015, 16, 547. [Google Scholar] [CrossRef] [Green Version]
- López-Castejón, G.; Sepulcre, M.P.; Mulero, I.; Pelegrín, P.; Meseguer, J.; Mulero, V. Molecular and functional characterization of gilthead seabream Sparus aurata caspase-1: The first identification of an inflammatory caspase in fish. Mol. Immunol. 2008, 45, 49–57. [Google Scholar] [CrossRef]
- Li, T.; Shan, S.; Wang, L.; Yang, G.; Zhu, J. Identification of a fish-specific NOD-like receptor subfamily C (NLRC) gene from common carp (Cyprinus carpio L.): Characterization, ontogeny and expression analysis in response to immune stimulation. Fish Shellfish Immunol. 2018, 82, 371–377. [Google Scholar] [CrossRef] [PubMed]
- Morimoto, T.; Biswas, G.; Kono, T.; Sakai, M.; Hikima, J. Immune responses in the Japanese pufferfish (Takifugu rubripes) head kidney cells stimulated with particulate silica. Fish Shellfish Immunol. 2016, 49, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, Q.; Yin, H.; Li, S. Cadmium exposure induces pyroptosis of lymphocytes in carp pronephros and spleens by activating NLRP3. Ecotoxicol. Environ. Saf. 2020, 202, 110903. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Planillo, R.; Kuffa, P.; Martínez-Colón, G.; Smith, B.L.; Rajendiran, T.M.; Núñez, G. K+ efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate Matter. Immunity 2013, 38, 1142–1153. [Google Scholar] [CrossRef] [Green Version]
- Franchi, L.; Kanneganti, T.D.; Dubyak, G.R.; Núñez, G. Differential requirement of P2X7 receptor and intracellular K+ for caspase-1 activation induced by intracellular and extracellular bacteria. J. Biol. Chem. 2007, 282, 18810–18818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hewinson, J.; Moore, S.F.; Glover, C.; Watts, A.G.; MacKenzie, A.B. A key role for redox signaling in rapid P2X 7 receptor-induced IL-1 beta processing in human monocytes. J. Immunol. 2008, 180, 8410–8420. [Google Scholar] [CrossRef] [Green Version]
- Lister, M.F.; Sharkey, J.; Sawatzky, D.A.; Hodgkiss, J.P.; Davidson, D.J.; Rossi, A.G.; Finlayson, K. The role of the purinergic P2X7 receptor in inflammation. J. Inflamm. 2007, 4, 5. [Google Scholar] [CrossRef] [Green Version]
- Pelegrin, P.; Barroso-Gutierrez, C.; Surprenant, A. P2X 7 receptor differentially couples to distinct release pathways for IL-1β in mouse macrophage. J. Immunol. 2008, 180, 7147–7157. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Li, X.; Coddou, C.; Geng, X.; Wei, J.; Sun, J. Molecular characterization and expression analysis of ATP-gated P2X7 receptor involved in Japanese flounder (Paralichthys olivaceus) innate immune response. PLoS ONE 2014, 9, e96625. [Google Scholar] [CrossRef] [PubMed]
- Angosto, D.; López-Castejó, G.; López-Muñ, A.; Sepulcre, M.P.; Arizcun, M.; Meseguer, J.; Mulero, V. Evolution of inflammasome functions in vertebrates: Inflammasome and caspase-1 trigger fish macrophage cell death but are dispensable for the processing of IL-1β. Innate Immun. 2012, 18, 815–824. [Google Scholar] [CrossRef] [PubMed]
- Cerezuela, R.; Meseguer, J.; Esteban, M.Á. Effects of dietary inulin, Bacillus subtilis and microalgae on intestinal gene expression in gilthead seabream (Sparus aurata L.). Fish Shellfish Immunol. 2013, 34, 843–848. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Yang, D.; Wang, Z.; Zheng, X.; Zhang, Y.; Liu, Q. EvpP inhibits neutrophils recruitment via Jnk-caspy inflammasome signaling in vivo. Fish Shellfish Immunol. 2019, 92, 851–860. [Google Scholar] [CrossRef] [PubMed]
- Vojtech, L.N.; Scharping, N.; Woodson, J.C.; Hansen, J.D. Roles of inflammatory caspases during processing of zebrafish interleukin-1β in Francisella noatunensis infection. Infect. Immun. 2012, 80, 2878–2885. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Kong, L.; Gao, Y.; Wu, C.; Xu, T. Characterization of NLR-A subfamily members in miiuy croaker and comparative genomics revealed NLRX1 underwent duplication and lose in actinopterygii. Fish Shellfish Immunol. 2015, 47, 397–406. [Google Scholar] [CrossRef]
- Li, J.; Chu, Q.; Xu, T. A genome-wide survey of expansive NLR-C subfamily in miiuy croaker and characterization of the NLR-B30.2 genes. Dev. Comp. Immunol. 2016, 61, 116–125. [Google Scholar] [CrossRef]
- Kumaresan, V.; Ravichandran, G.; Nizam, F.; Dhayanithi, N.B.; Arasu, M.V.; Al-Dhabi, N.A.; Harikrishnan, R.; Arockiaraj, J. Multifunctional murrel caspase 1, 2, 3, 8 and 9: Conservation, uniqueness and their pathogen-induced expression pattern. Fish Shellfish Immunol. 2016, 49, 493–504. [Google Scholar] [CrossRef]
- Davis, J.M.; Clay, H.; Lewis, J.L.; Ghori, N.; Herbomel, P.; Ramakrishnan, L.; Masumoto, J.; Zhou, W.; Chen, F.F.; Su, F.; et al. Molecular characterization reveals involvement of four caspases in the antibacterial immunity of tongue sole (Cynoglossus semilaevis). Fish Shellfish Immunol. 2016, 7, 116–125. [Google Scholar] [CrossRef]
- Terra, J.K.; Cote, C.K.; France, B.; Jenkins, A.L.; Bozue, J.A.; Welkos, S.L.; LeVine, S.M.; Bradley, K.A. Cutting Edge: Resistance to Bacillus anthracis infection mediated by a lethal toxin sensitive allele of Nalp1b/Nlrp1b. J. Immunol. 2010, 184, 17–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kebaier, C.; Chamberland, R.R.; Allen, I.C.; Gao, X.; Broglie, P.M.; Hall, J.D.; Jania, C.; Doerschuk, C.M.; Tilley, S.L.; Duncan, J.A. Staphylococcus aureus α-hemolysin mediates virulence in a murine model of severe pneumonia through activation of the NLRP3 inflammasome. J. Infect. Dis. 2012, 205, 807–817. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Zhao, S.; Gao, X.; Wang, R.; Liu, J.; Zhou, X.; Zhou, Y. The roles of inflammasomes in host defense against Mycobacterium tuberculosis. Pathogens 2021, 10, 120. [Google Scholar] [CrossRef] [PubMed]
- McElvania Tekippe, E.; Allen, I.C.; Hulseberg, P.D.; Sullivan, J.T.; McCann, J.R.; Sandor, M.; Braunstein, M.; Ting, J.P.Y. Granuloma formation and host defense in chronic Mycobacterium tuberculosis infection requires PYCARD/ASC but not NLRP3 or caspase-1. PLoS ONE 2010, 5, e12320. [Google Scholar] [CrossRef] [Green Version]
- Kleinnijenhuis, J.; Joosten, L.A.B.; van de Veerdonk, F.L.; Savage, N.; van Crevel, R.; Kullberg, B.J.; van der Ven, A.; Ottenhfoff, T.H.M.; Dinarello, C.A.C.A.; van der Meer, J.W.M.; et al. Transcriptional and inflammasome-mediated pathways for the induction of IL-1β production by Mycobacterium tuberculosis. Eur. J. Immunol. 2009, 39, 1914–1922. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.X.; Lu, J.F.; Rolhion, N.; Holden, D.W.; Nie, P.; Zhou, Y.; Yu, X.J. Edwardsiella tarda-induced cytotoxicity depends on its type III secretion system and flagellin. Infect. Immun. 2014, 82, 3436–3445. [Google Scholar] [CrossRef] [Green Version]
- Xu, W.; Gu, Z.; Zhang, L.; Zhang, Y.; Liu, Q.; Yang, D. Edwardsiella piscicida virulence effector trxlp promotes the NLRC4 inflammasome activation during infection. Microb. Pathog. 2018, 123, 496–504. [Google Scholar] [CrossRef]
- Chen, H.; Yang, D.; Han, F.; Tan, J.; Zhang, L.; Xiao, J.; Zhang, Y.; Liu, Q. The bacterial T6SS effector EvpP prevents NLRP3 inflammasome activation by inhibiting the Ca2+-dependent MAPK-Jnk pathway. Cell Host Microbe 2017, 21, 47–58. [Google Scholar] [CrossRef] [Green Version]
- Kaphingst, K.A.; Persky, S.; Lachance, C. Listeria monocytogenes is sensed by NLRP3 and AIM2 inflammasome. Eur. J. Immunol. 2010, 40, 1545–1551. [Google Scholar] [CrossRef] [Green Version]
- Vladimer, G.I.; Marty-Rolx, R.; Ghosh, S.; Wang, D.; Lien, E. Inflammasomes and host defenses against bacterial infections. Curr. Opin. Microbiol. 2013, 16, 23–31. [Google Scholar] [CrossRef] [Green Version]
- Barlan, A.U.; Danthi, P.; Wiethoff, C.M. Lysosomal localization and mechanism of membrane penetration influence nonenveloped virus activation of the NLRP3 inflammasome. Virology 2011, 412, 306–314. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, T.S.; de Sá, K.S.G.; Ishimoto, A.Y.; Becerra, A.; Oliveira, S.; Almeida, L.; Gonçalves, A.V.; Perucello, D.B.; Andrade, W.A.; Castro, R.; et al. Inflammasomes are activated in response to SARS-CoV-2 infection and are associated with COVID-19 severity in patients. J. Exp. Med. 2020, 218, e20201707. [Google Scholar] [CrossRef]
- Lupfer, C.; Kanneganti, T.-D. The expanding role of NLRs in antiviral immunity. Immunol. Rev. 2013, 255, 13–24. [Google Scholar] [CrossRef] [Green Version]
- Lowes, D.J.; Hevener, K.E.; Peters, B.M. Second-generation antidiabetic sulfonylureas inhibit Candida albicans and candidalysin-mediated activation of the NLRP3 inflammasome. Antimicrob. Agents Chemother. 2020, 64, e01777-19. [Google Scholar] [CrossRef]
- De Carvalho, R.V.H.; Silva, A.L.N.; Santos, L.L.; Andrade, W.A.; de Sá, K.S.G.; Zamboni, D.S. Macrophage priming is dispensable for NLRP3 inflammasome activation and restriction of Leishmania amazonensis replication. J. Leukoc. Biol. 2019, 106, 631–640. [Google Scholar] [CrossRef] [PubMed]
- Mohanty, B.R.; Sahoo, P.K. Edwardsiellosis in fish: A brief review. J. Biosci. 2007, 32, 1331–1344. [Google Scholar] [CrossRef]
- Hu, T.; Chen, R.; Zhang, L.; Wang, Z.; Yang, D.; Zhang, Y.; Liu, X.; Liu, Q. Balanced role of T3SS and T6SS in contribution to the full virulence of Edwardsiella piscicida. Fish Shellfish Immunol. 2019, 93, 871–878. [Google Scholar] [CrossRef]
- McCoy, A.J.; Koizumi, Y.; Higa, N.; Suzuki, T. Differential regulation of caspase-1 activation via NLRP3/NLRC4 inflammasomes mediated by aerolysin and type III secretion system during Aeromonas veronii infection. J. Immunol. 2010, 185, 7077–7084. [Google Scholar] [CrossRef] [Green Version]
- McCoy, A.J.; Koizumi, Y.; Toma, C.; Higa, N.; Dixit, V.; Taniguchi, S.; Tschopp, J.; Suzuki, T. Cytotoxins of the human pathogen Aeromonas hydrophila trigger, via the NLRP3 inflammasome, caspase-1 activation in macrophages. Eur. J. Immunol. 2010, 40, 2797–2803. [Google Scholar] [CrossRef] [PubMed]
- Morimoto, N.; Okamura, Y.; Maekawa, S.; Wang, H.; Aoki, T.; Kono, T.; Sakai, M.; Hikima, J. ASC-deficiency impairs host defense against Aeromonas hydrophila infection in Japanese medaka, Oryzias latipes. Fish Shellfish Immunol. 2020, 105, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Sha, Z.; Abernathy, J.W.; Wang, S.; Li, P.; Kucuktas, H.; Liu, H.; Peatman, E.; Liu, Z. NOD-like subfamily of the nucleotide-binding domain and leucine-rich repeat containing family receptors and their expression in channel catfish. Dev. Comp. Immunol. 2009, 33, 991–999. [Google Scholar] [CrossRef]

| Species | Formation Type | Gene Name | Ensembl Gene ID | Domain Structure | |
|---|---|---|---|---|---|
| Mammal | Human (Homo sapiens) | Typical-type | CASP1 | ENSG00000137752 |  |
| Mouse (Mus musculus) | Typical-type | Casp1 | ENSMUSG00000025888 |  | |
| Bird | Chicken (Gallus gallus) | Typical-type | CASP1 | ENSGALG00000001049 |  |
| Reptile | Common wall lizard (Podarcis muralis) | Typical-type | CASP1 | ENSPMRG00000020733 |  |
| Amphibian | Tropical clawed frog (Xenopus tropicalis) | Typical-type | casp1 | ENSXETG00000007792 |  |
| Fish | Japanese medaka (Oryzias latipes) | Typical-type | casp1 | ENSORLG00000006320 |  |
| Japanese pufferfish (Takifugu rubripes) | Typical-type | casp1 | ENSTRUG00000007971 |  | |
| Gilthead seabream (Sparus aurata) | Typical-type | casp1 | ENSSAUG00010008488 |  | |
| Turbot (Scophtalmus maximus) | Typical-type | casp1 | ENSSMAG00000013017 |  | |
| Zebrafish (Danio rerio) | Cyprinid-type | caspa | ENSDARG00000008165 |  | |
| caspb | ENSDARG00000052039 |  | |||
| caspbl | ENSDARG00000094433 |  | |||
| Common carp (Cyprinus carpio) | Cyprinid-type | caspa | ENSCCRG00000035668 |  | |
| caspb | ENSCCRG00000040063 |  |
| Species | Target | Infection Models | Stimulation Models | Reference |
|---|---|---|---|---|
| Gilthead seabream (Sparus aurata) | casp1 gene | Vibrio anguillarum Bacillus subtilis | Vibrio anguillarum DNA MDP | [72,82,83] |
| Casp1 activity | Salmonella enterica sv. Typhimurium | osmotic pressure | ||
| Japanese flounder (Paralichthys olivaceus) | asc gene | Edwardsiella tarda | LPS poly (I:C) zymosan extracellular ATP | [36,39,46,64,65] |
| casp1 gene | E. tarda E. piscicida | LPS poly (I:C) extracellular ATP | ||
| nlrb gene | E. ictaluri | |||
| nlrc gene | E. ictaluri | LPS | ||
| nlrc3 gene | E. tarda | LPS poly (I:C) extracellular ATP | ||
| casp1 and nlrp3 genes | E. piscicida | |||
| Casp1 activity | E. piscicida (EIB202) | extracellular ATP nigericin MSU | ||
| Zebrafish (Danio rerio) | asc and caspa genes | Rhabdovirus spring ciremia of carp virus (SVCV)E. tarda | [21,27,29,35,62,84,85] | |
| nlrc3l1 gene | E. piscicida | |||
| nlrp1, nlrp3, and caspb genes | E. tarda | |||
| CaspyA activity | Francisella noatunensis E. piscicida_ΔevpP | H2O2 | ||
| CaspyB activity | E. piscicida (0909I) | CTB+LPS | ||
| Miiuy croaker (Miichthys miiuy) | nlrc3 gene | V. anguillarum | poly (I:C) | [86,87] |
| nlrc35, nlrc39, and nlrc40 genes | V. anguillarum | LPS poly (I:C) | ||
| Rainbow trout (Oncorhynchus mykiss) | nlrc3 gene | LPS poly (I:C) | [59,66] | |
| Casp1 activity | LPS or zymosan + A. hydrophila | |||
| Asian seabass (Lates calcarifer) | nlrc3 gene | V. alginolyticus Streptococcus aureus | LPS poly (I:C) PGN | [67] |
| Goldfish (Carassius auratus) | asc gene | nigericin | [43] | |
| nlrc3l gene | ATP | |||
| Japanese pufferfish (Takifugu rubripes) | nlr-c1-8 and 10-12 genes | Lactobacillus paracasei spp. paracasei | [68,74] | |
| nlr-c1-5, 7-10, 12 and 13 genes | LPS | |||
| nlr-c1, 5, 7, 10 and 12 genes | nigericin nigericin+LPS | |||
| asc and casp1 genes | particulate silica | |||
| nlrc5-13, nlrc3 genes | particulate silica | |||
| Striped murrel (Channa striata) | casp1 gene | Aphanomyces invadans Aeromonas hydrophila | [88] | |
| Tongue sole (Cynoglossus semilaevis) | casp1 gene | E. tarda | [89] | |
| Turbot (Scophtalmus maximus) | asc gene | E. piscicida | [47] | |
| nlrc3a and nlrc3b genes | V. anguillarum St. iniae | |||
| Common carp (Cyprinus carpio) | casp1 and nlrp3 genes | cadmium | [73,75] | |
| nlrc gene | V. anguillarum | poly (I:C) flagellin PGN | ||
| Nile tilapia (Oreochromis niloticus) | nlrc3 gene | St. agalactiae | [40] | |
| Orange-spotted grouper (Epinephelus coioides) | asc and casp1 genes | ATP | [44] | |
| Casp1 activity | ATP | |||
| Japanese medaka (Oryzias latipes) | asc1 gene | Ae. hydrophila | [42] | |
| asc2 gene | nigericin | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morimoto, N.; Kono, T.; Sakai, M.; Hikima, J.-i. Inflammasomes in Teleosts: Structures and Mechanisms That Induce Pyroptosis during Bacterial Infection. Int. J. Mol. Sci. 2021, 22, 4389. https://doi.org/10.3390/ijms22094389
Morimoto N, Kono T, Sakai M, Hikima J-i. Inflammasomes in Teleosts: Structures and Mechanisms That Induce Pyroptosis during Bacterial Infection. International Journal of Molecular Sciences. 2021; 22(9):4389. https://doi.org/10.3390/ijms22094389
Chicago/Turabian StyleMorimoto, Natsuki, Tomoya Kono, Masahiro Sakai, and Jun-ichi Hikima. 2021. "Inflammasomes in Teleosts: Structures and Mechanisms That Induce Pyroptosis during Bacterial Infection" International Journal of Molecular Sciences 22, no. 9: 4389. https://doi.org/10.3390/ijms22094389
APA StyleMorimoto, N., Kono, T., Sakai, M., & Hikima, J.-i. (2021). Inflammasomes in Teleosts: Structures and Mechanisms That Induce Pyroptosis during Bacterial Infection. International Journal of Molecular Sciences, 22(9), 4389. https://doi.org/10.3390/ijms22094389






