RETRACTED: Formation of Proto-Kranz in C3 Rice Induced by Spike-Stalk Injection Method
Abstract
1. Introduction
2. Results
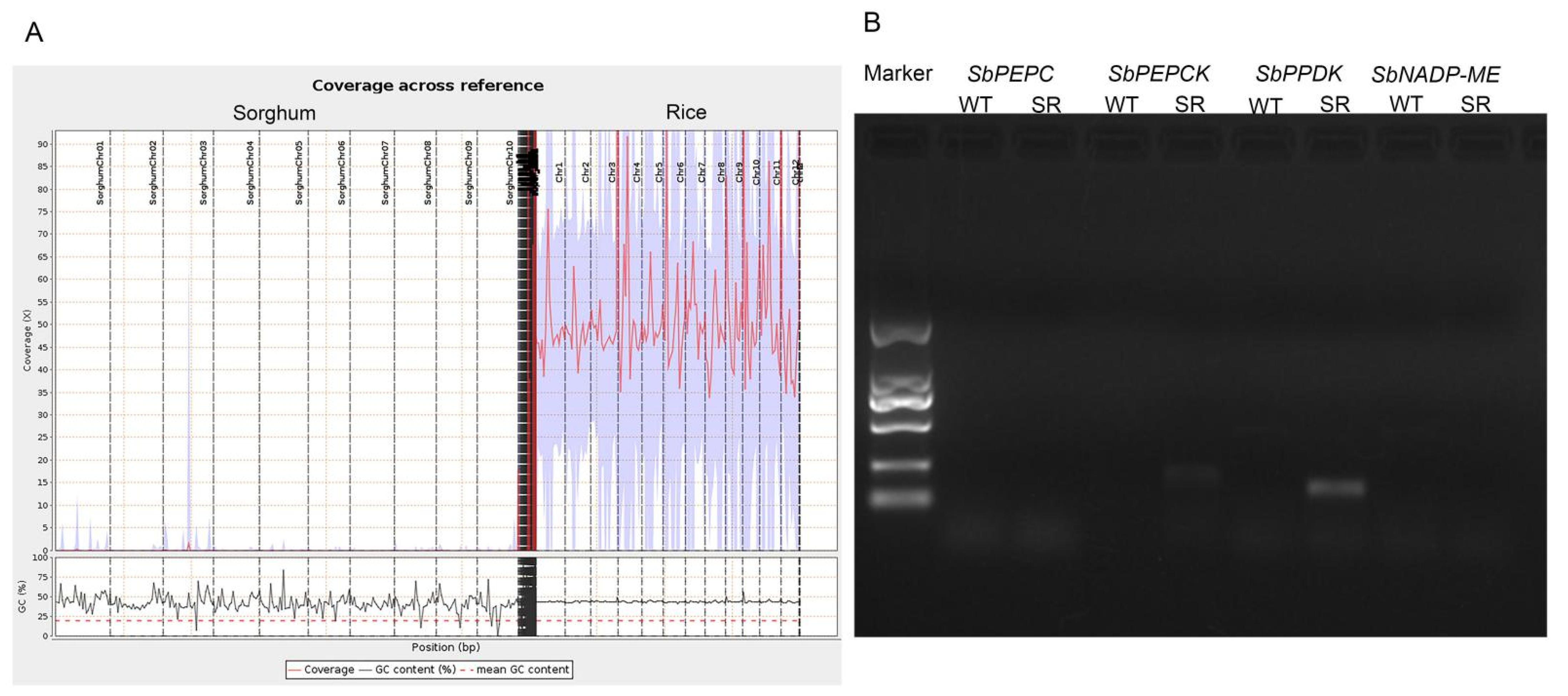
2.1. Genome Resequencing of Rice SR
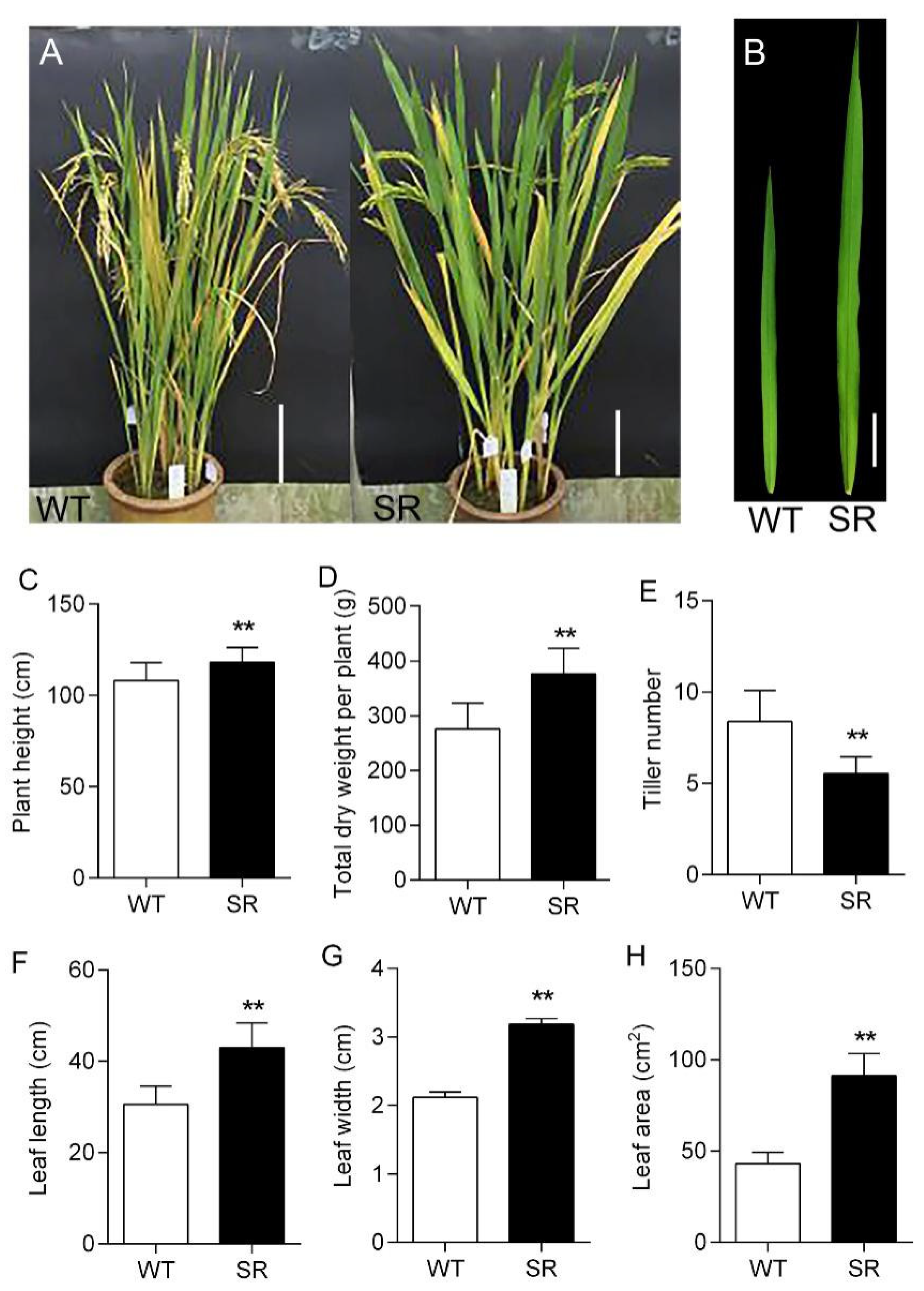
2.2. Increased Vegetative Biomass in Rice SR
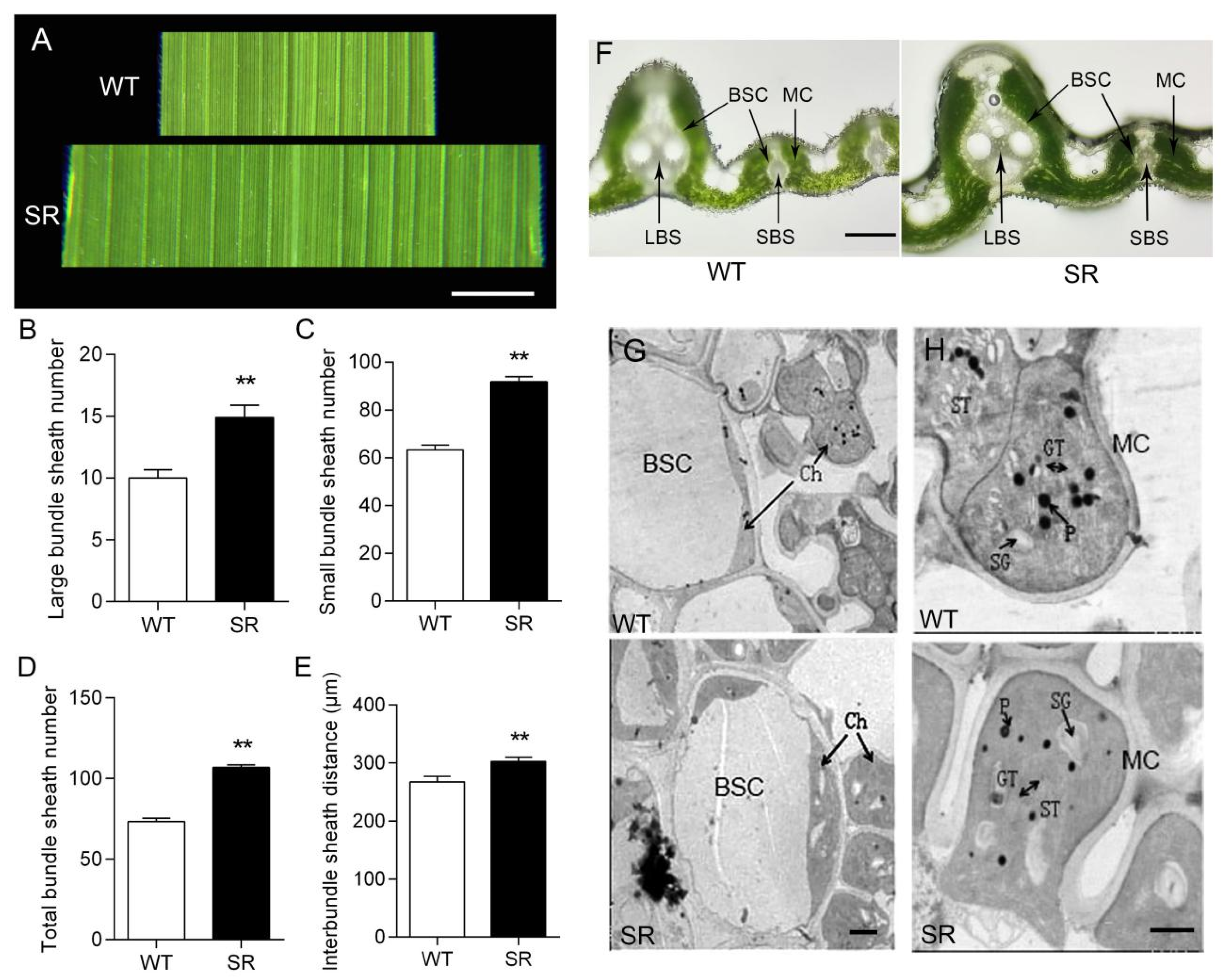
2.3. Proto-Kranz Anatomy and Delayed Leaf Senescence in Rice SR
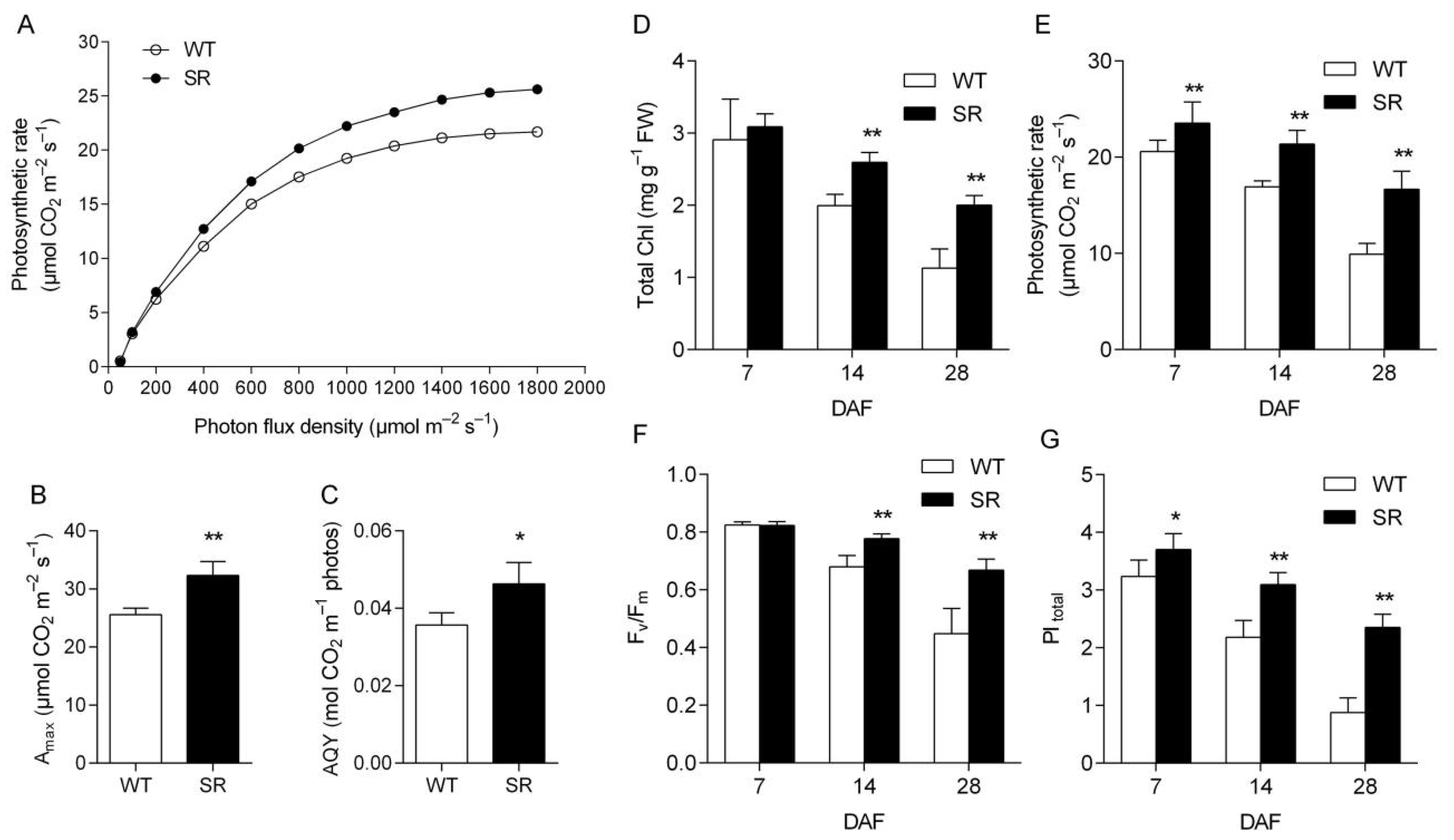
2.4. Increased Photosynthetic Capacity in Rice SR
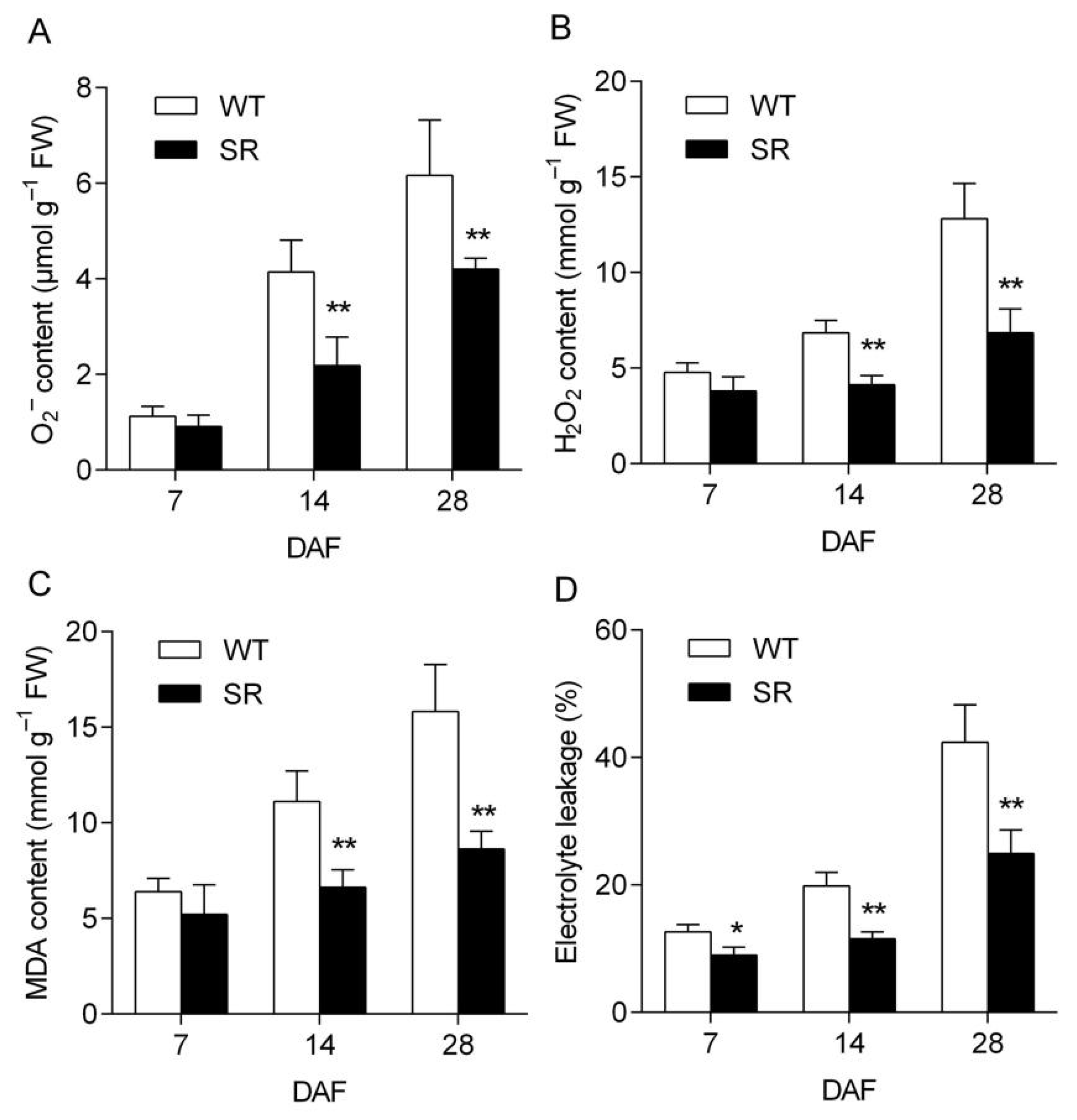
2.5. Reduced ROS Accumulation and Oxidative Damage in the Leaves of Rice SR during Natural Senescence
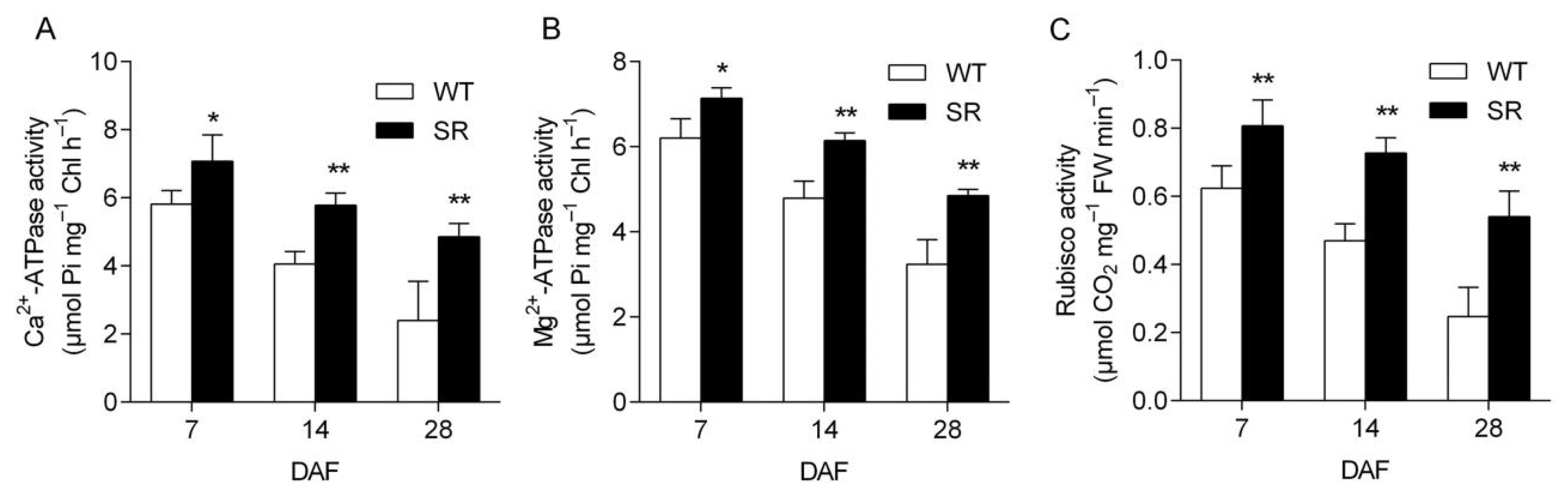
2.6. Elevated Activities of Ca2+-ATPase, Mg2+-ATPase, and Rubisco in Rice SR during Natural Senescence
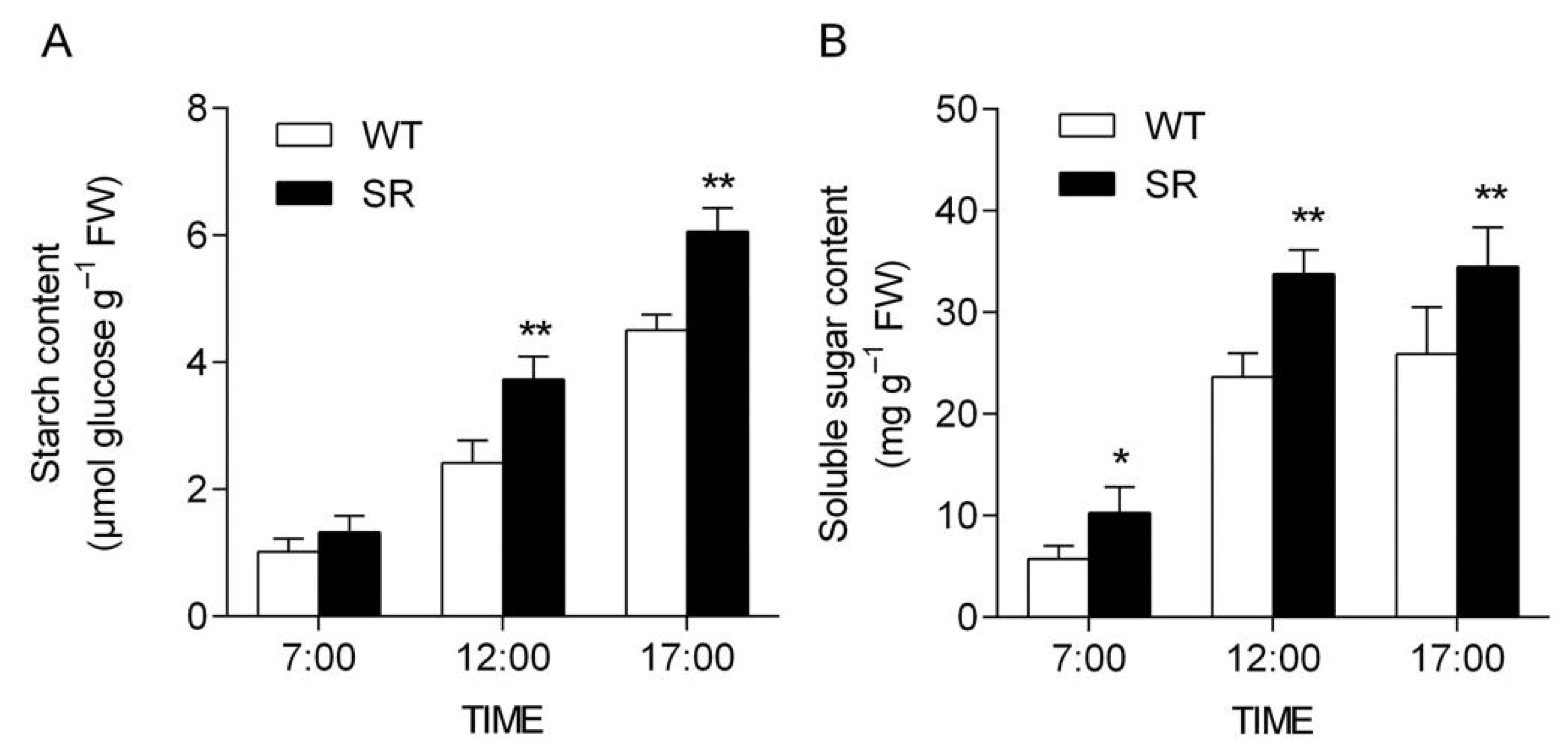
2.7. Enhanced Carbohydrate Assimilation in Rice SR
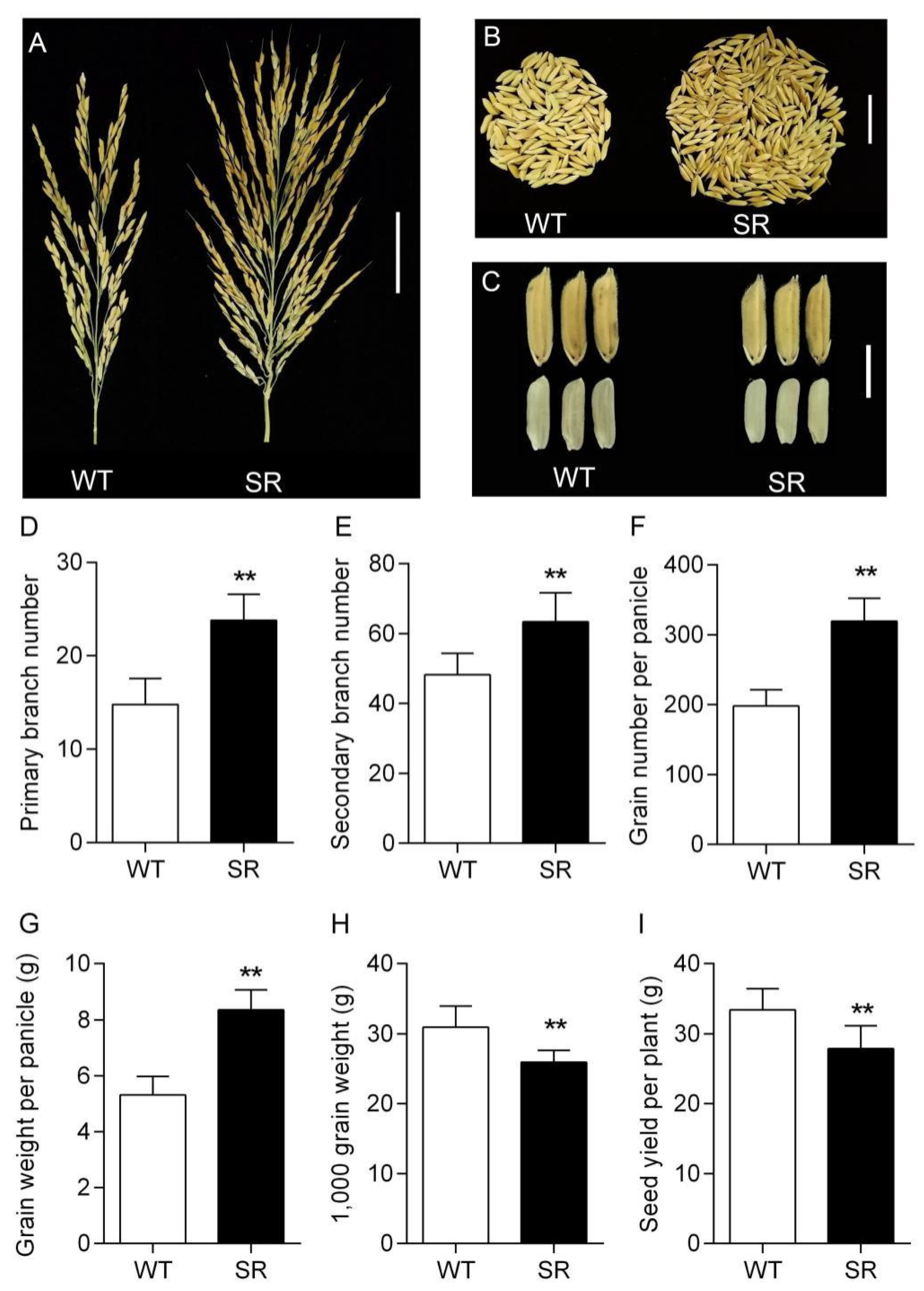
2.8. Large and Heavy Panicle in Rice SR
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. DNA Extraction and Genome Resequencing
4.3. RNA Extraction and Reverse Transcription Polymerase Chain Reaction (RT-PCR)
4.4. Photosynthetic Measurements
4.5. Detection of Reactive Oxygen Species (ROS)
4.6. Lipid Peroxidation and Electrolyte Leakage Determinations
4.7. Starch and Soluble Sugar Content Measurements
4.8. Ca2+-ATPase, Mg2+-ATPase, and Rubisco Activity Determinations
4.9. Immunoblot Analysis
4.10. Transmission Electron Microscopy (TEM)
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sage, R.F.; Sage, T.L.; Kocacinar, F. Photorespiration and the evolution of C4 photosynthesis. Annu. Rev. Plant Biol. 2012, 63, 19–47. [Google Scholar] [CrossRef]
- Leakey, A.D. Rising atmospheric carbon dioxide concentration and the future of C4 crops for food and fuel. Proc. Biol. Sci. 2009, 276, 2333–2343. [Google Scholar]
- Bräutigam, A.; Gowik, U. Photorespiration connects C3 and C4 photosynthesis. J. Exp. Bot. 2016, 67, 2953–2962. [Google Scholar] [CrossRef]
- Kapralov, M.V.; Kubien, D.S.; Andersson, I.; Filatov, D.A. Changes in rubisco kinetics during the evolution of C4 photosynthesis in flaveria (Asteraceae) are associated with positive selection on genes encoding the enzyme. Mol. Biol. Evol. 2010, 4, 1491–1503. [Google Scholar] [CrossRef]
- Schuler, M.L.; Mantegazza, O.; Weber, A.P.M. Engineering C4 photosynthesis into C3 chassis in the synthetic biology age. Plant J. 2016, 87, 51–65. [Google Scholar] [CrossRef]
- Sedelnikova, O.V.; Hughes, T.E.; Langdale, J.A. Understanding the genetic basis of C4 Kranz anatomy with a view to engineering C3 crops. Annu. Rev. Genet. 2018, 52, 249–270. [Google Scholar] [CrossRef]
- Long, S.P.; Marshall-Colon, A.; Zhu, X.G. Meeting the global food demand of the future by engineering crop photosynthesis and yield potential. Cell 2015, 161, 56–66. [Google Scholar] [CrossRef] [PubMed]
- von Caemmerer, S.; Quick, W.P.; Furbank, R.T. The development of C(4)rice: Current progress and future challenges. Science 2012, 336, 1671–1672. [Google Scholar] [CrossRef]
- Khoshravesh, R.; Stinson, C.R.; Stata, M.; Busch, F.A.; Sage, R.F.; Ludwig, M.; Sage, T.L. C3–C4intermediacy in grasses: Organelle enrichment and distribution, glycine decarboxylase expression, and the rise of C2photosynthesis. J. Exp. Bot. 2016, 67, 3065–3078. [Google Scholar] [CrossRef] [PubMed]
- Muhaidat, R.; Sage, T.L.; Frohlich, M.W.; Dengler, N.G.; Sage, R.F. Characterization of C3-C4 intermediate species in the genus Heliotropium L. (Boraginaceae): Anatomy, ultrastructure and enzyme activity. Plant Cell Environ. 2011, 34, 1723–1736. [Google Scholar] [CrossRef] [PubMed]
- Sage, T.L.; Busch, F.A.; Johnson, D.C.; Friesen, P.C.; Stinson, C.R.; Stata, M.; Sultmanis, S.; Rahman, B.A.; Rawsthorne, S.; Sage, R.F. Initial events during the evolution of C4 photosynthesis in C3 species of Flaveria. Plant Physiol. 2013, 163, 1266–1276. [Google Scholar] [CrossRef]
- Stata, M.; Sage, T.L.; Hoffmann, N.; Covshoff, S.; Ka-Shu, W.G.; Sage, R.F. Mesophyll chloroplast investment in C3, C4 and C2 species of the Genus Flaveria. Plant Cell Physiol. 2016, 57, 904–918. [Google Scholar] [CrossRef]
- Vicentini, A.; Barber, J.C.; Aliscioni, S.S.; Giussani, L.M.; Kellogg, E.A. The age of the grasses and clusters of origins of C-4 photosynthesis. Glob. Chang. Biol. 2008, 14, 2963–2977. [Google Scholar] [CrossRef]
- Dengler, N.G.; Dengler, R.E.; Donelly, P.M.; Hattersley, P.W. Quantitative leaf anatomy of C3 and C4 grasses (Poaceae): Bundle sheath and mesophyll surface area relationships. Ann. Bot. 1994, 73, 241–255. [Google Scholar] [CrossRef]
- Ueno, O.; Kawano, Y.; Wakayama, M.; Takeda, T. Leaf vascular systems in C(3) and C(4) grasses: A two-dimensional analysis. Ann. Bot. 2006, 97, 611–621. [Google Scholar] [CrossRef]
- Dengler, N.G.; Nelson, T. Leaf structure and development in C4 plants. In C4 Plant Biology; Sage, R.F., Monson, R.K., Eds.; Academic Press: San Diego, CA, USA, 1999; pp. 133–172. [Google Scholar]
- Pena, D.L.; Lorz, H.; Schell, J. Transgenic rye plant obtained by injecting DNA into young floral tillers. Nature 1987, 325, 274–276. [Google Scholar] [CrossRef]
- Zhao, B.; Jia, J.; Yang, H.; Li, C.; Zhan, Q.; Wang, B.; Zhou, K.; Yuan, L. RAPD analysis of new rice strains developed through the method of spike-stalk-injection DNA from wild relative. Acta Agron. Sin. 2000, 26, 424–430. [Google Scholar]
- Hu, Y.; Mao, B.; Xia, Y.; Peng, Y.; Zhang, D.; Tang, L.; Shao, Y.; Li, Y.; Zhao, B. Spike-stalk injection method causes extensive phenotypic and genotypic variations for rice germplasm. Front. Plant Sci. 2020, 11, 575373. [Google Scholar] [CrossRef]
- Feller, U.; Anders, I.; Ma, T. Rubiscolytics: Fate of Rubisco after its enzymatic function in a cell is terminated. J. Exp. Bot. 2008, 59, 1615–1624. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Dong, Y.; Liu, B. Isolation of Zizania latifolia species_specific DNA sequences and their utility in identification of Z. latifolia DNA introgressed into rice. Acta Bot. Sin. 2000, 42, 324–326. [Google Scholar]
- Xing, Q.; Zhao, B.R.; Xu, K.; Yang, H.H.; Liu, X.; Wang, S.W.; Jin, D.M.; Yuan, L.P.; Wang, B. Test of agronomic characteristics and amplified fragment length polymorphism analysis of new rice germplasm developed from transformation of genomic DNA of distant relatives. Plant Mol. Biol. Rep. 2004, 22, 155–164. [Google Scholar] [CrossRef]
- Langdale, J.A. C4 Cycles: Past, present, and future research on C4 photosynthesis. Plant Cell 2011, 23, 3879–3892. [Google Scholar] [CrossRef]
- Lin, J.F.; Wu, S.H. Molecular events in senescing Arabidopsis leaves. Plant J. 2004, 39, 612–628. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Vlad, D.; Langdale, J.A. Finding the genes to build C4 rice. Curr. Opin. Plant Biol. 2016, 31, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Khoshravesh, R.; Karki, S.; Tapia, R.; Balahadia, C.P.; Bandyopadhyay, A.; Quick, W.P.; Furbank, R.; Sage, T.L.; Langdale, J.A. Re-creation of a key step in the evolutionary switch from C3 to C4 leaf anatomy. Curr. Biol. 2017, 27, 3278–3287. [Google Scholar] [CrossRef] [PubMed]
- Takai, T.; Adachi, S.; Taguchi-Shiobara, F.; Sanoh-Arai, Y.; Iwasawa, N.; Yoshinaga, S.; Hirose, S.; Taniguchi, Y.; Yamanouchi, U.; Wu, J.-Z.; et al. A natural variant of NAL1, selected in high-yield rice breeding programs, pleiotropically increases photosynthesis rate. Sci. Rep. 2013, 3, 2149. [Google Scholar] [CrossRef]
- Wilson, P.J.; Thompson, K.; Hodgson, J.G. Specific leaf area and leaf dry matter content as alternative predictors of plant strategies. New Phytol. 2002, 143, 155–162. [Google Scholar] [CrossRef]
- Ishikawa, C.; Hatanaka, T.; Misoo, S.; Miyake, C.; Fukayama, H. Functional incorporation of sorghum small subunit increases the catalytic turnover rate of Rubisco in transgenic rice. Plant Physiol. 2011, 156, 1603–1611. [Google Scholar] [CrossRef]
- Carmo-Silva, E.; Andraloic, P.; Scales, J.; Driever, S.; Mead, A.; Lawson, T.; Raines, C.; Parry, M. Phenotyping of field-grown wheat in the UK highlights contribution of light response of photosynthesis and flag leaf longevity to grain yield. J. Exp. Bot. 2017, 68, 3473–3486. [Google Scholar] [CrossRef]
- Ermakova, M.; Arrivault, S.; Giuliani, R.; Danila, F.; Alonso-Cantabrana, H.; Vlad, D.; Ishihara, H.; Feil, R.; Guenther, M.; Borghi, G.L.; et al. Installation of C4 photosynthetic pathway enzymes in rice using a single construct. Plant Biotechnol. J. 2020, 19, 575–588. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome project data processing subgroup. The sequence alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Farquhar, G.D.; von Caemmerer, S.; Berry, J.A. A biochemical model of photosynthetic CO2 assimilation in leaves of C3 species. Planta 1980, 149, 78–90. [Google Scholar] [CrossRef] [PubMed]
- Lichtenthaler, H.K. Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. Methods Enzymol. 1987, 148, 350–382. [Google Scholar]
- Jiang, D.X.; Hou, J.J.; Gao, W.W.; Tong, X.; Li, M.; Chu, X.; Chen, G.X. Exogenous spermidine alleviates the adverse effects of aluminum toxicity on photosystem II through improved antioxidant system and endogenous polyamine contents. Ecotoxicol. Environ. Saf. 2020, 207, 111265. [Google Scholar] [CrossRef]
- Draper, H.H.; Hadley, M. Malondialdehyde determination as index of lipid peroxidation. Meth. Enzymol. 1990, 186, 421–431. [Google Scholar]
- Liu, H.X.; Zeng, S.X.; Wang, Y.R.; Li, P.; Chen, D.F.; Guo, J.Y. The effect of low temperature on superoxide dismutase in various organelles of cucumber seedling cotyledons with different cold tolerance. Acta Phytophysiol. Sin. 1985, 1, 48–57. [Google Scholar]
- Refat, A.B.; Shotaro, O.; Tijen, D.; Takuya, F.; Ikuo, S.; Tobias, I.B.; Hideaki, M.; Yoko, Y. Aluminium reduces sugar uptake in tobacco cell cultures: A potential cause of inhibited elongation but not of toxicity. J. Exp. Bot. 2010, 61, 1597–1610. [Google Scholar]
- Kügler, M.; Jansch, L.; Kruft, V.; Schmitz, U.K.; Braun, H.-P. Analysis of the chloroplast protein complexes by blue-native polyacrylamide gel electrophoresis (BN-PAGE). Photosynth. Res. 1997, 53, 35–44. [Google Scholar] [CrossRef]
- Wang, G.P.; Zhang, X.Y.; Li, F.; Luo, Y.; Wang, W. Overaccumulation of glycine betaine enhances tolerance to drought and heat stress in wheat leaves in the protection of photosynthesis. Photosynthetica 2010, 48, 117–126. [Google Scholar] [CrossRef]
- Bota, J.; Medrano, H.; Flexas, J. Is photosynthesis limited by decreased Rubisco activity and RuBP content under progressive water stress? New Phytol. 2004, 162, 671–681. [Google Scholar] [CrossRef]
- Parry, M.A.J.; Andralojc, P.J.; Parmar, S.; Keys, A.J.; Habash, D.; Paul, M.J.; Alred, R.; Quick, W.P.; Servaites, J.C. Regulation of Rubisco inhibitors in the light. Plant Cell Environ. 1997, 20, 528–534. [Google Scholar] [CrossRef]
- Balasaheb, V.S.; Robert, E.S.; Spencer, W.; Oula, G. Shade compromises the photosynthetic efficiency of NADP-ME less than that of PEP-CK and NAD-ME C4 grasses. J. Exp. Bot. 2018, 69, 3053–3068. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, D.; Wang, F.; Zhang, H.; Gao, W.; Tong, X.; Lv, C.; Chen, G. RETRACTED: Formation of Proto-Kranz in C3 Rice Induced by Spike-Stalk Injection Method. Int. J. Mol. Sci. 2021, 22, 4305. https://doi.org/10.3390/ijms22094305
Jiang D, Wang F, Zhang H, Gao W, Tong X, Lv C, Chen G. RETRACTED: Formation of Proto-Kranz in C3 Rice Induced by Spike-Stalk Injection Method. International Journal of Molecular Sciences. 2021; 22(9):4305. https://doi.org/10.3390/ijms22094305
Chicago/Turabian StyleJiang, Dexing, Feng Wang, Haizi Zhang, Wenwen Gao, Xi Tong, Chuangen Lv, and Guoxiang Chen. 2021. "RETRACTED: Formation of Proto-Kranz in C3 Rice Induced by Spike-Stalk Injection Method" International Journal of Molecular Sciences 22, no. 9: 4305. https://doi.org/10.3390/ijms22094305
APA StyleJiang, D., Wang, F., Zhang, H., Gao, W., Tong, X., Lv, C., & Chen, G. (2021). RETRACTED: Formation of Proto-Kranz in C3 Rice Induced by Spike-Stalk Injection Method. International Journal of Molecular Sciences, 22(9), 4305. https://doi.org/10.3390/ijms22094305




