Inhalational Anesthetics Inhibit Neuroglioma Cell Proliferation and Migration via miR-138, -210 and -335
Abstract
1. Introduction
2. Results
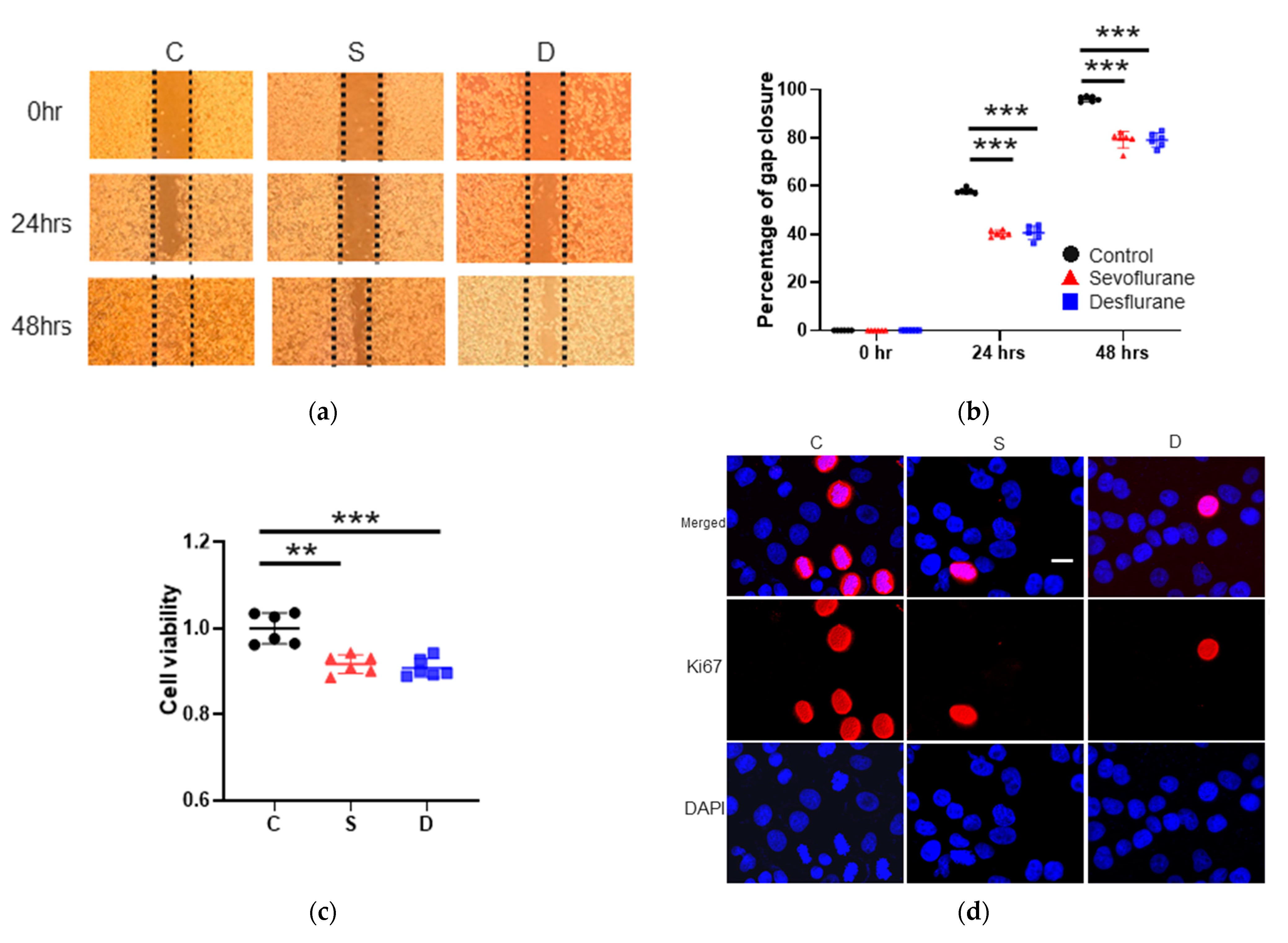
2.1. Sevoflurane and Desflurane Exposure Inhibited H4 Cell Proliferation and Migration
2.1.1. Cell Migration
2.1.2. Cell Proliferation Test
2.2. Sevoflurane Exposure Increased miR-210 Expressions and Desflurane Exposure Enhanced miR-138 and -335 Expressions
miRNA Changes after Anesthesia
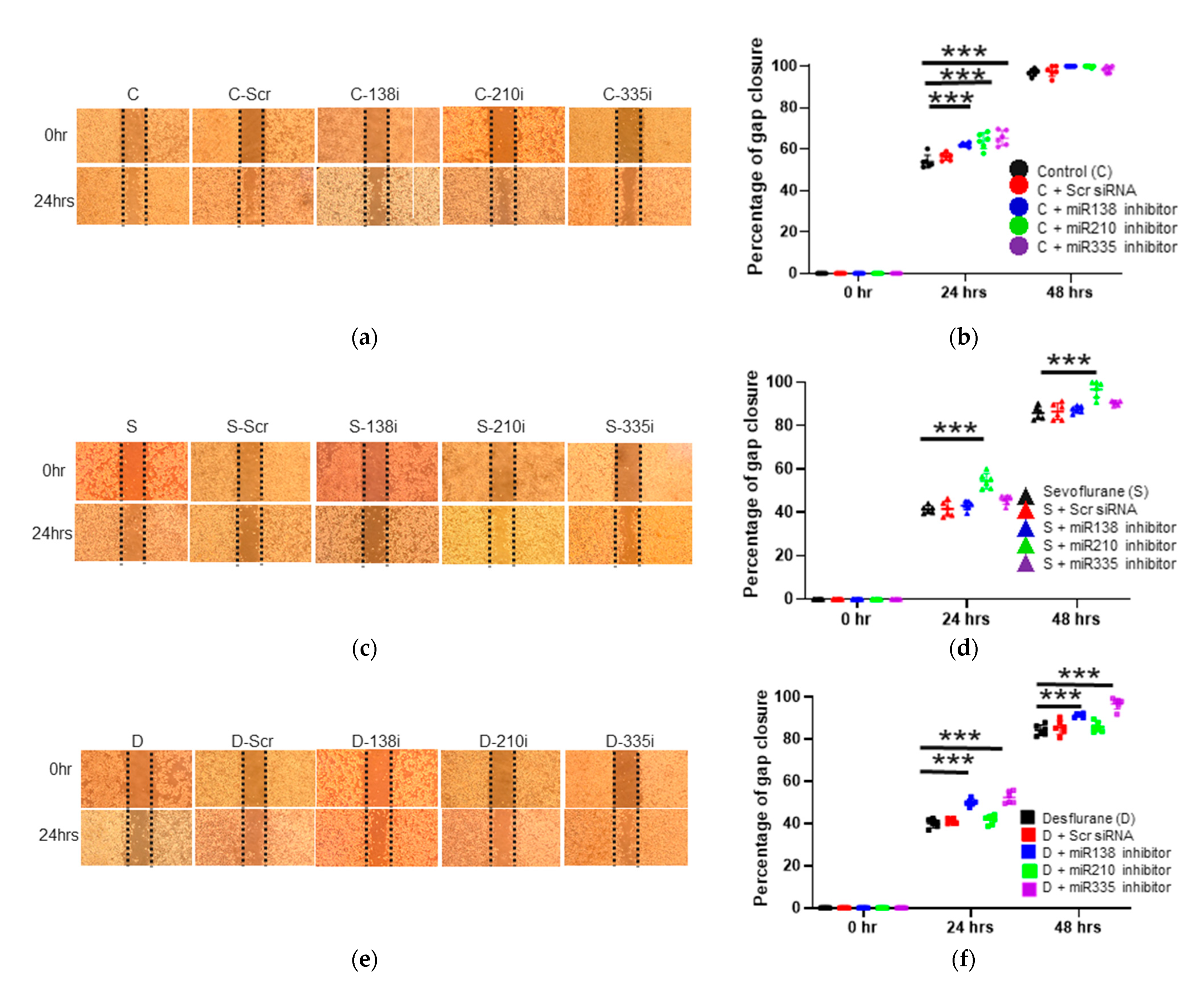
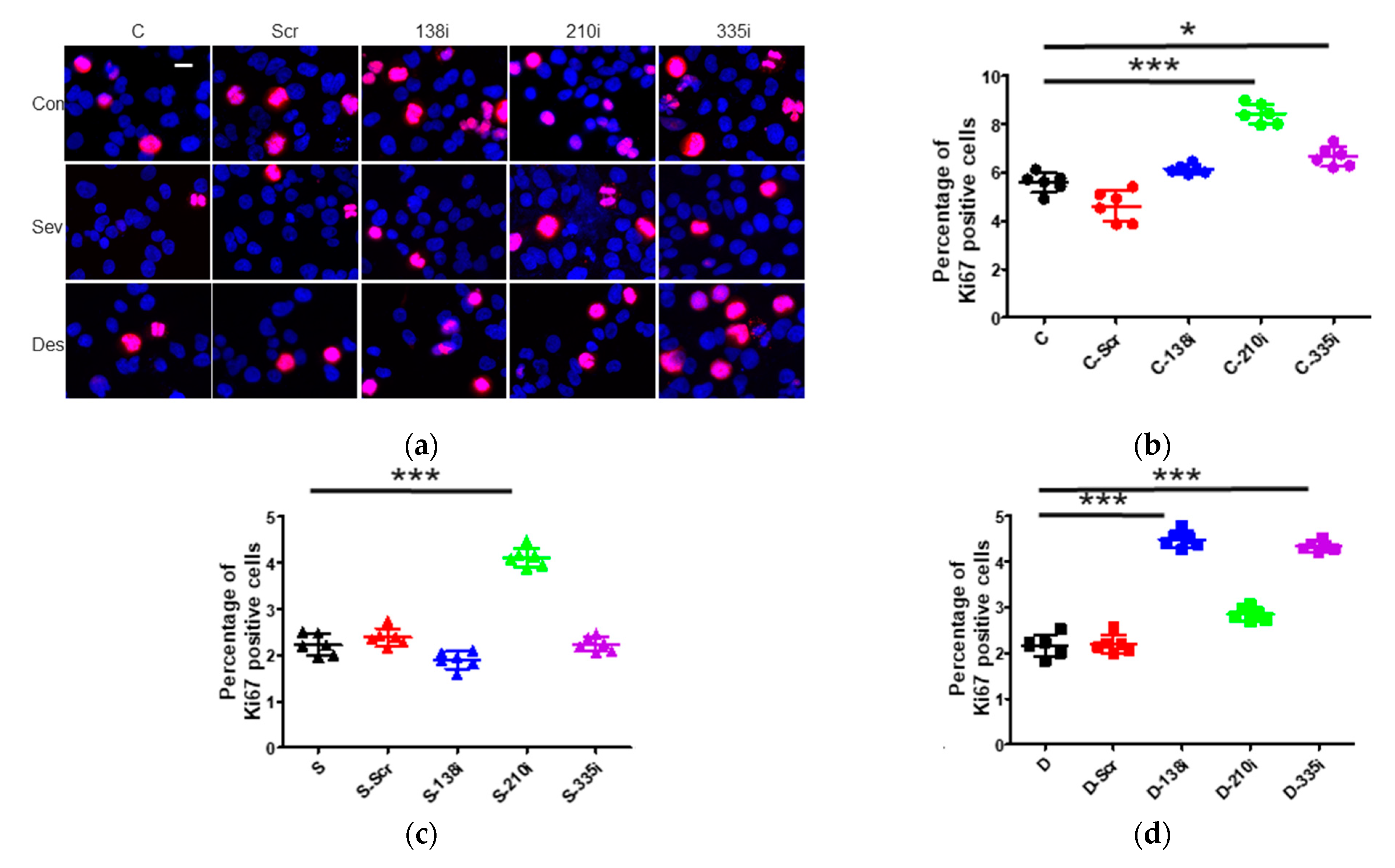
2.3. The Inhibition of miR-138, -210 and -335 Reversed the Sevoflurane- and Desflurane-Induced Suppression of Proliferation and Migration
2.3.1. Cell Migration Ability after the Inhibitor Administration of miR-138, -210 and -335
2.3.2. Cell Migration Ability after the Inhibitor Administration of miR-138, -210 and -335 with Sevoflurane Exposure
2.3.3. Cell Migration Ability after the Inhibitor Administration of miR-138, -210 and -335 with Desflurane Exposure
2.3.4. Cell Proliferation after the Inhibitor Administration of miR-138, -210 and -335
2.3.5. Cell Proliferation after the Inhibitor Administration of miR-138, -210 and -335 with Sevoflurane or Desflurane Exposure
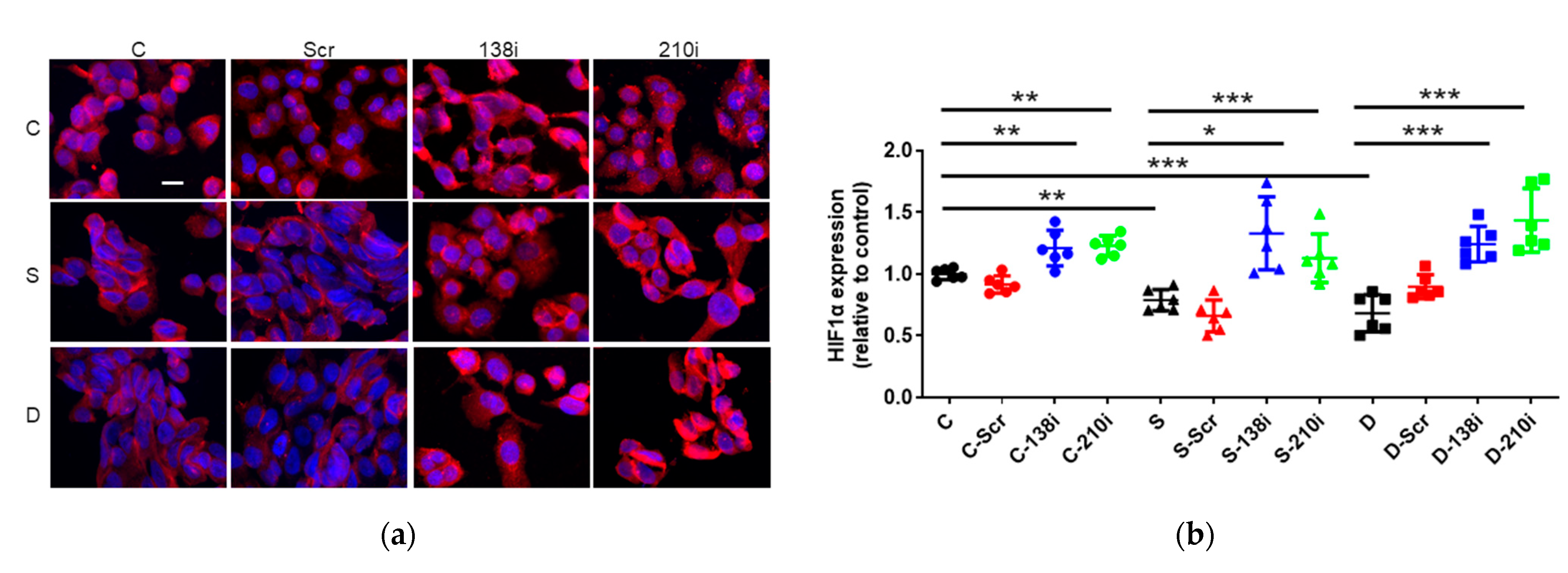
2.4. Sevoflurane and Desflurane Exposure Attenuated HIF-1α Protein Expression Which Was Reverted by miR-138 and -210 Inhibitor Treatment
The Inhibition of miR-138 and -210 enhanced HIF-1α Expression at Any Conditions
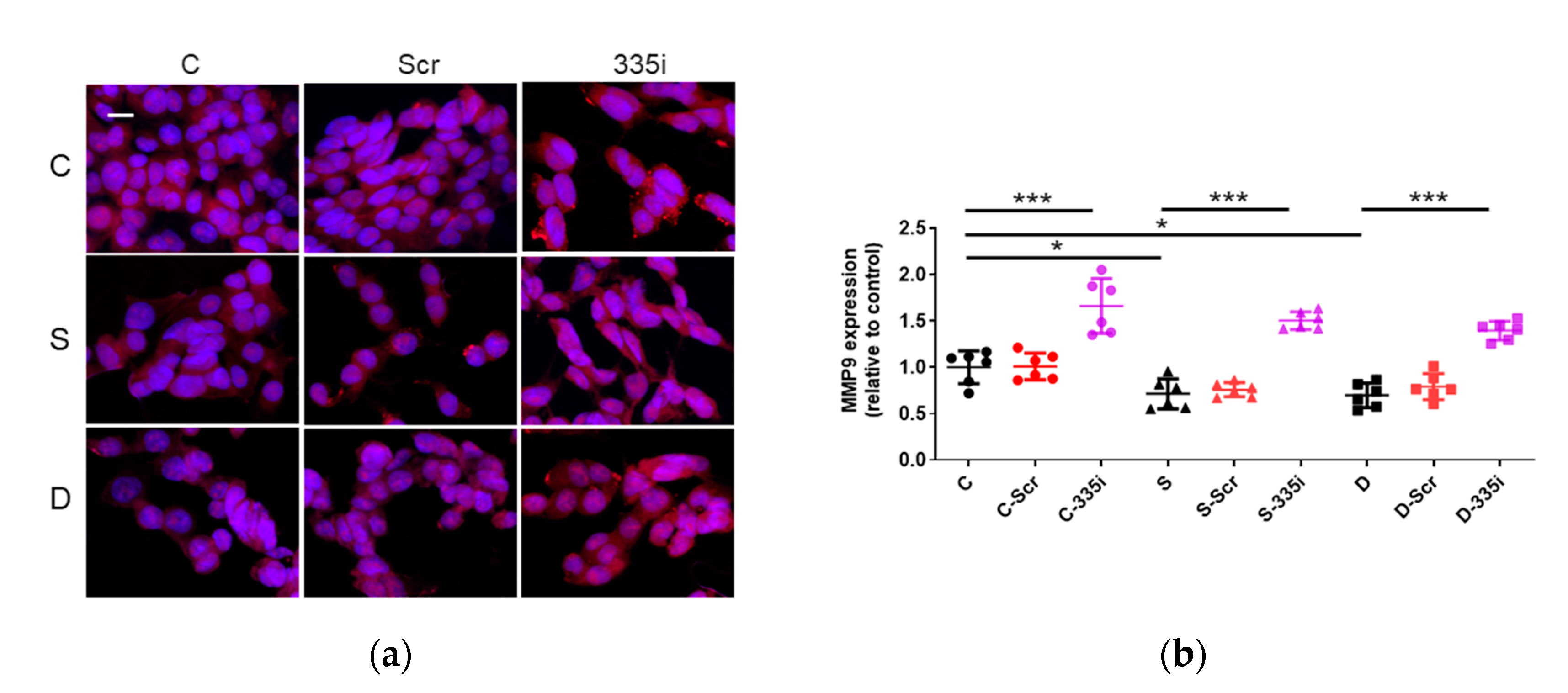
2.5. Desflurane Exposure Attenuated MMP9 Protein Expression Which Was Reverted by miR-335 Inhibitor Treatment
The Inhibition of miR-335 Enhanced MMP9 Expression
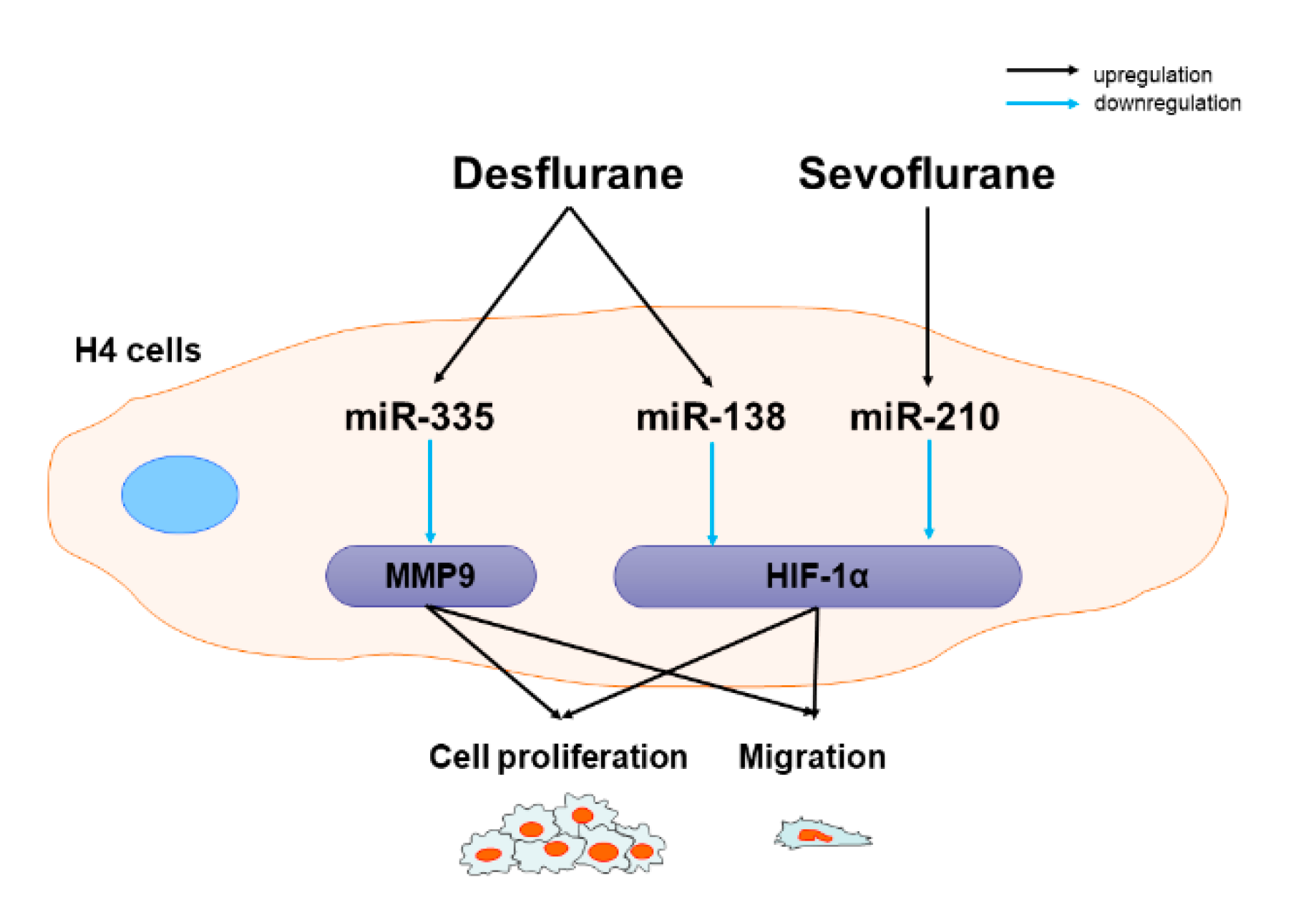
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Inhalational Anaesthetic Exposure
4.3. Wound Healing Assay
4.4. Cell Proliferation Test
4.5. RNA Extraction and Reverse Transcription
4.6. qRT-PCR
4.7. miRNA Inhibitor Transfection
4.8. Immunofluorescent Staining
4.9. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Mehlen, P.; Puisieux, A. Metastasis: A question of life or death. Nat. Rev. Cancer 2006, 6, 449–458. [Google Scholar] [CrossRef]
- Perry, N.J.S.; Buggy, D.; Ma, D. Can anesthesia influence cancer outcomes after surgery? JAMA Surg. 2019, 154, 279–280. [Google Scholar] [CrossRef]
- Gao, C.; He, X.F.; Xu, Q.R.; Xu, Y.J.; Shen, J. Sevoflurane downregulates insulin-like growth factor-1 to inhibit cell proliferation, invasion and trigger apoptosis in glioma through the PI3K/AKT signaling pathway. Anticancer Drugs 2019, 30, e0744. [Google Scholar] [CrossRef]
- Hurmath, F.K.; Mittal, M.; Ramaswamy, P.; Umamaheswara Rao, G.S.; Dalavaikodihalli Nanjaiah, N. Sevoflurane and thiopental preconditioning attenuates the migration and activity of MMP-2 in U87MG glioma cells. Neurochem. Int. 2016, 94, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Benzonana, L.L.; Zhao, H.; Watts, H.R.; Perry, N.J.; Bevan, C.; Brown, R.; Ma, D. Prostate cancer cell malignancy via modulation of HIF-1alpha pathway with isoflurane and propofol alone and in combination. Br. J. Cancer 2014, 111, 1338–1349. [Google Scholar] [CrossRef] [PubMed]
- Soni, S.; Padwad, Y.S. HIF-1 in cancer therapy: Two decade long story of a transcription factor. Acta Oncol. 2017, 56, 503–515. [Google Scholar] [CrossRef] [PubMed]
- Wigerup, C.; Pahlman, S.; Bexell, D. Therapeutic targeting of hypoxia and hypoxia-inducible factors in cancer. Pharmacol. Ther. 2016, 164, 152–169. [Google Scholar] [CrossRef]
- Minet, E.; Michel, G.; Remacle, J.; Michiels, C. Role of HIF-1 as a transcription factor involved in embryonic development, cancer progression and apoptosis (review). Int. J. Mol. Med. 2000, 5, 253–259. [Google Scholar] [CrossRef]
- Liang, H.; Gu, M.; Yang, C.; Wang, H.; Wen, X.; Zhou, Q. Sevoflurane inhibits invasion and migration of lung cancer cells by inactivating the p38 MAPK signaling pathway. J. Anesth. 2012, 26, 381–392. [Google Scholar] [CrossRef] [PubMed]
- Ala-Aho, R.; Kahari, V.M. Collagenases in cancer. Biochimie 2005, 87, 273–286. [Google Scholar] [CrossRef]
- Jansson, M.D.; Lund, A.H. MicroRNA and cancer. Mol. Oncol. 2012, 6, 590–610. [Google Scholar] [CrossRef]
- Ishikawa, M.; Tanaka, S.; Arai, M.; Genda, Y.; Sakamoto, A. Differences in microRNA changes of healthy rat liver between sevoflurane and propofol anesthesia. Anesthesiology 2012, 117, 1245–1252. [Google Scholar] [CrossRef]
- Jiang, S.; Liu, Y.; Huang, L.; Zhang, F.; Kang, R. Effects of propofol on cancer development and chemotherapy: Potential mechanisms. Eur. J. Pharmacol. 2018, 831, 46–51. [Google Scholar] [CrossRef]
- Qiu, H.; Chen, F.; Chen, M. MicroRNA-138 negatively regulates the hypoxia-inducible factor 1alpha to suppress melanoma growth and metastasis. Biol. Open 2019, 8, 42937. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Flach, H.; Onizawa, M.; Wei, L.; McManus, M.T.; Weiss, A. Negative regulation of Hif1a expression and TH17 differentiation by the hypoxia-regulated microRNA miR-210. Nat. Immunol. 2014, 15, 393–401. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; Cao, H.; Chen, Z.; Ma, Z.; Wan, X.; Peng, R.; Jiang, B. PAX6, a novel target of miR-335, inhibits cell proliferation and invasion in glioma cells. Mol. Med. Rep. 2014, 10, 399–404. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ishikawa, M.; Iwasaki, M.; Zhao, H.; Saito, J.; Hu, C.; Sun, Q.; Sakamoto, A.; Ma, D. Sevoflurane and desflurane exposure enhanced cell proliferation and migration in ovarian cancer cells via miR-210 and miR-138 downregulation. Int. J. Mol. Sci. 2021, 22, 1826. [Google Scholar] [CrossRef] [PubMed]
- Huitink, J.M.; Heimerikxs, M.; Nieuwland, M.; Loer, S.A.; Brugman, W.; Velds, A.; Sie, D.; Kerkhoven, R.M. Volatile anesthetics modulate gene expression in breast and brain tumor cells. Anesth. Analg. 2010, 111, 1411–1415. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, G.; Bacon, C.L.; Odumeru, O.; Fagan, C.; Fitzpatrick, T.; Gallagher, H.C.; Moriarty, D.C.; Regan, C.M. Antiproliferative actions of inhalational anesthetics: Comparisons to the valproate teratogen. Int. J. Dev. Neurosci. 2000, 18, 39–45. [Google Scholar] [CrossRef]
- Ciechanowicz, S.; Zhao, H.; Chen, Q.; Cui, J.; Mi, E.; Mi, E.; Lian, Q.; Ma, D. Differential effects of sevoflurane on the metastatic potential and chemosensitivity of non-small-cell lung adenocarcinoma and renal cell carcinoma in vitro. Br. J. Anaesth. 2018, 120, 368–375. [Google Scholar] [CrossRef]
- Goto, G.; Hori, Y.; Ishikawa, M.; Tanaka, S.; Sakamoto, A. Changes in the gene expression levels of microRNAs in the rat hippocampus by sevoflurane and propofol anesthesia. Mol. Med. Rep. 2014, 9, 1715–1722. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, S.; Ishikawa, M.; Arai, M.; Genda, Y.; Sakamoto, A. Changes in microRNA expression in rat lungs caused by sevoflurane anesthesia: A TaqMan(R) low-density array study. Biomed. Res. 2012, 33, 255–263. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Wu, Y.; Wang, J.; He, J.; Han, X. Sevoflurane inhibits the migration and invasion of colorectal cancer cells through regulating ERK/MMP-9 pathway by up-regulating miR-203. Eur. J. Pharmacol. 2019, 850, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Yi, W.; Li, D.; Guo, Y.; Zhang, Y.; Huang, B.; Li, X. Sevoflurane inhibits the migration and invasion of glioma cells by upregulating microRNA-637. Int. J. Mol. Med. 2016, 38, 1857–1863. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Shen, J.; Meng, Z.X.; He, X.F. Sevoflurane Inhibits Glioma Cells Proliferation and Metastasis through miRNA-124-3p/ROCK1 Axis. Pathol. Oncol. Res. 2020, 26, 947–954. [Google Scholar] [CrossRef]
- Yin, D.; Ogawa, S.; Kawamata, N.; Leiter, A.; Ham, M.; Li, D.; Doan, N.B.; Said, J.W.; Black, K.L.; Phillip Koeffler, H. miR-34a functions as a tumor suppressor modulating EGFR in glioblastoma multiforme. Oncogene 2013, 32, 1155–1163. [Google Scholar] [CrossRef]
- Guo, N.L.; Zhang, J.X.; Wu, J.P.; Xu, Y.H. Isoflurane promotes glucose metabolism through up-regulation of miR-21 and suppresses mitochondrial oxidative phosphorylation in ovarian cancer cells. Biosci. Rep. 2017, 37, BSR20170818. [Google Scholar] [CrossRef]
- Lu, Y.; Li, Y.; Wang, Z.; Xie, S.; Wang, Q.; Lei, X.; Ruan, Y.; Li, J. Downregulation of RGMA by HIF-1A/miR-210-3p axis promotes cell proliferation in oral squamous cell carcinoma. Biomed. Pharmacother. 2019, 112, 108608. [Google Scholar] [CrossRef]
- Qin, Q.; Furong, W.; Baosheng, L. Multiple functions of hypoxia-regulated miR-210 in cancer. J. Exp. Clin. Cancer Res. 2014, 33, 50. [Google Scholar] [CrossRef]
- Zuo, J.; Wen, M.; Lei, M.; Peng, X.; Yang, X.; Liu, Z. MiR-210 links hypoxia with cell proliferation regulation in human Laryngocarcinoma cancer. J. Cell Biochem. 2015, 116, 1039–1049. [Google Scholar] [CrossRef]
- Qiu, S.; Huang, D.; Yin, D.; Li, F.; Li, X.; Kung, H.F.; Peng, Y. Suppression of tumorigenicity by microRNA-138 through inhibition of EZH2-CDK4/6-pRb-E2F1 signal loop in glioblastoma multiforme. Biochim. Biophys. Acta 2013, 1832, 1697–1707. [Google Scholar] [CrossRef]
- Yuan, M.; Zhao, S.; Chen, R.; Wang, G.; Bie, Y.; Wu, Q.; Cheng, J. MicroRNA-138 inhibits tumor growth and enhances chemosensitivity in human cervical cancer by targeting H2AX. Exp. Ther. Med. 2020, 19, 630–638. [Google Scholar] [CrossRef]
- Yeh, Y.M.; Chuang, C.M.; Chao, K.C.; Wang, L.H. MicroRNA-138 suppresses ovarian cancer cell invasion and metastasis by targeting SOX4 and HIF-1alpha. Int. J. Cancer 2013, 133, 867–878. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Liu, R.; Wang, J.; Yan, Z.; Qian, S.; Zhang, W. RNAi knockdown of hypoxia-inducible factor-1alpha decreased the proliferation, migration, and invasion of hypoxic hepatocellular carcinoma cells. Cell Biochem. Biophys. 2015, 71, 1677–1684. [Google Scholar] [CrossRef] [PubMed]
- Ban, H.S.; Uno, M.; Nakamura, H. Suppression of hypoxia-induced HIF-1alpha accumulation by VEGFR inhibitors: Different profiles of AAL993 versus SU5416 and KRN633. Cancer Lett. 2010, 296, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Talks, K.L.; Turley, H.; Gatter, K.C.; Maxwell, P.H.; Pugh, C.W.; Ratcliffe, P.J.; Harris, A.L. The expression and distribution of the hypoxia-inducible factors HIF-1alpha and HIF-2alpha in normal human tissues, cancers, and tumor-associated macrophages. Am. J. Pathol. 2000, 157, 411–421. [Google Scholar] [CrossRef]
- Semenza, G.L. Hypoxia-inducible factors: Mediators of cancer progression and targets for cancer therapy. Trends Pharmacol. Sci. 2012, 33, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Monti, E.; Gariboldi, M.B. HIF-1 as a target for cancer chemotherapy, chemosensitization and chemoprevention. Curr. Mol. Pharmacol. 2011, 4, 62–77. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.H.; Jiang, G.; Zheng, J.Z.; Lu, Z.; Hunter, T.; Vogt, P.K. Phosphatidylinositol 3-kinase signaling controls levels of hypoxia-inducible factor 1. Cell Growth Differ. 2001, 12, 363–369. [Google Scholar]
- Liang, H.; Wang, H.B.; Liu, H.Z.; Wen, X.J.; Zhou, Q.L.; Yang, C.X. The effects of combined treatment with sevoflurane and cisplatin on growth and invasion of human adenocarcinoma cell line A549. Biomed. Pharmacother. 2013, 67, 503–509. [Google Scholar] [CrossRef]
- Lengyel, E.; Schmalfeldt, B.; Konik, E.; Spathe, K.; Harting, K.; Fenn, A.; Berger, U.; Fridman, R.; Schmitt, M.; Prechtel, D.; et al. Expression of latent matrix metalloproteinase 9 (MMP-9) predicts survival in advanced ovarian cancer. Gynecol. Oncol. 2001, 82, 291–298. [Google Scholar] [CrossRef] [PubMed]
- Nabeshima, K.; Inoue, T.; Shimao, Y.; Sameshima, T. Matrix metalloproteinases in tumor invasion: Role for cell migration. Pathol. Int. 2002, 52, 255–264. [Google Scholar] [CrossRef] [PubMed]
- D’Costa, Z.; Jones, K.; Azad, A.; van Stiphout, R.; Lim, S.Y.; Gomes, A.L.; Kinchesh, P.; Smart, S.C.; Gillies McKenna, W.; Buffa, F.M.; et al. Gemcitabine-induced TIMP1 Attenuates therapy response and promotes tumor growth and liver metastasis in pancreatic cancer. Cancer Res. 2017, 77, 5952–5962. [Google Scholar] [CrossRef] [PubMed]
- Musumeci, G.; Magro, G.; Cardile, V.; Coco, M.; Marzagalli, R.; Castrogiovanni, P.; Imbesi, R.; Graziano, A.C.; Barone, F.; Di Rosa, M.; et al. Characterization of matrix metalloproteinase-2 and-9, ADAM-10 and N-cadherin expression in human glioblastoma multiforme. Cell Tissue Res. 2015, 362, 45–60. [Google Scholar] [CrossRef] [PubMed]
- Hanemaaijer, R.; Verheijen, J.H.; Maguire, T.M.; Visser, H.; Toet, K.; McDermott, E.; O’Higgins, N.; Duffy, M.J. Increased gelatinase-A and gelatinase-B activities in malignant vs. benign breast tumors. Int. J. Cancer 2000, 86, 204–207. [Google Scholar] [CrossRef]
- Malmstrom, R.E.; Alexandersson, A.; Balmer, K.C.; Weilitz, J. In vivo characterization of the novel neuropeptide Y Y1 receptor antagonist H 409/22. J. Cardiovasc. Pharmacol. 2000, 36, 516–525. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Yu, X.; Sun, S.; Zhang, X.; Yang, W.; Zhang, J.; Zhang, X.; Jiang, Z. Increased expression of MMP-2 and MMP-9 indicates poor prognosis in glioma recurrence. Biomed. Pharmacother. 2019, 118, 109369. [Google Scholar] [CrossRef]
- Muller-Edenborn, B.; Roth-Z’graggen, B.; Bartnicka, K.; Borgeat, A.; Hoos, A.; Borsig, L.; Beck-Schimmer, B. Volatile anesthetics reduce invasion of colorectal cancer cells through down-regulation of matrix metalloproteinase-9. Anesthesiology 2012, 117, 293–301. [Google Scholar] [CrossRef] [PubMed]
- Gong, M.; Ma, J.; Guillemette, R.; Zhou, M.; Yang, Y.; Yang, Y.; Hock, J.M.; Yu, X. miR-335 inhibits small cell lung cancer bone metastases via IGF-IR and RANKL pathways. Mol. Cancer Res. 2014, 12, 101–110. [Google Scholar] [CrossRef]
- Yan, Z.; Xiong, Y.; Xu, W.; Gao, J.; Cheng, Y.; Wang, Z.; Chen, F.; Zheng, G. Identification of hsa-miR-335 as a prognostic signature in gastric cancer. PLoS ONE 2012, 7, e40037. [Google Scholar] [CrossRef]
- Cao, J.; Cai, J.; Huang, D.; Han, Q.; Yang, Q.; Li, T.; Ding, H.; Wang, Z. miR-335 represents an invasion suppressor gene in ovarian cancer by targeting Bcl-w. Oncol. Rep. 2013, 30, 701–706. [Google Scholar] [CrossRef] [PubMed]
- Hafez, M.M.; Hassan, Z.K.; Zekri, A.R.; Gaber, A.A.; Al Rejaie, S.S.; Sayed-Ahmed, M.M.; Al Shabanah, O. MicroRNAs and metastasis-related gene expression in Egyptian breast cancer patients. Asian Pac. J. Cancer Prev. 2012, 13, 591–598. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, M.; Zhao, H.; Jaffer, T.; Unwith, S.; Benzonana, L.; Lian, Q.; Sakamoto, A.; Ma, D. Volatile anaesthetics enhance the metastasis related cellular signalling including CXCR2 of ovarian cancer cells. Oncotarget 2016, 7, 26042–26056. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Verkman, A.S. Increased migration and metastatic potential of tumor cells expressing aquaporin water channels. FASEB J. 2006, 20, 1892–1894. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ishikawa, M.; Iwasaki, M.; Zhao, H.; Saito, J.; Hu, C.; Sun, Q.; Sakamoto, A.; Ma, D. Inhalational Anesthetics Inhibit Neuroglioma Cell Proliferation and Migration via miR-138, -210 and -335. Int. J. Mol. Sci. 2021, 22, 4355. https://doi.org/10.3390/ijms22094355
Ishikawa M, Iwasaki M, Zhao H, Saito J, Hu C, Sun Q, Sakamoto A, Ma D. Inhalational Anesthetics Inhibit Neuroglioma Cell Proliferation and Migration via miR-138, -210 and -335. International Journal of Molecular Sciences. 2021; 22(9):4355. https://doi.org/10.3390/ijms22094355
Chicago/Turabian StyleIshikawa, Masashi, Masae Iwasaki, Hailin Zhao, Junichi Saito, Cong Hu, Qizhe Sun, Atsuhiro Sakamoto, and Daqing Ma. 2021. "Inhalational Anesthetics Inhibit Neuroglioma Cell Proliferation and Migration via miR-138, -210 and -335" International Journal of Molecular Sciences 22, no. 9: 4355. https://doi.org/10.3390/ijms22094355
APA StyleIshikawa, M., Iwasaki, M., Zhao, H., Saito, J., Hu, C., Sun, Q., Sakamoto, A., & Ma, D. (2021). Inhalational Anesthetics Inhibit Neuroglioma Cell Proliferation and Migration via miR-138, -210 and -335. International Journal of Molecular Sciences, 22(9), 4355. https://doi.org/10.3390/ijms22094355








