MiRNAs and Muscle Regeneration: Therapeutic Targets in Duchenne Muscular Dystrophy
Abstract
1. Introduction
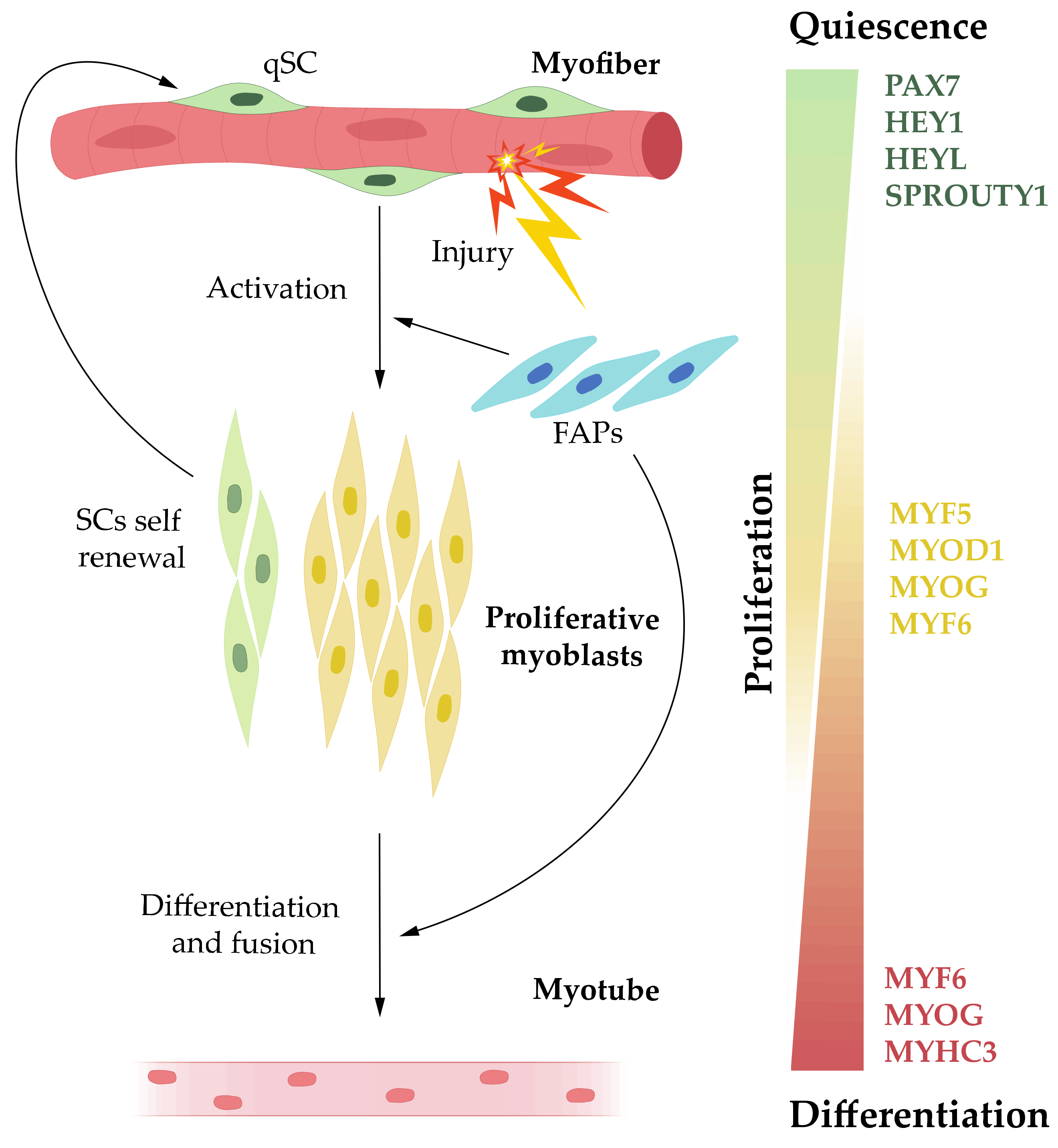
2. Muscle Regeneration
3. MicroRNAs in Muscle Biology
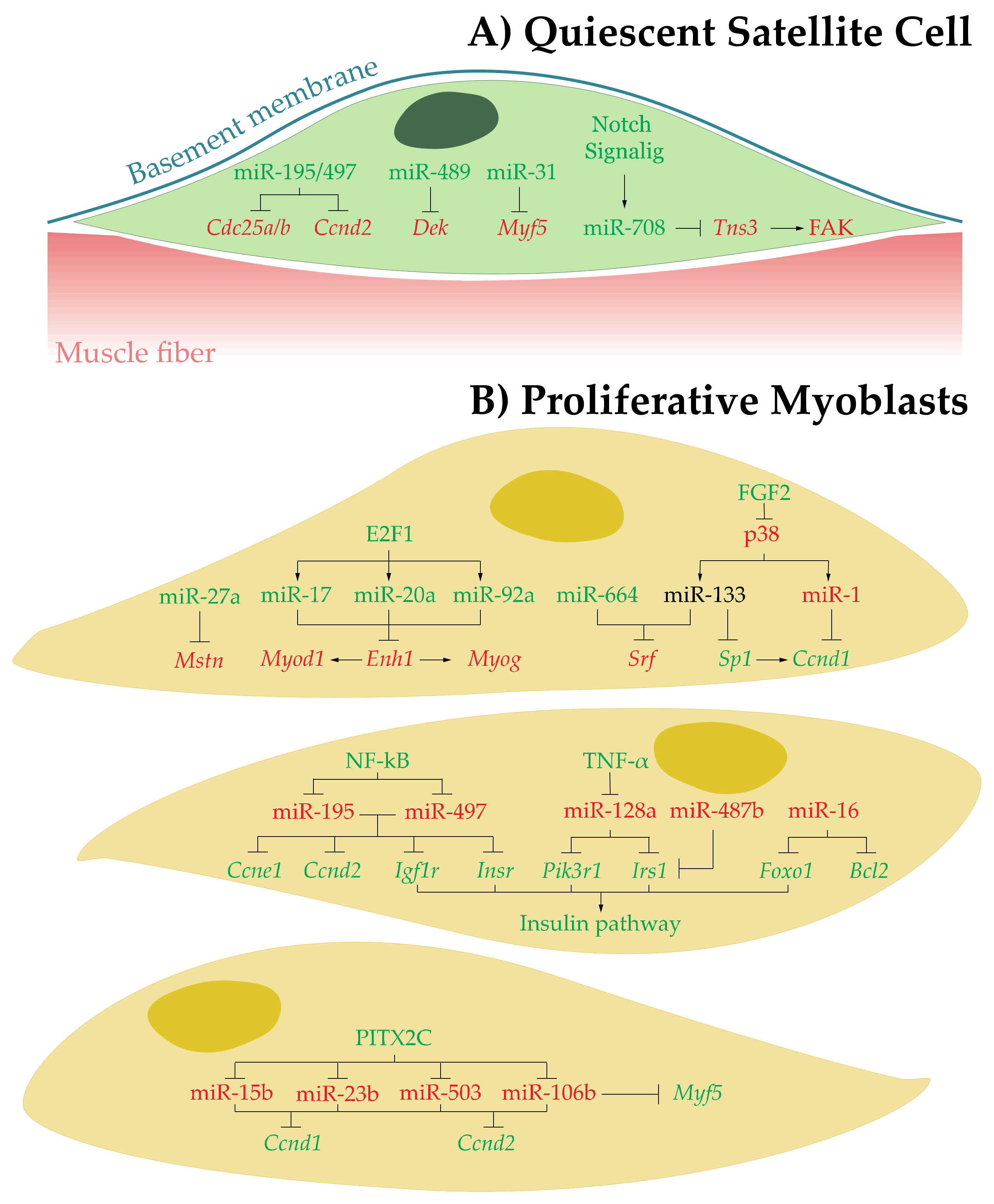
3.1. MicroRNAs in Quiescent SCs
3.2. MicroRNAs in Proliferative Myoblasts
3.2.1. Upregulated miRNAs
3.2.2. Downregulated miRNAs
| microRNAs | Targets | Function | References |
|---|---|---|---|
| Quiescent Satellite Cell State | |||
| miR-195/497↑ | Cdc25a/b and Ccnd2 | Promotes SCs quiescence by inducing cell cycle arrest | [45] |
| miR-489↑ | Dek | Regulates SCs quiescence | [46] |
| miR-31↑ | Myf5 | Prevents MYF5 protein accumulation and premature activation of SCs | [47] |
| miR-708↑ | Tns3 | Regulates quiescence and self-renewal by active repression of SCs migration | [48] |
| Proliferative Myoblast State | |||
| miR-27 ↑ | Mstn | Enhances and/or promotes myoblast proliferation | [51] |
| miR-17, miR-20a, and miR-92a↑ | Enh1 | Enhance and/or promotes myoblast proliferation | [53] |
| miR-664↑ and miR-133↑ | Srf | Enhances and/or promotes myoblast proliferation by inducing cell cycle genes expression | [54,55] |
| miR-133↓ | Sp1 | Inhibit myoblast proliferation | [57] |
| miR-1↓ | Ccnd1 | Inhibit myoblast proliferation | [57] |
| miR-195 and miR-497↓ | Igf1r, Insr, Ccne1 and Ccnd2 | Inhibit myoblast proliferation | [61] |
| miR-128a↓ | Irs1 and Pik3r1 | Inhibits myoblast proliferation | [67] |
| miR-487b↓ | Irs1 | Inhibits myoblast proliferation | [68] |
| miR-16↓ | Foxo1 and Bcl2 | Inhibits myoblast proliferation and promotes myoblast apoptosis | [71] |
| miR-15b, miR-23b, miR-106b, and miR-503↓ | Ccnd1 and Ccnd2 | Inhibits myoblast proliferation | [72] |
| miR-106b↓ | Myf5 | Prevents commitment to myogenic cell fate | [72] |
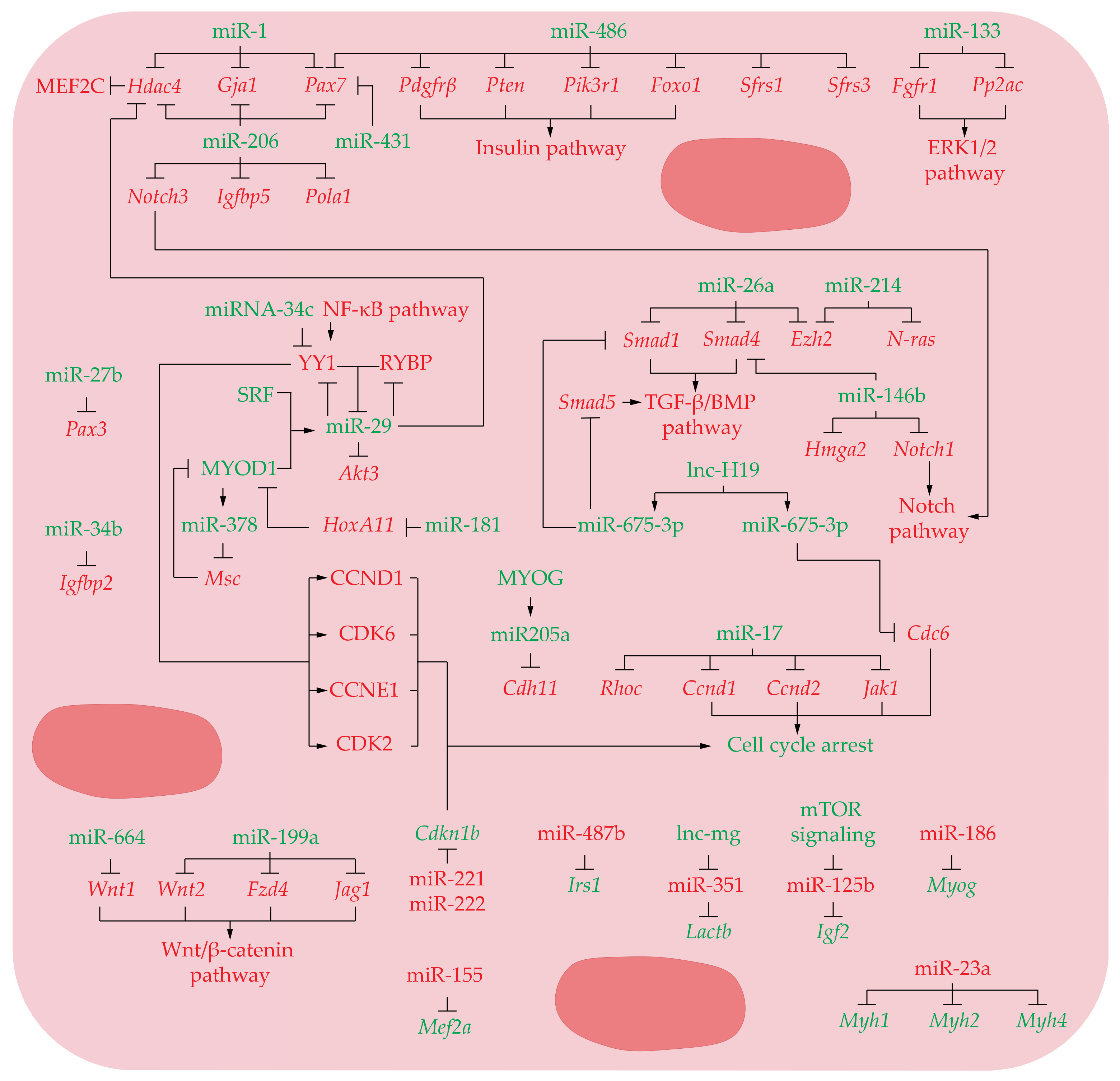
3.3. MicroRNAs in Differentiating Myoblast
3.3.1. Upregulated miRNAs
3.3.2. Downregulated miRNAs
| Differentiating Myoblasts State | |||
|---|---|---|---|
| miR-1↑, miR-206↑, and miR-29↑ | Hdac4 | Promotes myoblast differentiation | [54,77] |
| miR-1↑ and miR-206↑ | Gja1 | Promote myoblast fusion | [74] |
| miR-1↑ and miR-206↑ | Pax7 | Promote satellite cell differentiation and restrict their proliferative potential | [17] |
| miR-206↑ | Pax7 | Activates myoblast differentiation | [75] |
| miR-206↑ | Pax7, Notch3, and Igfbp5 | Stimulates SC differentiation and skeletal muscle regeneration | [76] |
| miR-206↑ | Pola1 | Promotes myoblast differentiation by inducing a cell cycle arrest | [78] |
| miR-486↑ | Pax7 | Promote initial muscle differentiation | [75] |
| miR-486↑ | Pdgfrβ, Pten, Pik3r1, Foxo1, Sfrs1, and Sfrs3 | Promotes myoblast differentiation by inhibiting PTEN/AKT pathway and splicing factors | [80] |
| miR-431↑ | Pax7 | Mediates satellite cell heterogeneity and promotes muscle differentiation | [79] |
| miR-133↑ | Fgfr1 and Pp2ac | Promotes muscle precursor cells differentiation | [82] |
| miR-27b↑ | Pax3 | Ensures rapid and robust entry into the myogenic differentiation program | [83] |
| miR-29↑ | Rybp and Yy1 | Ensures proper myoblast differentiation into myotubes | [85,86] |
| miR-29↑ | Akt3 | Reduces proliferation and facilitate differentiation of precursor muscle cells | [87] |
| miR-26a↑ | Smad1 and Smad4 | Promotes myoblast differentiation | [88] |
| miR-26a↑ and miR-214↑ | Ezh2 | Induces muscle cell differentiation | [91,92] |
| miR-214↑ | N-ras | Promotes myogenic differentiation by facilitating exit from mitosis | [93] |
| miR-146b↑ | Smad4, Notch1, and Hmga2 | Promotes myogenic differentiation | [94] |
| miR-675-3p↑ | Smad1 and Smad5 | Promotes myogenic differentiation downregulates the BMP pathway | [99] |
| miR-675-5p↑ | Cdc6 | Promotes myogenic differentiation by repression of DNA replication | [99] |
| miR-181↑ | HoxA11 | Promotes myogenic differentiation | [108] |
| miR-378↑ | Msc | Promotes myogenic differentiation | [109] |
| miR-205a↑ | Cdh11 | Inhibits myoblast proliferation and promote myoblast differentiation | [110] |
| miR-17↑ | Ccnd1, Ccnd2, Jak1, and Rhoc | Promotes differentiation of precursor muscle cells by inducing cell cycle arrest and extracellular matrix expression | [100] |
| miR-34b↑ | Igfbp2 | Represses proliferation and promotes differentiation of myoblasts | [106] |
| miR-34c↑ | Yy1 | Represses proliferation and promotes differentiation of myoblasts by leading to G0/G1 arrest | [107] |
| miR-664↑ | Wnt1 | Downregulates Wnt/β-catenin signaling pathway to allow normal myogenic differentiation | [55] |
| miR-199a↑ | Wnt2, Fzd4, and Jag1 | Downregulates Wnt/β-catenin signaling pathway to allow normal myogenic differentiation | [111] |
| miR-155↓ | Mef2a | Represses myoblast differentiation | [115] |
| miR-487b↓ | Irs1 | Suppresses the proliferation and differentiation of myoblasts by repressing PI3K/Akt and MAPK/Erk pathways | [68] |
| miR-221 and 222↓ | Cdkn1b | Inhibits myoblast differentiation by allowing myoblast to proliferate and avoiding to acquire myotube morphology | [112] |
| miR-351↓ | Lactb | Represses myoblast differentiation | [113] |
| miR-125b↓ | Igf2 | Represses myoblast differentiation | [118] |
| miR-186↓ | Myog | Represses myoblast differentiation | [119] |
| miR-23a↓ | Myh1, Myh2, and Myh4 | Prevents myogenic differentiation by inhibiting myosin genes expression | [120] |
4. Regulatory Role of miRNAs in Muscle Regeneration as a Therapeutic Target in DMD
| Muscle Regeneration Event | Target microRNA | Molecular Approach | Experimental System | Reference |
|---|---|---|---|---|
| Duchenne Muscular Dystrophy | ||||
| Fibrosis and Inflammation | miR-29 | miRNA Mimic | Human DMD myoblast | [130] |
| miRNA Mimic | mdx mice | [129] | ||
| miR-29 + micro-dystrophin overexpression by AAV | mdx/utrn+/− mice | [131] | ||
| miR-206 | AntagomiR Sponge | mdx mice | [132] | |
| miR-21 | AntagomiR | Human DMD fibroblasts Mdx mice | [130] | |
| AntagomiR | Senescence mdx mice | [133] | ||
| miR-146a | KO | mdx mice | [134] | |
| Muscle differentiation | miR-29 | miRNA Mimic | Human DMD myoblast | [130] |
| miRNA Mimic | mdx mice | [129] | ||
| miR-31 | AntagomiR or Sponges + exon skipping | Human DMD myoblasts | [135] | |
| miR-431 | Transgenic overexpression | mdx mice | [79] | |
| miR-127 | siRNA mimic Transgenic overexpression | C2C12 myoblasts mdx mice | [136] | |
Current and Future Aplicable Technologies for miRNAs Modulation in DMD
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Baghdadi, M.B.; Tajbakhsh, S. Regulation and Phylogeny of Skeletal Muscle Regeneration. Dev. Biol. 2018, 433, 200–209. [Google Scholar] [CrossRef]
- Sutcu, H.H.; Ricchetti, M. Loss of Heterogeneity, Quiescence, and Differentiation in Muscle Stem Cells. Stem Cell Investig. 2018, 5. [Google Scholar] [CrossRef]
- Sacco, A.; Puri, P.L. Regulation of Muscle Satellite Cell Function in Tissue Homeostasis and Aging. Cell Stem Cell 2015, 16, 585–587. [Google Scholar] [CrossRef]
- Bentzinger, C.F.; Wang, Y.X.; Rudnicki, M.A. Building Muscle: Molecular Regulation of Myogenesis. Cold Spring Harb. Perspect. Biol. 2012, 4, a008342. [Google Scholar] [CrossRef] [PubMed]
- Dumont, N.A.; Bentzinger, C.F.; Sincennes, M.C.; Rudnicki, M.A. Satellite Cells and Skeletal Muscle Regeneration. Compr. Physiol. 2015, 5, 1027–1059. [Google Scholar] [CrossRef]
- Dumont, N.A.; Wang, Y.X.; Rudnicki, M.A. Intrinsic and Extrinsic Mechanisms Regulating Satellite Cell Function. Development 2015, 142, 1572–1581. [Google Scholar] [CrossRef] [PubMed]
- Musarò, A. The Basis of Muscle Regeneration. Adv. Biol. 2014, 2014, 1–16. [Google Scholar] [CrossRef]
- Perandini, L.A.; Chimin, P.; da Silva Lutkemeyer, D.; Câmara, N.O.S. Chronic Inflammation in Skeletal Muscle Impairs Satellite Cells Function during Regeneration: Can Physical Exercise Restore the Satellite Cell Niche? FEBS J. 2018, 285, 1973–1984. [Google Scholar] [CrossRef] [PubMed]
- Berardi, E.; Annibali, D.; Cassano, M.; Crippa, S.; Sampaolesi, M. Molecular and Cell-Based Therapies for Muscle Degenerations: A Road under Construction. Front. Physiol. 2014, 5, 119. [Google Scholar] [CrossRef]
- Chal, J.; Oginuma, M.; Al Tanoury, Z.; Gobert, B.; Sumara, O.; Hick, A.; Bousson, F.; Zidouni, Y.; Mursch, C.; Moncuquet, P.; et al. Differentiation of Pluripotent Stem Cells to Muscle Fiber to Model Duchenne Muscular Dystrophy. Nat. Biotechnol. 2015, 33, 962–969. [Google Scholar] [CrossRef] [PubMed]
- Dumont, N.A.; Wang, Y.X.; von Maltzahn, J.; Pasut, A.; Bentzinger, C.F.; Brun, C.E.; Rudnicki, M.A. Dystrophin Expression in Muscle Stem Cells Regulates Their Polarity and Asymmetric Division. Nat. Med. 2015, 21, 1455–1463. [Google Scholar] [CrossRef] [PubMed]
- Glaich, O.; Parikh, S.; Bell, R.E.; Mekahel, K.; Donyo, M.; Leader, Y.; Shayevitch, R.; Sheinboim, D.; Yannai, S.; Hollander, D.; et al. DNA Methylation Directs MicroRNA Biogenesis in Mammalian Cells. Nat. Commun. 2019, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Broughton, J.P.; Lovci, M.T.; Huang, J.L.; Yeo, G.W.; Pasquinelli, A.E. Pairing beyond the Seed Supports MicroRNA Targeting Specificity. Mol. Cell 2016, 64, 320–333. [Google Scholar] [CrossRef]
- Vasudevan, S.; Steitz, J.A. AU-Rich-Element-Mediated Upregulation of Translation by FXR1 and Argonaute 2. Cell 2007, 128, 1105–1118. [Google Scholar] [CrossRef] [PubMed]
- Truesdell, S.S.; Mortensen, R.D.; Seo, M.; Schroeder, J.C.; Lee, J.H.; Letonqueze, O.; Vasudevan, S. MicroRNA-Mediated MRNA Translation Activation in Quiescent Cells and Oocytes Involves Recruitment of a Nuclear MicroRNP. Sci. Rep. 2012, 2, 1–12. [Google Scholar] [CrossRef]
- Bukhari, S.I.A.; Truesdell, S.S.; Lee, S.; Kollu, S.; Classon, A.; Boukhali, M.; Jain, E.; Mortensen, R.D.; Yanagiya, A.; Sadreyev, R.I.; et al. A Specialized Mechanism of Translation Mediated by FXR1a-Associated MicroRNP in Cellular Quiescence. Mol. Cell 2016, 61, 760–773. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.F.; Tao, Y.; Li, J.; Deng, Z.; Yan, Z.; Xiao, X.; Wang, D.Z. MicroRNA-1 and MicroRNA-206 Regulate Skeletal Muscle Satellite Cell Proliferation and Differentiation by Repressing Pax7. J. Cell Biol. 2010, 190, 867–879. [Google Scholar] [CrossRef]
- Mauro A Satellite Cell of Skeletal Muscle Fibers. J. Biophys. Biochem. Cytol. 1961, 9, 493–495. [CrossRef]
- Shea, K.L.; Xiang, W.; LaPorta, V.S.; Licht, J.D.; Keller, C.; Basson, M.A.; Brack, A.S. Sprouty1 Regulates Reversible Quiescence of a Self-Renewing Adult Muscle Stem Cell Pool during Regeneration. Cell Stem Cell 2010, 6, 117–129. [Google Scholar] [CrossRef]
- Fukada, S.I.; Yamaguchi, M.; Kokubo, H.; Ogawa, R.; Uezumi, A.; Yoneda, T.; Matev, M.M.; Motohashi, N.; Ito, T.; Zolkiewska, A.; et al. Hesr1 and Hesr3 Are Essential to Generate Undifferentiated Quiescent Satellite Cells and to Maintain Satellite Cell Numbers. Development 2011. [Google Scholar] [CrossRef]
- Bjornson, C.R.R.; Cheung, T.H.; Liu, L.; Tripathi, P.V.; Steeper, K.M.; Rando, T.A. Notch Signaling Is Necessary to Maintain Quiescence in Adult Muscle Stem Cells. Stem Cells 2012, 30, 232–242. [Google Scholar] [CrossRef]
- Philippos, M.; Sambasivan, R.; Castel, D.; Rocheteau, P.; Bizzarro, V.; Tajbakhsh, S. A Critical Requirement for Notch Signaling in Maintenance of the Quiescent Skeletal Muscle Stem Cell State. Stem Cells 2012, 30, 243–252. [Google Scholar] [CrossRef]
- Rodgers, J.T.; King, K.Y.; Brett, J.O.; Cromie, M.J.; Charville, G.W.; Maguire, K.K.; Brunson, C.; Mastey, N.; Liu, L.; Tsai, C.-R.; et al. MTORC1 Controls the Adaptive Transition of Quiescent Stem Cells from G0 to G(Alert). Nature 2014, 510, 393–396. [Google Scholar] [CrossRef]
- Stark, D.A.; Karvas, R.M.; Siege, A.L.; Cornelison, D.D.W. Eph/Ephrin Interactions Modulate Muscle Satellite Cell Motility and Patterning. Development 2011. [Google Scholar] [CrossRef] [PubMed]
- Bentzinger, C.F.; von Maltzahn, J.; Dumont, N.A.; Stark, D.A.; Wang, Y.X.; Nhan, K.; Frenette, J.; Cornelison, D.D.W.; Rudnicki, M.A. Wnt7a Stimulates Myogenic Stem Cell Motility and Engraftment Resulting in Improved Muscle Strength. J. Cell Biol. 2014. [Google Scholar] [CrossRef] [PubMed]
- Davie, J.K.; Cho, J.H.; Meadows, E.; Flynn, J.M.; Knapp, J.R.; Klein, W.H. Target Gene Selectivity of the Myogenic Basic Helix-Loop-Helix Transcription Factor Myogenin in Embryonic Muscle. Dev. Biol. 2007. [Google Scholar] [CrossRef][Green Version]
- Von Maltzahn, J.; Bentzinger, C.F.; Rudnicki, M.A. Characteristics of Satellite Cells and Multipotent Adult Stem Cells in the Skeletal Muscle. In Stem Cells and Cancer; Springer: Berlin/Heidelberg, Germany, 2014; Volume 12, pp. 63–73. [Google Scholar]
- Wosczyna, M.N.; Rando, T.A. A Muscle Stem Cell Support Group: Coordinated Cellular Responses in Muscle Regeneration. Physiol. Behav. 2019, 176, 139–148. [Google Scholar] [CrossRef]
- Joe, A.W.B.; Yi, L.; Natarajan, A.; le Grand, F.; So, L.; Wang, J.; Rudnicki, M.A.; Rossi, F.M.V. Muscle Injury Activates Resident Fibro/Adipogenic Progenitors That Facilitate Myogenesis. Physiol. Behav. 2010, 176, 139–148. [Google Scholar] [CrossRef]
- Uezumi, A.; Ito, T.; Morikawa, D.; Shimizu, N.; Yoneda, T.; Segawa, M.; Yamaguchi, M.; Ogawa, R.; Matev, M.M.; Miyagoe-Suzuki, Y.; et al. Fibrosis and Adipogenesis Originate from a Common Mesenchymal Progenitor in Skeletal Muscle. J. Cell Sci. 2011, 124, 3654–3664. [Google Scholar] [CrossRef]
- Fiore, D.; Judson, R.N.; Low, M.; Lee, S.; Zhang, E.; Hopkins, C.; Xu, P.; Lenzi, A.; Rossi, F.M.V.; Lemos, D.R. Pharmacological Blockage of Fibro/Adipogenic Progenitor Expansion and Suppression of Regenerative Fibrogenesis Is Associated with Impaired Skeletal Muscle Regeneration. Stem Cell Res. 2016, 17, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Uezumi, A.; Fukada, S.I.; Yamamoto, N.; Takeda, S.; Tsuchida, K. Mesenchymal Progenitors Distinct from Satellite Cells Contribute to Ectopic Fat Cell Formation in Skeletal Muscle. Nat. Cell Biol. 2010. [Google Scholar] [CrossRef] [PubMed]
- Heredia, J.E.; Mukundan, L.; Chen, F.M.; Mueller, A.A.; Deo, R.C.; Locksley, R.M.; Rando, T.A.; Chawla, A. Type 2 Innate Signals Stimulate Fibro/Adipogenic Progenitors to Facilitate Muscle Regeneration. Cell 2013, 153, 376–388. [Google Scholar] [CrossRef]
- Pillon, N.J.; Bilan, P.J.; Fink, L.N.; Klip, A. Cross-Talk between Skeletal Muscle and Immune Cells: Muscle-Derived Mediators and Metabolic Implications. Am. J. Physiol. Endocrinol. Metab. 2013, 304, E453–E465. [Google Scholar] [CrossRef] [PubMed]
- Mozzetta, C.; Consalvi, S.; Saccone, V.; Tierney, M.; Diamantini, A.; Mitchell, K.J.; Marazzi, G.; Borsellino, G.; Battistini, L.; Sassoon, D.; et al. Fibroadipogenic Progenitors Mediate the Ability of HDAC Inhibitors to Promote Regeneration in Dystrophic Muscles of Young, but Not Old Mdx Mice. EMBO Mol. Med. 2013, 5, 626–639. [Google Scholar] [CrossRef]
- Paneru, B.D.; Al-Tobasei, R.; Kenney, B.; Leeds, T.D.; Salem, M. RNA-Seq Reveals MicroRNA Expression Signature and Genetic Polymorphism Associated with Growth and Muscle Quality Traits in Rainbow Trout. Sci. Rep. 2017, 7, 1–15. [Google Scholar] [CrossRef]
- Castel, D.; Baghdadi, M.B.; Mella, S.; Gayraud-Morel, B.; Marty, V.; Cavaillé, J.; Antoniewski, C.; Tajbakhsh, S. Small-RNA Sequencing Identifies Dynamic MicroRNA Deregulation during Skeletal Muscle Lineage Progression. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef]
- Coenen-Stass, A.M.L.; Sork, H.; Gatto, S.; Godfrey, C.; Bhomra, A.; Krjutškov, K.; Hart, J.R.; Westholm, J.O.; O’Donovan, L.; Roos, A.; et al. Comprehensive RNA-Sequencing Analysis in Serum and Muscle Reveals Novel Small RNA Signatures with Biomarker Potential for DMD. Mol. Ther. Nucleic Acids 2018, 13, 1–15. [Google Scholar] [CrossRef]
- Liu, Z.; Li, C.; Li, X.; Yao, Y.; Ni, W.; Zhang, X.; Cao, Y.; Hazi, W.; Wang, D.; Quan, R.; et al. Expression Profiles of MicroRNAs in Skeletal Muscle of Sheep by Deep Sequencing. Asian Australas. J. Anim. Sci. 2019, 32, 757–766. [Google Scholar] [CrossRef]
- Sempere, L.F.; Freemantle, S.; Pitha-Rowe, I.; Moss, E.; Dmitrovsky, E.; Ambros, V. Expression Profiling of Mammalian MicroRNAs Uncovers a Subset of Brain-Expressed MicroRNAs with Possible Roles in Murine and Human Neuronal Differentiation. Genome Biol. 2004, 5. [Google Scholar] [CrossRef]
- Van Rooij, E.; Sutherland, L.B.; Qi, X.; Richardson, J.A.; Hill, J.; Olson, E.N. Control of Stress-Dependent Cardiac Growth and Gene Expression by a MicroRNA. Science 2007, 316, 575–579. [Google Scholar] [CrossRef] [PubMed]
- Van Rooij, E.; Quiat, D.; Johnson, B.A.; Sutherland, L.B.; Qi, X.; Richardson, J.A.; Kelm, R.J.; Olson, E.N. A Family of MicroRNAs Encoded by Myosin Genes Governs Myosin Expression and Muscle Performance. Dev. Cell 2009, 17, 662–673. [Google Scholar] [CrossRef]
- Small, E.M.; O’Rourke, J.R.; Moresi, V.; Sutherland, L.B.; McAnally, J.; Gerard, R.D.; Richardson, J.A.; Olson, E.N. Regulation of PI3-Kinase/Akt Signaling by Muscle-Enriched MicroRNA-486. Proc. Natl. Acad. Sci. USA 2010, 107, 4218–4223. [Google Scholar] [CrossRef]
- Diniz, G.P.; Wang, D.Z. Regulation of Skeletal Muscle by Micro RNAs. Compr. Physiol. 2016, 6, 1279–1294. [Google Scholar] [CrossRef]
- Sato, T.; Yamamoto, T.; Sehara-Fujisawa, A. MiR-195/497 Induce Postnatal Quiescence of Skeletal Muscle Stem Cells. Nat. Commun. 2014, 5, 4597. [Google Scholar] [CrossRef] [PubMed]
- Cheung, T.H.; Quach, N.L.; Charville, G.W.; Liu, L.; Park, L.; Edalati, A.; Yoo, B.; Hoang, P.; Rando, T.A. Maintenance of Muscle Stem-Cell Quiescence by MicroRNA-489. Nature 2012, 482, 524–528. [Google Scholar] [CrossRef]
- Crist, C.G.; Montarras, D.; Buckingham, M. Muscle Satellite Cells Are Primed for Myogenesis but Maintain Quiescence with Sequestration of Myf5 MRNA Targeted by MicroRNA-31 in MRNP Granules. Cell Stem Cell 2012, 11, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Baghdadi, M.B.; Firmino, J.; Soni, K.; Evano, B.; di Girolamo, D.; Mourikis, P.; Castel, D.; Tajbakhsh, S. Notch-Induced MiR-708 Antagonizes Satellite Cell Migration and Maintains Quiescence. Cell Stem Cell 2018, 23, 859–868.e5. [Google Scholar] [CrossRef]
- Khodadoust, M.S.; Verhaegen, M.; Kappes, F.; Riveiro-Falkenbach, E.; Cigudosa, J.C.; Kim, D.S.L.; Chinnaiyan, A.M.; Markovitz, D.M.; Soengas, M.S. Melanoma Proliferation and Chemoresistance Controlled by the DEK Oncogene. Cancer Res. 2009, 69, 6405–6413. [Google Scholar] [CrossRef]
- Mendes Soares, L.M.; Zanier, K.; Mackereth, C.; Sattler, M.; Valcárcel, J. Intron Removal Requires Proofreading of U2AF/3′ Splice Site Recognition by DEK. Science 2006, 312, 1961–1965. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Chen, X.; Yu, B.; He, J.; Chen, D. MicroRNA-27a Promotes Myoblast Proliferation by Targeting Myostatin. Biochem. Biophys. Res. Commun. 2012, 423, 265–269. [Google Scholar] [CrossRef]
- Joulia, D.; Bernardi, H.; Garandel, V.; Rabenoelina, F.; Vernus, B.; Cabello, G. Mechanisms Involved in the Inhibition of Myoblast Proliferation and Differentiation by Myostatin. Exp. Cell Res. 2003, 286, 263–275. [Google Scholar] [CrossRef]
- Qiu, H.; Liu, N.; Luo, L.; Zhong, J.; Tang, Z.; Kang, K.; Qu, J.; Peng, W.; Liu, L.; Li, L.; et al. MicroRNA-17-92 Regulates Myoblast Proliferation and Differentiation by Targeting the ENH1/Id1 Signaling Axis. Cell Death Differ. 2016, 23, 1658–1669. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.F.; Mandel, E.M.; Thomson, J.M.; Wu, Q.; Callis, T.E.; Hammond, S.M.; Conlon, F.L.; Wang, D.Z. The Role of MicroRNA-1 and MicroRNA-133 in Skeletal Muscle Proliferation and Differentiation. Nat. Genet. 2006, 38, 228–233. [Google Scholar] [CrossRef]
- Cai, R.; Qimuge, N.; Ma, M.; Wang, Y.; Tang, G.; Zhang, Q.; Sun, Y.; Chen, X.; Yu, T.; Dong, W.; et al. MicroRNA-664-5p Promotes Myoblast Proliferation and Inhibits Myoblast Differentiation by Targeting Serum Response Factor and Wnt1. J. Biol. Chem. 2018, 293, 19177–19190. [Google Scholar] [CrossRef]
- Wallace, M.A.; della Gatta, P.A.; Ahmad Mir, B.; Kowalski, G.M.; Kloehn, J.; McConville, M.J.; Russell, A.P.; Lamon, S. Overexpression of Striated Muscle Activator of Rho Signaling (STARS) Increases C2C12 Skeletal Muscle Cell Differentiation. Front. Physiol. 2016, 7, 7. [Google Scholar] [CrossRef]
- Zhang, D.; Li, X.; Chen, C.; Li, Y.; Zhao, L.; Jing, Y.; Liu, W.; Wang, X.; Zhang, Y.; Xia, H.; et al. Attenuation of P38-Mediated MiR-1/133 Expression Facilitates Myoblast Proliferation during the Early Stage of Muscle Regeneration. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Bakkar, N.; Wang, J.; Ladner, K.J.; Wang, H.; Dahlman, J.M.; Carathers, M.; Acharyya, S.; Rudnicki, M.A.; Hollenbach, A.D.; Guttridge, D.C. IKK/NF-ΚB Regulates Skeletal Myogenesis via a Signaling Switch to Inhibit Differentiation and Promote Mitochondrial Biogenesis. J. Cell Biol. 2008, 180, 787–802. [Google Scholar] [CrossRef]
- Langen, R.C.J.; Schols, A.M.W.J.; Kelders, M.C.J.M.; Wouters, E.F.M.; Janssen-Heininger, Y.M.W. Inflammatory Cytokines Inhibit Myogenic Differentiation through Activation of Nuclear Factor-κΒ. FASEB J. 2001, 15, 1169–1180. [Google Scholar] [CrossRef] [PubMed]
- Guttridge, D.C.; Albanese, C.; Reuther, J.Y.; Pestell, R.G.; Baldwin, A.S. NF-ΚB Controls Cell Growth and Differentiation through Transcriptional Regulation of Cyclin D1. Mol. Cell. Biol. 1999, 19, 5785–5799. [Google Scholar] [CrossRef]
- Wei, W.; Zhang, W.Y.; Bai, J.B.; Zhang, H.X.; Zhao, Y.Y.; Li, X.Y.; Zhao, S.H. The NF-ΚB-Modulated MicroRNAs MiR-195 and MiR-497 Inhibit Myoblast Proliferation by Targeting Igf1r, Insr and Cyclin Genes. J. Cell Sci. 2016, 129, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Alter, J.; Rozentzweig, D.; Bengal, E. Inhibition of Myoblast Differentiation by Tumor Necrosis Factor Ais Mediated by C-Jun N-Terminal Kinase 1 and Leukemia Inhibitory Factor. J. Biol. Chem. 2008, 283, 23224–23234. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.E.; Jin, B.; Li, Y.P. TNF-α Regulates Myogenesis and Muscle Regeneration by Activating P38 MAPK. Am. J. Physiol. Cell Physiol. 2007, 292. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.P. TNF-α Is a Mitogen in Skeletal Muscle. Am. J. Physiol. Cell Physiol. 2003, 285. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-P.; Schwartz, R.J. TNF-α Regulates Early Differentiation of C2C12 Myoblasts in an Autocrine Fashion. FASEB J. 2001, 15, 1413–1415. [Google Scholar] [CrossRef] [PubMed]
- Warren, G.L.; Hulderman, T.; Jensen, N.; McKinstry, M.; Mishra, M.; Luster, M.I.; Simeonova, P.P. Physiological Role of Tumor Necrosis Factor Alpha in Traumatic Muscle Injury. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2002, 16, 1630–1632. [Google Scholar] [CrossRef]
- Motohashi, N.; Alexander, M.S.; Shimizu-Motohashi, Y.; Myers, J.A.; Kawahara, G.; Kunkel, L.M. Regulation of IRS1/Akt Insulin Signaling by MicroRNA-128a during Myogenesis. J. Cell Sci. 2013, 126, 2678–2691. [Google Scholar] [CrossRef]
- Wang, J.; Tan, J.; Qi, Q.; Yang, L.; Wang, Y.; Zhang, C.; Hu, L.; Chen, H.; Fang, X. MiR-487b-3p Suppresses the Proliferation and Differentiation of Myoblasts by Targeting IRS1 in Skeletal Muscle Myogenesis. Int. J. Biol. Sci. 2018, 14, 760–774. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, W.A.; Ahad, A.; Ahsan, H. The Mystery of BCL2 Family: Bcl-2 Proteins and Apoptosis: An Update. Arch. Toxicol. 2015, 89, 289–317. [Google Scholar] [CrossRef] [PubMed]
- Flück, M.; Carson, J.A.; Gordon, S.E.; Ziemiecki, A.; Booth, F.W. Focal Adhesion Proteins FAK and Paxillin Increase in Hypertrophied Skeletal Muscle. Am. J. Physiol. Cell Physiol. 1999, 277. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.; Ouyang, H.; Abdalla, B.A.; Xu, H.; Nie, Q.; Zhang, X. MiR-16 Controls Myoblast Proliferation and Apoptosis through Directly Suppressing Bcl2 and FOXO1 Activities. Biochim. Biophys. Acta Gene Regul. Mech. 2017, 1860, 674–684. [Google Scholar] [CrossRef] [PubMed]
- Lozano-Velasco, E.; Vallejo, D.; Esteban, F.J.; Doherty, C.; Hernández-Torres, F.; Franco, D.; Aránega, A.E. A Pitx2 -MicroRNA Pathway Modulates Cell Proliferation in Myoblasts and Skeletal-Muscle Satellite Cells and Promotes Their Commitment to a Myogenic Cell Fate. Mol. Cell. Biol. 2015, 35, 2892–2909. [Google Scholar] [CrossRef]
- Lu, J.; McKinsey, T.A.; Zhang, C.L.; Olson, E.N. Regulation of Skeletal Myogenesis by Association of the MEF2 Transcription Factor with Class II Histone Deacetylases. Mol. Cell 2000, 6, 233–244. [Google Scholar] [CrossRef]
- Anderson, C.; Catoe, H.; Werner, R. MIR-206 Regulates Connexin43 Expression during Skeletal Muscle Development. Nucleic Acids Res. 2006, 34, 5863–5871. [Google Scholar] [CrossRef]
- Dey, B.K.; Gagan, J.; Dutta, A. MiR-206 and -486 Induce Myoblast Differentiation by Downregulating Pax7. Mol. Cell. Biol. 2011, 31, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Williams, A.H.; Maxeiner, J.M.; Bezprozvannaya, S.; Shelton, J.M.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. MicroRNA-206 Promotes Skeletal Muscle Regeneration and Delays Progression of Duchenne Muscular Dystrophy in Mice. J. Clin. Investig. 2012, 122, 2054–2065. [Google Scholar] [CrossRef]
- Winbanks, C.E.; Wang, B.; Beyer, C.; Koh, P.; White, L.; Kantharidis, P.; Gregorevic, P. TGF-β Regulates MiR-206 and MiR-29 to Control Myogenic Differentiation through Regulation of HDAC4. J. Biol. Chem. 2011, 286, 13805–13814. [Google Scholar] [CrossRef] [PubMed]
- Hak, K.K.; Yong, S.L.; Sivaprasad, U.; Malhotra, A.; Dutta, A. Muscle-Specific MicroRNA MiR-206 Promotes Muscle Differentiation. J. Cell Biol. 2006, 174, 677–687. [Google Scholar] [CrossRef]
- Wu, R.; Li, H.; Zhai, L.; Zou, X.; Meng, J.; Zhong, R.; Li, C.; Wang, H.; Zhang, Y.; Zhu, D. MicroRNA-431 Accelerates Muscle Regeneration and Ameliorates Muscular Dystrophy by Targeting Pax7 in Mice. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Alexander, M.S.; Casar, J.C.; Motohashi, N.; Myers, J.A.; Eisenberg, I.; Gonzalez, R.T.; Estrella, E.A.; Kang, P.B.; Kawahara, G.; Kunkel, L.M. Regulation of DMD Pathology by an Ankyrin-Encoded MiRNA. Skelet. Muscle 2011, 1, 27. [Google Scholar] [CrossRef]
- Jones, N.C.; Fedorov, Y.V.; Rosenthal, R.S.; Olwin, B.B. ERK1/2 Is Required for Myoblast Proliferation but Is Dispensable for Muscle Gene Expression and Cell Fusion. J. Cell. Physiol. 2001, 186, 104–115. [Google Scholar] [CrossRef]
- Feng, Y.; Niu, L.L.; Wei, W.; Zhang, W.Y.; Li, X.Y.; Cao, J.H.; Zhao, S.H. A Feedback Circuit between MiR-133 and the ERK1/2 Pathway Involving an Exquisite Mechanism for Regulating Myoblast Proliferation and Differentiation. Cell Death Dis. 2013, 4, e934. [Google Scholar] [CrossRef]
- Crist, C.G.; Montarras, D.; Pallafacchina, G.; Rocancourt, D.; Cumano, A.; Conway, S.J.; Buckingham, M. Muscle Stem Cell Behavior Is Modified by MicroRNA-27 Regulation of Pax3 Expression. Proc. Natl. Acad. Sci. USA 2009, 106, 13383–13387. [Google Scholar] [CrossRef]
- Relaix, F.; Montarras, D.; Zaffran, S.; Gayraud-Morel, B.; Rocancourt, D.; Tajbakhsh, S.; Mansouri, A.; Cumano, A.; Buckingham, M. Pax3 and Pax7 Have Distinct and Overlapping Functions in Adult Muscle Progenitor Cells. J. Cell Biol. 2006, 172, 91–102. [Google Scholar] [CrossRef]
- Wang, H.; Garzon, R.; Sun, H.; Ladner, K.J.; Singh, R.; Dahlman, J.; Cheng, A.; Hall, B.M.; Qualman, S.J.; Chandler, D.S.; et al. NF-ΚB-YY1-MiR-29 Regulatory Circuitry in Skeletal Myogenesis and Rhabdomyosarcoma. Cancer Cell 2008, 14, 369–381. [Google Scholar] [CrossRef]
- Zhou, L.; Wang, L.; Lu, L.; Jiang, P.; Sun, H.; Wang, H. A Novel Target of MicroRNA-29, Ring1 and YY1-Binding Protein (Rybp), Negatively Regulates Skeletal Myogenesis. J. Biol. Chem. 2012, 287, 25255–25265. [Google Scholar] [CrossRef]
- Wei, W.; He, H.B.; Zhang, W.Y.; Zhang, H.X.; Bai, J.B.; Liu, H.Z.; Cao, J.H.; Chang, K.C.; Li, X.Y.; Zhao, S.H. MiR-29 Targets Akt3 to Reduce Proliferation and Facilitate Differentiation of Myoblasts in Skeletal Muscle Development. Cell Death Dis. 2013, 4, e668. [Google Scholar] [CrossRef] [PubMed]
- Dey, B.K.; Gagan, J.; Yan, Z.; Dutta, A. MiR-26a Is Required for Skeletal Muscle Differentiation and Regeneration in Mice. Genes Dev. 2012, 26, 2180–2191. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Noulet, F.; Edom-Vovard, F.; le Grand, F.; Duprez, D. Bmp Signaling at the Tips of Skeletal Muscles Regulates the Number of Fetal Muscle Progenitors and Satellite Cells during Development. Dev. Cell 2010, 18, 643–654. [Google Scholar] [CrossRef]
- Caretti, G.; di Padova, M.; Micales, B.; Lyons, G.E.; Sartorelli, V. The Polycomb Ezh2 Methyltransferase Regulates Muscle Gene Expression and Skeletal Muscle Differentiation. Genes Dev. 2004, 18, 2627–2638. [Google Scholar] [CrossRef] [PubMed]
- Chung, F.W.; Tellam, R.L. MicroRNA-26a Targets the Histone Methyltransferase Enhancer of Zeste Homolog 2 during Myogenesis. J. Biol. Chem. 2008, 283, 9836–9843. [Google Scholar] [CrossRef]
- Juan, A.H.; Kumar, R.M.; Marx, J.G.; Young, R.A.; Sartorelli, V. Mir-214-Dependent Regulation of the Polycomb Protein Ezh2 in Skeletal Muscle and Embryonic Stem Cells. Mol. Cell 2009, 36, 61–74. [Google Scholar] [CrossRef]
- Liu, J.; Luo, X.J.; Xiong, A.W.; di Zhang, Z.; Yue, S.; Zhu, M.S.; Cheng, S.Y. MicroRNA-214 Promotes Myogenic Differentiation by Facilitating Exit from Mitosis via down-Regulation of Proto-Oncogene N-Ras. J. Biol. Chem. 2010, 285, 26599–26607. [Google Scholar] [CrossRef] [PubMed]
- Khanna, N.; Ge, Y.; Chen, J. MicroRNA-146b Promotes Myogenic Differentiation and Modulates Multiple Gene Targets in Muscle Cells. PLoS ONE 2014, 9, e100657. [Google Scholar] [CrossRef] [PubMed]
- Conboy, I.M.; Rando, T.A. The Regulation of Notch Signaling Controls Satellite Cell Activation and Cell Fate Determination in Postnatal Myogenesis. Dev. Cell 2002, 3, 397–409. [Google Scholar] [CrossRef]
- Wilson-Rawls, J.; Molkentin, J.D.; Black, B.L.; Olson, E.N. Activated Notch Inhibits Myogenic Activity of the MADS-Box Transcription Factor Myocyte Enhancer Factor 2C. Mol. Cell. Biol. 1999, 19, 2853–2862. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Gilbert, J.A.; Zhang, Y.; Zhang, M.; Qiu, Q.; Ramanujan, K.; Shavlakadze, T.; Eash, J.K.; Scaramozza, A.; Goddeeris, M.M.; et al. An HMGA2-IGF2BP2 Axis Regulates Myoblast Proliferation and Myogenesis. Dev. Cell 2012, 23, 1176–1188. [Google Scholar] [CrossRef] [PubMed]
- Sweta, S.; Dudnakova, T.; Sudheer, S.; Baker, A.H.; Bhushan, R. Importance of Long Non-Coding RNAs in the Development and Disease of Skeletal Muscle and Cardiovascular Lineages. Front. Cell Dev. Biol. 2019, 7, 1–19. [Google Scholar] [CrossRef]
- Dey, B.K.; Pfeifer, K.; Dutta, A. The H19 Long Noncoding RNA Gives Rise to MicroRNAs MiR-675-3p and MiR-675-5p to Promote Skeletal Muscle Differentiation and Regeneration. Genes Dev. 2014, 28, 491–501. [Google Scholar] [CrossRef] [PubMed]
- Kong, D.; He, M.; Yang, L.; Zhou, R.; Yan, Y.Q.; Liang, Y.; Teng, C.B. MiR-17 and MiR-19 Cooperatively Promote Skeletal Muscle Cell Differentiation. Cell. Mol. Life Sci. 2019, 76, 5041–5054. [Google Scholar] [CrossRef]
- Jang, Y.-N.; Baik, E.J. JAK-STAT Pathway and Myogenic Differentiation. JAK-STAT 2013, 2, e23282. [Google Scholar] [CrossRef]
- Trenerry, M.K.; Gatta, P.A.D.; Cameron-Smith, D. JAK/STAT Signaling and Human in Vitro Myogenesis. BMC Physiol. 2011, 11. [Google Scholar] [CrossRef][Green Version]
- Guan, X.; Chen, S.; Zhao, Y. The Role of RhoC in Malignant Tumor Invasion, Metastasis and Targeted Therapy. Histol. Histopathol. 2018, 33, 255–260. [Google Scholar]
- Clark, E.A.; Golub, T.R.; Lander, E.S.; Hynes, R.O. Genomic Analysis of Metastasis Reveals an Essential Role for RhoC. Nature 2000, 406, 532–535. [Google Scholar] [CrossRef]
- Sharples, A.P.; Al-Shanti, N.; Hughes, D.C.; Lewis, M.P.; Stewart, C.E. The Role of Insulin-like-Growth Factor Binding Protein 2 (IGFBP2) and Phosphatase and Tensin Homologue (PTEN) in the Regulation of Myoblast Differentiation and Hypertrophy. Growth Horm. IGF Res. 2013, 23, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, X.; Li, Z.; Abdalla, B.A.; Chen, Y.; Nie, Q. MiR-34b-5p Mediates the Proliferation and Differentiation of Myoblasts by Targeting IGFBP2. Cells 2019, 8, 360. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Liu, C.; Su, Y.; Zhang, K.; Zhang, Y.; Chen, M.; Ge, M.; Gu, L.; Lu, T.; Li, N.; et al. MiRNA-34c Inhibits Myoblasts Proliferation by Targeting YY1. Cell Cycle 2017, 16, 1661–1672. [Google Scholar] [CrossRef]
- Naguibneva, I.; Ameyar-Zazoua, M.; Polesskaya, A.; Ait-Si-Ali, S.; Groisman, R.; Souidi, M.; Cuvellier, S.; Harel-Bellan, A. The MicroRNA MiR-181 Targets the Homeobox Protein Hox-A11 during Mammalian Myoblast Differentiation. Nat. Cell Biol. 2006, 8, 278–284. [Google Scholar] [CrossRef]
- Gagan, J.; Dey, B.K.; Layer, R.; Yan, Z.; Dutta, A. MicroRNA-378 Targets the Myogenic Repressor MyoR during Myoblast Differentiation. J. Biol. Chem. 2011, 286, 19431–19438. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Ouyang, H.; Chen, X.; Yu, J.; Abdalla, B.A.; Chen, B.; Nie, Q. Gga-MiR-205a Affecting Myoblast Proliferation and Differentiation by Targeting CDH11. Front. Genet. 2018, 9, 414. [Google Scholar] [CrossRef]
- Alexander, M.S.; Kawahara, G.; Motohashi, N.; Casar, J.C.; Eisenberg, I.; Myers, J.A.; Gasperini, M.J.; Estrella, E.A.; Kho, A.T.; Mitsuhashi, S.; et al. MicroRNA-199a Is Induced in Dystrophic Muscle and Affects WNT Signaling, Cell Proliferation, and Myogenic Differentiation. Cell Death Differ. 2013, 20, 1194–1208. [Google Scholar] [CrossRef]
- Cardinalli, B.; Castellani, L.; Fasanaro, P.; Basso, A.; Alemà, S.; Martelli, F.; Falcone, G. Microrna-221 and Microrna-222 Modulate Differentiation and Maturation of Skeletal Muscle Cells. PLoS ONE 2009, 4. [Google Scholar] [CrossRef]
- Du, J.; Zhang, P.; Zhao, X.; He, J.; Xu, Y.; Zou, Q.; Luo, J.; Shen, L.; Gu, H.; Tang, Q.; et al. MicroRNA-351-5p Mediates Skeletal Myogenesis by Directly Targeting Lactamase-β and Is Regulated by Lnc-Mg. FASEB J. 2019, 33, 1911–1926. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Liu, J.; Xiao, J.; Yang, L.; Cai, M.; Shen, H.; Chen, X.; Ma, Y.; Hu, S.; Wang, Z.; et al. Lnc-Mg Is a Long Non-Coding RNA That Promotes Myogenesis. Nat. Commun. 2017, 8, 14718. [Google Scholar] [CrossRef] [PubMed]
- Seok, H.Y.; Tatsuguchi, M.; Callis, T.E.; He, A.; Pu, W.T.; Wang, D.Z. MiR-155 Inhibits Expression of the MEF2A Protein to Repress Skeletal Muscle Differentiation. J. Biol. Chem. 2011, 286, 35339–35346. [Google Scholar] [CrossRef] [PubMed]
- Florini, J.R.; Magri, K.A.; Ewton, D.Z.; James, P.L.; Grindstaff, K.; Rotwein, P.S. “Spontaneous” Differentiation of Skeletal Myoblasts Is Dependent upon Autocrine Secretion of Insulin-like Growth Factor-II. J. Biol. Chem. 1991, 266, 15917–15923. [Google Scholar] [CrossRef]
- Florini, J.R.; Ewton, D.Z.; Coolican, S.A. Growth Hormone and the Insulin-Like Growth Factor System in Myogenesis. Endocr. Rev. 1996, 17, 481–517. [Google Scholar] [CrossRef]
- Ge, Y.; Sun, Y.; Chen, J. IGF-II Is Regulated by MicroRNA-125b in Skeletal Myogenesis. J. Cell Biol. 2011, 192, 69–81. [Google Scholar] [CrossRef]
- Antoniou, A.; Mastroyiannopoulos, N.P.; Uney, J.B.; Phylactou, L.A. MiR-186 Inhibits Muscle Cell Differentiation through Myogenin Regulation. J. Biol. Chem. 2014, 289, 3923–3935. [Google Scholar] [CrossRef]
- Wang, L.; Chen, X.; Zheng, Y.; Li, F.; Lu, Z.; Chen, C.; Liu, J.; Wang, Y.; Peng, Y.; Shen, Z.; et al. MiR-23a Inhibits Myogenic Differentiation through down Regulation of Fast Myosin Heavy Chain Isoforms. Exp. Cell Res. 2012, 318, 2324–2334. [Google Scholar] [CrossRef] [PubMed]
- Emery, A.E.H. The Muscular Dystrophies. Lancet 2002, 359, 687–695. [Google Scholar] [CrossRef]
- Mercuri, E.; Muntoni, F. Muscular Dystrophies. Lancet 2013, 381, 845–860. [Google Scholar] [CrossRef]
- Bushby, K.M.D.; Thambyayah, M.; Gardner-Medwin, D. Prevalence and Incidence of Becker Muscular Dystrophy. Lancet 1991, 337, 1022–1024. [Google Scholar] [CrossRef]
- Mah, J.K.; Korngut, L.; Dykeman, J.; Day, L.; Pringsheim, T.; Jette, N. A Systematic Review and Meta-Analysis on the Epidemiology of Duchenne and Becker Muscular Dystrophy. Neuromuscul. Disord. 2014, 24, 482–491. [Google Scholar] [CrossRef]
- Sun, C.; Serra, C.; Lee, G.; Wagner, K.R. Stem Cell-Based Therapies for Duchenne Muscular Dystrophy. Exp. Neurol. 2020, 323, 113086. [Google Scholar] [CrossRef] [PubMed]
- Dumont, N.A.; Rudnicki, M.A. Targeting Muscle Stem Cell Intrinsic Defects to Treat Duchenne Muscular Dystrophy. NPJ Regen. Med. 2016, 1. [Google Scholar] [CrossRef]
- Perry, M.M.; Muntoni, F. Noncoding RNAs and Duchenne Muscular Dystrophy. Epigenomics 2016. [Google Scholar] [CrossRef]
- Kirby, T.J.; Chaillou, T.; McCarthy, J.J. The Role of MicroRNAs in Skeletal Muscle Health and Disease. Front. Biosci. Landmark 2015, 20, 37–77. [Google Scholar]
- Wang, L.; Zhou, L.; Jiang, P.; Lu, L.; Chen, X.; Lan, H.; Guttridge, D.C.; Sun, H.; Wang, H. Loss of MiR-29 in Myoblasts Contributes to Dystrophic Muscle Pathogenesis. Mol. Ther. 2012, 20, 1222–1233. [Google Scholar] [CrossRef]
- Zanotti, S.; Gibertini, S.; Curcio, M.; Savadori, P.; Pasanisi, B.; Morandi, L.; Cornelio, F.; Mantegazza, R.; Mora, M. Opposing Roles of MiR-21 and MiR-29 in the Progression of Fibrosis in Duchenne Muscular Dystrophy. Biochim. Biophys. Acta Mol. Basis Dis. 2015, 1852, 1451–1464. [Google Scholar] [CrossRef]
- Heller, K.N.; Mendell, J.T.; Mendell, J.R.; Rodino-Klapac, L.R. MicroRNA-29 Overexpression by Adeno-Associated Virus Suppresses Fibrosis and Restores Muscle Function in Combination with Micro-Dystrophin. JCI Insight 2017, 2. [Google Scholar] [CrossRef]
- Bulaklak, K.; Xiao, B.; Qiao, C.; Li, J.; Patel, T.; Jin, Q.; Li, J.; Xiao, X. MicroRNA-206 Downregulation Improves Therapeutic Gene Expression and Motor Function in Mdx Mice. Mol. Ther. Nucleic Acids 2018, 12, 283–293. [Google Scholar] [CrossRef]
- Ardite, E.; Perdiguero, E.; Vidal, B.; Gutarra, S.; Serrano, A.L.; Muñoz-Cánoves, P. PAI-1-Regulated MiR-21 Defines a Novel Age-Associated Fibrogenic Pathway in Muscular Dystrophy. J. Cell Biol. 2012, 196, 163–175. [Google Scholar] [CrossRef]
- Bronisz-Budzyńska, I.; Chwalenia, K.; Mucha, O.; Podkalicka, P.; Bukowska-Strakova, K.; Józkowicz, A.; Łoboda, A.; Kozakowska, M.; Dulak, J. MiR-146a Deficiency Does Not Aggravate Muscular Dystrophy in Mdx Mice. Skelet. Muscle 2019, 9, 22. [Google Scholar] [CrossRef] [PubMed]
- Cacchiarelli, D.; Incitti, T.; Martone, J.; Cesana, M.; Cazzella, V.; Santini, T.; Sthandier, O.; Bozzoni, I. MiR-31 Modulates Dystrophin Expression: New Implications for Duchenne Muscular Dystrophy Therapy. EMBO Rep. 2011, 12, 136–141. [Google Scholar] [CrossRef]
- Zhai, L.; Wu, R.; Han, W.; Zhang, Y.; Zhu, D. MiR-127 Enhances Myogenic Cell Differentiation by Targeting S1PR3. Cell Death Dis. 2017, 8, e2707. [Google Scholar] [CrossRef]
- Wang, Z. The Guideline of the Design and Validation of MiRNA Mimics. Methods Mol. Biol. 2011, 676, 211–223. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.J.; Saltzman, W.M.; Slack, F.J. Canonical and Non-Canonical Barriers Facing AntimiR Cancer Therapeutics. Curr. Med. Chem. 2013, 20, 3582–3593. [Google Scholar] [CrossRef]
- Davis, S.; Lollo, B.; Freier, S.; Esau, C. Improved Targeting of MiRNA with Antisense Oligonucleotides. Nucleic Acids Res. 2006, 34, 2294–2304. [Google Scholar] [CrossRef] [PubMed]
- Ørom, U.A.; Kauppinen, S.; Lund, A.H. LNA-Modified Oligonucleotides Mediate Specific Inhibition of MicroRNA Function. Gene 2006, 372, 137–141. [Google Scholar] [CrossRef]
- Haraguchi, T.; Nakano, H.; Tagawa, T.; Ohki, T.; Ueno, Y.; Yoshida, T.; Iba, H. A Potent 2’-O-Methylated RNA-Based MicroRNA Inhibitor with Unique Secondary Structures. Nucleic Acids Res. 2012, 40, e58. [Google Scholar] [CrossRef] [PubMed]
- Bono, N.; Ponti, F.; Mantovani, D.; Candiani, G. Non-Viral in Vitro Gene Delivery: It Is Now Time to Set the Bar! Pharmaceutics 2020, 12, 183. [Google Scholar] [CrossRef] [PubMed]
- Scherr, M.; Venturini, L.; Battmer, K.; Schaller-Schoenitz, M.; Schaefer, D.; Dallmann, I.; Ganser, A.; Eder, M. Lentivirus-Mediated Antagomir Expression for Specific Inhibition of MiRNA Function. Nucleic Acids Res. 2007, 35, e149. [Google Scholar] [CrossRef]
- Ebert, M.S.; Neilson, J.R.; Sharp, P.A. MicroRNA Sponges: Competitive Inhibitors of Small RNAs in Mammalian Cells. Nat. Methods 2007, 4, 721–726. [Google Scholar] [CrossRef]
- Haraguchi, T.; Ozaki, Y.; Iba, H. Vectors Expressing Efficient RNA Decoys Achieve the Long-Term Suppression of Specific MicroRNA Activity in Mammalian Cells. Nucleic Acids Res. 2009, 37, 43. [Google Scholar] [CrossRef] [PubMed]
- van Dongen, S.; Abreu-Goodger, C.; Enright, A.J. Detecting MicroRNA Binding and SiRNA Off-Target Effects from Expression Data. Nat. Methods 2008, 5, 1023–1025. [Google Scholar] [CrossRef] [PubMed]
- Loinger, A.; Shemla, Y.; Simon, I.; Margalit, H.; Biham, O. Competition between Small RNAs: A Quantitative View. Biophys. J. 2012, 102, 1712–1721. [Google Scholar] [CrossRef]
- Yu, B.; Zhao, X.; Lee, J.L.; Lee, R.J. Targeted Delivery Systems for Oligonucleotide Therapeutics. AAPS J. 2009, 11, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Czauderna, F.; Fechtner, M.; Dames, S.; Aygün, H.; Klippel, A.; Pronk, G.J.; Giese, K.; Kaufmann, J. Structural Variations and Stabilising Modifications of Synthetic SiRNAs in Mammalian Cells. Nucleic Acids Res. 2003, 31, 2705–2716. [Google Scholar] [CrossRef]
- Raemdonck, K.; Vandenbroucke, R.E.; Demeester, J.; Sanders, N.N.; de Smedt, S.C. Maintaining the Silence: Reflections on Long-Term RNAi. Drug Discov. Today 2008, 13, 917–931. [Google Scholar] [CrossRef]
- Uludag, H.; Ubeda, A.; Ansari, A. At the Intersection of Biomaterials and Gene Therapy: Progress in Non-Viral Delivery of Nucleic Acids. Front. Bioeng. Biotechnol. 2019, 7, 131. [Google Scholar] [CrossRef] [PubMed]
- Sanada, F.; Taniyama, Y.; Kanbara, Y.; Otsu, R.; Ikeda-Iwabu, Y.; Carracedo, M.; Rakugi, H.; Morishita, R. Gene Therapy in Peripheral Artery Disease. Expert Opin. Biol. Ther. 2015, 15, 381–390. [Google Scholar] [CrossRef]
- Shimamura, M.; Nakagami, H.; Taniyama, Y.; Morishita, R. Gene Therapy for Peripheral Arterial Disease. Expert Opin. Biol. Ther. 2014, 14, 1175–1184. [Google Scholar] [CrossRef] [PubMed]
- Mathiyalagan, P.; Sahoo, S. Exosomes-based gene therapy for MicroRNA delivery. In Methods in Molecular Biology; Humana Press Inc.: New York, NY, USA, 2017; Volume 1521, pp. 139–152. [Google Scholar] [CrossRef]
- Schade, A.; Delyagina, E.; Scharfenberg, D.; Skorska, A.; Lux, C.; David, R.; Steinhoff, G. Innovative Strategy for MicroRNA Delivery in Human Mesenchymal Stem Cells via Magnetic Nanoparticles. Int. J. Mol. Sci. 2013, 14, 10710–10726. [Google Scholar] [CrossRef] [PubMed]
- Yin, P.T.; Shah, B.P.; Lee, K.-B. Combined Magnetic Nanoparticle-Based MicroRNA and Hyperthermia Therapy to Enhance Apoptosis in Brain Cancer Cells. Small 2014, 10, 4106–4112. [Google Scholar] [CrossRef]
- Tivnan, A.; Orr, W.S.; Gubala, V.; Nooney, R.; Williams, D.E.; McDonagh, C.; Prenter, S.; Harvey, H.; Domingo-Fernández, R.; Bray, I.M.; et al. Inhibition of Neuroblastoma Tumor Growth by Targeted Delivery of MicroRNA-34a Using Anti-Disialoganglioside GD2 Coated Nanoparticles. PLoS ONE 2012, 7, e38129. [Google Scholar] [CrossRef] [PubMed]
- Bertucci, A.; Prasetyanto, E.A.; Septiadi, D.; Manicardi, A.; Brognara, E.; Gambari, R.; Corradini, R.; de Cola, L. Combined Delivery of Temozolomide and Anti-MiR221 PNA Using Mesoporous Silica Nanoparticles Induces Apoptosis in Resistant Glioma Cells. Small 2015, 11, 5687–5695. [Google Scholar] [CrossRef] [PubMed]
- Ekin, A.; Karatas, O.F.; Culha, M.; Ozen, M. Designing a Gold Nanoparticle-Based Nanocarrier for MicroRNA Transfection into the Prostate and Breast Cancer Cells. J. Gene Med. 2014, 16, 331–335. [Google Scholar] [CrossRef]
- Ghosh, R.; Singh, L.C.; Shohet, J.M.; Gunaratne, P.H. A Gold Nanoparticle Platform for the Delivery of Functional MicroRNAs into Cancer Cells. Biomaterials 2013, 34, 807–816. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Namgung, R.; Kim, W.J.; Kim, J.I.; Park, I.K. Targeted Delivery of MicroRNA-145 to Metastatic Breast Cancer by Peptide Conjugated Branched PEI Gene Carrier. Macromol. Res. 2013, 21, 1201–1209. [Google Scholar] [CrossRef]
- Ibrahim, A.F.; Weirauch, U.; Thomas, M.; Grünweller, A.; Hartmann, R.K.; Aigner, A. MicroRNA Replacement Therapy for MiR-145 and MiR-33a Is Efficacious in a Model of Colon Carcinoma. Cancer Res. 2011, 71, 5214–5224. [Google Scholar] [CrossRef]
- Saraiva, C.; Talhada, D.; Rai, A.; Ferreira, R.; Ferreira, L.; Bernardino, L.; Ruscher, K. MicroRNA-124-Loaded Nanoparticles Increase Survival and Neuronal Differentiation of Neural Stem Cells in Vitro but Do Not Contribute to Stroke Outcome in Vivo. PLoS ONE 2018, 13, e0193609. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Li, R.T.; Qian, H.Q.; Wei, J.; Xie, L.; Shen, J.; Yang, M.; Qian, X.P.; Yu, L.X.; Jiang, X.Q.; et al. Targeted Delivery of MiR-200c/DOC to Inhibit Cancer Stem Cells and Cancer Cells by the Gelatinases-Stimuli Nanoparticles. Biomaterials 2013, 34, 7191–7203. [Google Scholar] [CrossRef] [PubMed]
- Chiou, G.Y.; Cherng, J.Y.; Hsu, H.S.; Wang, M.L.; Tsai, C.M.; Lu, K.H.; Chien, Y.; Hung, S.C.; Chen, Y.W.; Wong, C.I.; et al. Cationic Polyurethanes-Short Branch PEI-Mediated Delivery of Mir145 Inhibited Epithelial-Mesenchymal Transdifferentiation and Cancer Stem-like Properties and in Lung Adenocarcinoma. J. Control. Release 2012, 159, 240–250. [Google Scholar] [CrossRef]
- Yang, Y.P.; Chien, Y.; Chiou, G.Y.; Cherng, J.Y.; Wang, M.L.; Lo, W.L.; Chang, Y.L.; Huang, P.I.; Chen, Y.W.; Shih, Y.H.; et al. Inhibition of Cancer Stem Cell-like Properties and Reduced Chemoradioresistance of Glioblastoma Using MicroRNA145 with Cationic Polyurethane-Short Branch PEI. Biomaterials 2012, 33, 1462–1476. [Google Scholar] [CrossRef]
- Liang, G.; Zhu, Y.; Jing, A.; Wang, J.; Hu, F.; Feng, W.; Xiao, Z.; Chen, B. Cationic MicroRNA-Delivering Nanocarriers for Efficient Treatment of Colon Carcinoma in Xenograft Model. Gene Ther. 2016, 23, 829–838. [Google Scholar] [CrossRef] [PubMed]
- Blanco, E.; Shen, H.; Ferrari, M. Principles of Nanoparticle Design for Overcoming Biological Barriers to Drug Delivery. Nat. Biotechnol. 2015, 33, 941–951. [Google Scholar] [CrossRef]
- Cun, D.; Jensen, D.K.; Maltesen, M.J.; Bunker, M.; Whiteside, P.; Scurr, D.; Foged, C.; Nielsen, H.M. High Loading Efficiency and Sustained Release of SiRNA Encapsulated in PLGA Nanoparticles: Quality by Design Optimization and Characterization. Eur. J. Pharm. Biopharm. 2011, 77, 26–35. [Google Scholar] [CrossRef]
- Decuzzi, P.; Godin, B.; Tanaka, T.; Lee, S.Y.; Chiappini, C.; Liu, X.; Ferrari, M. Size and Shape Effects in the Biodistribution of Intravascularly Injected Particles. J. Control. Release 2010, 141, 320–327. [Google Scholar] [CrossRef]
- Kim, S.H.; Mok, H.; Jeong, J.H.; Kim, S.W.; Park, T.G. Comparative Evaluation of Target-Specific GFP Gene Silencing Efficiencies for Antisense ODN, Synthetic SiRNA, and SiRNA Plasmid Complexed with PEI-PEG-FOL Conjugate. Bioconj. Chem. 2006, 17, 241–244. [Google Scholar] [CrossRef]
- Xie, Y.; Murray-Stewart, T.; Wang, Y.; Yu, F.; Li, J.; Marton, L.J.; Casero, R.A.; Oupický, D. Self-Immolative Nanoparticles for Simultaneous Delivery of MicroRNA and Targeting of Polyamine Metabolism in Combination Cancer Therapy. J. Control. Release 2017, 246, 110–119. [Google Scholar] [CrossRef]
- Lee, T.J.; Yoo, J.Y.; Shu, D.; Li, H.; Zhang, J.; Yu, J.G.; Jaime-Ramirez, A.C.; Acunzo, M.; Romano, G.; Cui, R.; et al. RNA Nanoparticle-Based Targeted Therapy for Glioblastoma through Inhibition of Oncogenic MiR-21. Mol. Ther. 2017, 25, 1544–1555. [Google Scholar] [CrossRef]
- Nierenberg, D.; Khaled, A.R.; Flores, O. Formation of a Protein Corona Influences the Biological Identity of Nanomaterials. Rep. Pract. Oncol. Radiother. 2018, 23, 300–308. [Google Scholar] [CrossRef] [PubMed]
- Barbero, F.; Russo, L.; Vitali, M.; Piella, J.; Salvo, I.; Borrajo, M.L.; Busquets-Fité, M.; Grandori, R.; Bastús, N.G.; Casals, E.; et al. Formation of the Protein Corona: The Interface between Nanoparticles and the Immune System. Semin. Immunol. 2017, 34, 52–60. [Google Scholar] [CrossRef]
- Huang, D.; Yue, F.; Qiu, J.; Deng, M.; Kuang, S. Polymeric Nanoparticles Functionalized with Muscle-Homing Peptides for Targeted Delivery of Phosphatase and Tensin Homolog Inhibitor to Skeletal Muscle. Acta Biomater. 2020, 118, 196–206. [Google Scholar] [CrossRef]
- Del Valle, L.; Diaz, A.; Puiggalí, J. Hydrogels for Biomedical Applications: Cellulose, Chitosan, and Protein/Peptide Derivatives. Gels 2017, 3, 27. [Google Scholar] [CrossRef] [PubMed]
- Youngblood, R.L.; Truong, N.F.; Segura, T.; Shea, L.D. It’s All in the Delivery: Designing Hydrogels for Cell and Non-Viral Gene Therapies. Mol. Ther. 2018, 26, 2087–2106. [Google Scholar] [CrossRef] [PubMed]
- Carballo-Pedrares, N.; Fuentes-Boquete, I.; Díaz-Prado, S.; Rey-Rico, A. Hydrogel-Based Localized Nonviral Gene Delivery in Regenerative Medicine Approaches—An Overview. Pharmaceutics 2020, 12, 752. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.L.; Liu, Y.; Chung, J.J.; Wang, T.; Gaffey, A.C.; Lu, M.; Cavanaugh, C.A.; Zhou, S.; Kanade, R.; Atluri, P.; et al. Sustained MiRNA Delivery from an Injectable Hydrogel Promotes Cardiomyocyte Proliferation and Functional Regeneration after Ischaemic Injury. Nat. Biomed. Eng. 2017, 1, 983–992. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, X.; Jin, R.; Chen, L.; Dang, M.; Cao, H.; Dong, Y.; Cai, B.; Bai, G.; Gooding, J.J.; et al. Injectable Hydrogel with MSNs/MicroRNA-21-5p Delivery Enables Both Immunomodification and Enhanced Angiogenesis for Myocardial Infarction Therapy in Pigs. Sci. Adv. 2021, 7, 6740–6764. [Google Scholar] [CrossRef]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-Mediated Transfer of MRNAs and MicroRNAs Is a Novel Mechanism of Genetic Exchange between Cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Parolini, I.; Federici, C.; Raggi, C.; Lugini, L.; Palleschi, S.; de Milito, A.; Coscia, C.; Iessi, E.; Logozzi, M.; Molinari, A.; et al. Microenvironmental PH Is a Key Factor for Exosome Traffic in Tumor Cells. J. Biol. Chem. 2009, 284, 34211–34222. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, Y.; Liu, H.; Tang, W.H. Exosomes: Biogenesis, Biologic Function and Clinical Potential. Cell Biosci. 2019, 9, 19. [Google Scholar] [CrossRef]
- Nance, M.E.; Hakim, C.H.; Yang, N.N.; Duan, D. Nanotherapy for Duchenne Muscular Dystrophy. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnology 2018, 10. [Google Scholar] [CrossRef] [PubMed]
- Sandonà, M.; Consalvi, S.; Tucciarone, L.; de Bardi, M.; Scimeca, M.; Angelini, D.; Buffa, V.; D’Amico, A.; Bertini, E.; Cazzaniga, S.; et al. HDAC inhibitors tune miRNAs in extracellular vesicles of dystrophic muscle-resident mesenchymal cells. EMBO Rep. 2020, 21, e50863. [Google Scholar] [CrossRef]
- Minetti, G.C.; Colussi, C.; Adami, R.; Serra, C.; Mozzetta, C.; Parente, V.; Fortuni, S.; Straino, S.; Sampaolesi, M.; di Padova, M.; et al. Functional and Morphological Recovery of Dystrophic Muscles in Mice Treated with Deacetylase Inhibitors. Nat. Med. 2006, 12, 1147–1150. [Google Scholar] [CrossRef]
- Consalvi, S.; Saccone, V.; Giordani, L.; Minetti, G.; Mozzetta, C.; Puri, P.L. Histone Deacetylase Inhibitors in the Treatment of Muscular Dystrophies: Epigenetic Drugs for Genetic Diseases. Mol. Med. 2011, 17, 457–465. [Google Scholar] [CrossRef]
- Consalvi, S.; Mozzetta, C.; Bettica, P.; Germani, M.; Fiorentini, F.; del Bene, F.; Rocchetti, M.; Leoni, F.; Monzani, V.; Mascagni, P.; et al. Preclinical Studies in the Mdx Mouse Model of Duchenne Muscular Dystrophy with the Histone Deacetylase Inhibitor Givinostat. Mol. Med. 2013, 19, 79–87. [Google Scholar] [CrossRef]
- Saccone, V.; Consalvi, S.; Giordani, L.; Mozzetta, C.; Barozzi, I.; Sandoná, M.; Ryan, T.; Rojas-Muñoz, A.; Madaro, L.; Fasanaro, P.; et al. HDAC-Regulated MyomiRs Control BAF60 Variant Exchange and Direct the Functional Phenotype of Fibro-Adipogenic Progenitors in Dystrophic Muscles. Genes Dev. 2014, 28, 841–857. [Google Scholar] [CrossRef]
- Bettica, P.; Petrini, S.; D’Oria, V.; D’Amico, A.; Catteruccia, M.; Pane, M.; Sivo, S.; Magri, F.; Brajkovic, S.; Messina, S.; et al. Histological Effects of Givinostat in Boys with Duchenne Muscular Dystrophy. Neuromuscul. Disord. 2016, 26, 643–649. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Erviti, L.; Seow, Y.; Yin, H.; Betts, C.; Lakhal, S.; Wood, M.J.A. Delivery of SiRNA to the Mouse Brain by Systemic Injection of Targeted Exosomes. Nat. Biotechnol. 2011, 29, 341–345. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aránega, A.E.; Lozano-Velasco, E.; Rodriguez-Outeiriño, L.; Ramírez de Acuña, F.; Franco, D.; Hernández-Torres, F. MiRNAs and Muscle Regeneration: Therapeutic Targets in Duchenne Muscular Dystrophy. Int. J. Mol. Sci. 2021, 22, 4236. https://doi.org/10.3390/ijms22084236
Aránega AE, Lozano-Velasco E, Rodriguez-Outeiriño L, Ramírez de Acuña F, Franco D, Hernández-Torres F. MiRNAs and Muscle Regeneration: Therapeutic Targets in Duchenne Muscular Dystrophy. International Journal of Molecular Sciences. 2021; 22(8):4236. https://doi.org/10.3390/ijms22084236
Chicago/Turabian StyleAránega, Amelia Eva, Estefanía Lozano-Velasco, Lara Rodriguez-Outeiriño, Felicitas Ramírez de Acuña, Diego Franco, and Francisco Hernández-Torres. 2021. "MiRNAs and Muscle Regeneration: Therapeutic Targets in Duchenne Muscular Dystrophy" International Journal of Molecular Sciences 22, no. 8: 4236. https://doi.org/10.3390/ijms22084236
APA StyleAránega, A. E., Lozano-Velasco, E., Rodriguez-Outeiriño, L., Ramírez de Acuña, F., Franco, D., & Hernández-Torres, F. (2021). MiRNAs and Muscle Regeneration: Therapeutic Targets in Duchenne Muscular Dystrophy. International Journal of Molecular Sciences, 22(8), 4236. https://doi.org/10.3390/ijms22084236









