Transgenerational Effects of Di(2-Ethylhexyl) Phthalate on Anogenital Distance, Sperm Functions and DNA Methylation in Rat Offspring
Abstract
1. Introduction
2. Results
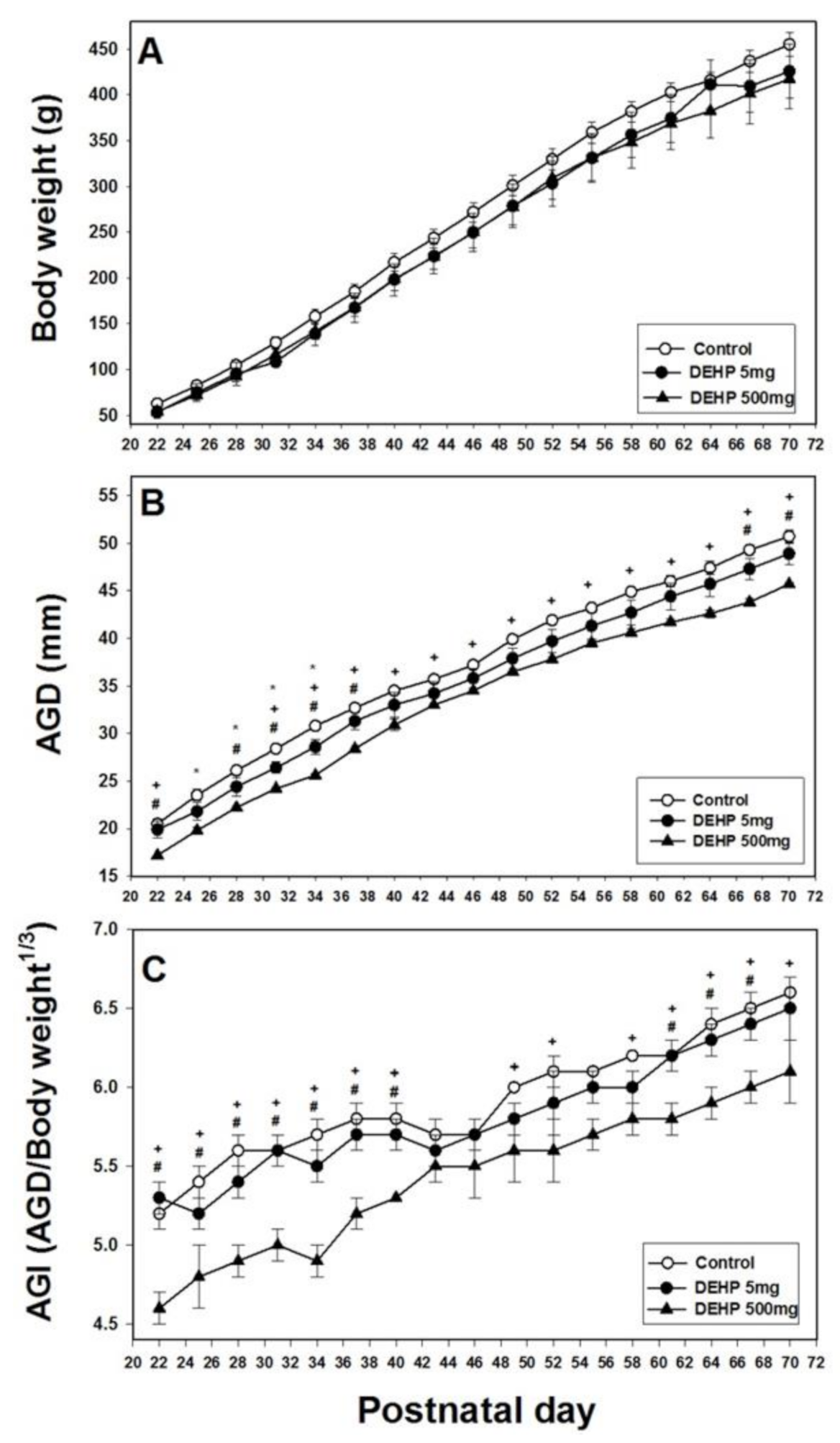
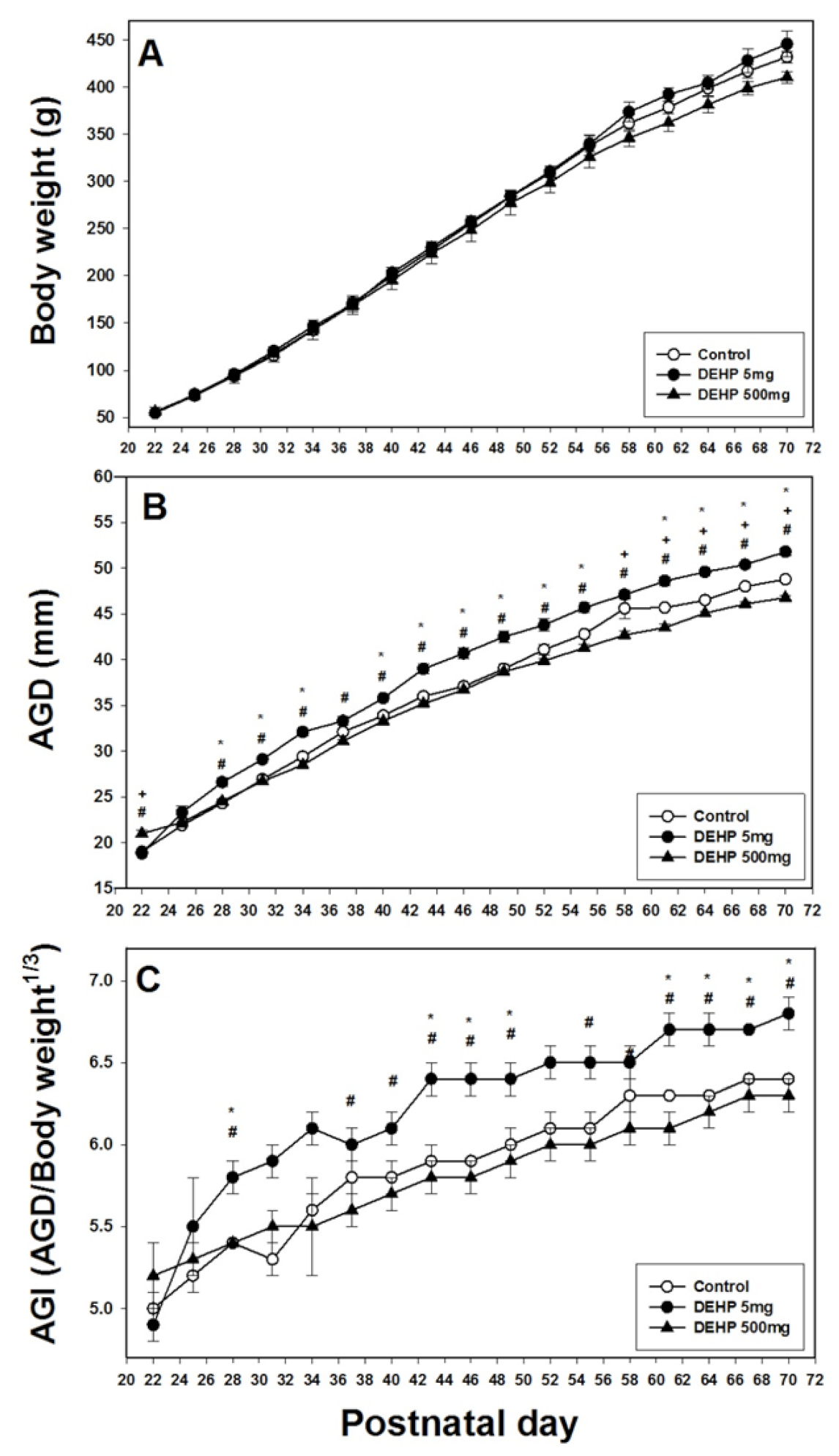
2.1. Effects of DEHP Exposure on Body Weight, AGD, and AGI
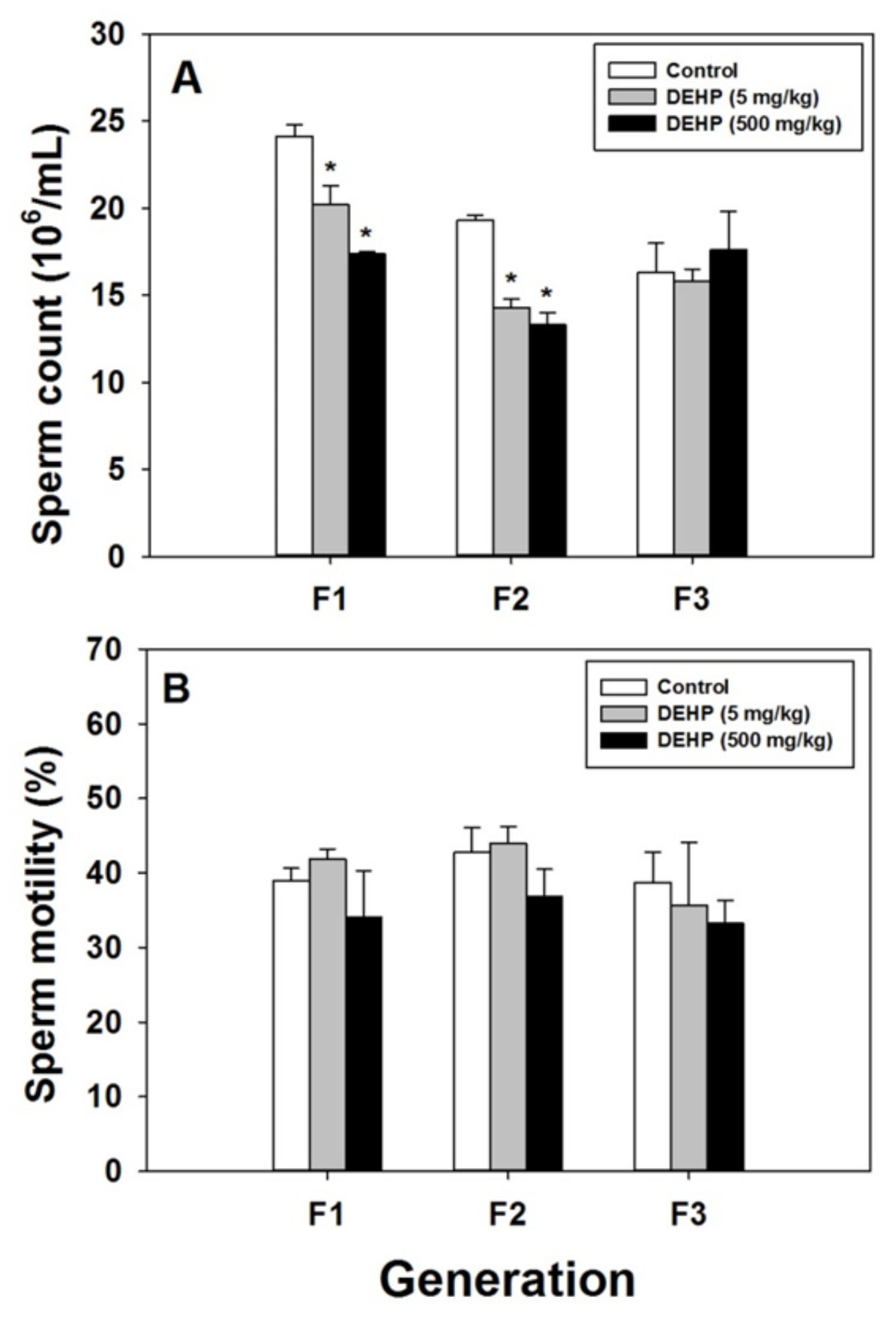
2.2. Effects of DEHP Exposure on Sperm Count and Motility
2.3. Effect of DEHP Exposure on Sperm Chromatin DNA
2.4. Hypermethylated and Hypomethylated Genes in Sperm DNA
2.5. Gene Symbol, Gene Name, Chromosome, and Ratio of Relative Fold Expression of Hypermethylated Genes
2.6. Gene Symbol, Gene Name, Chromosome, and Ratio of Relative Fold Expression of Hypomethylated Genes
3. Discussion
4. Materials and Methods
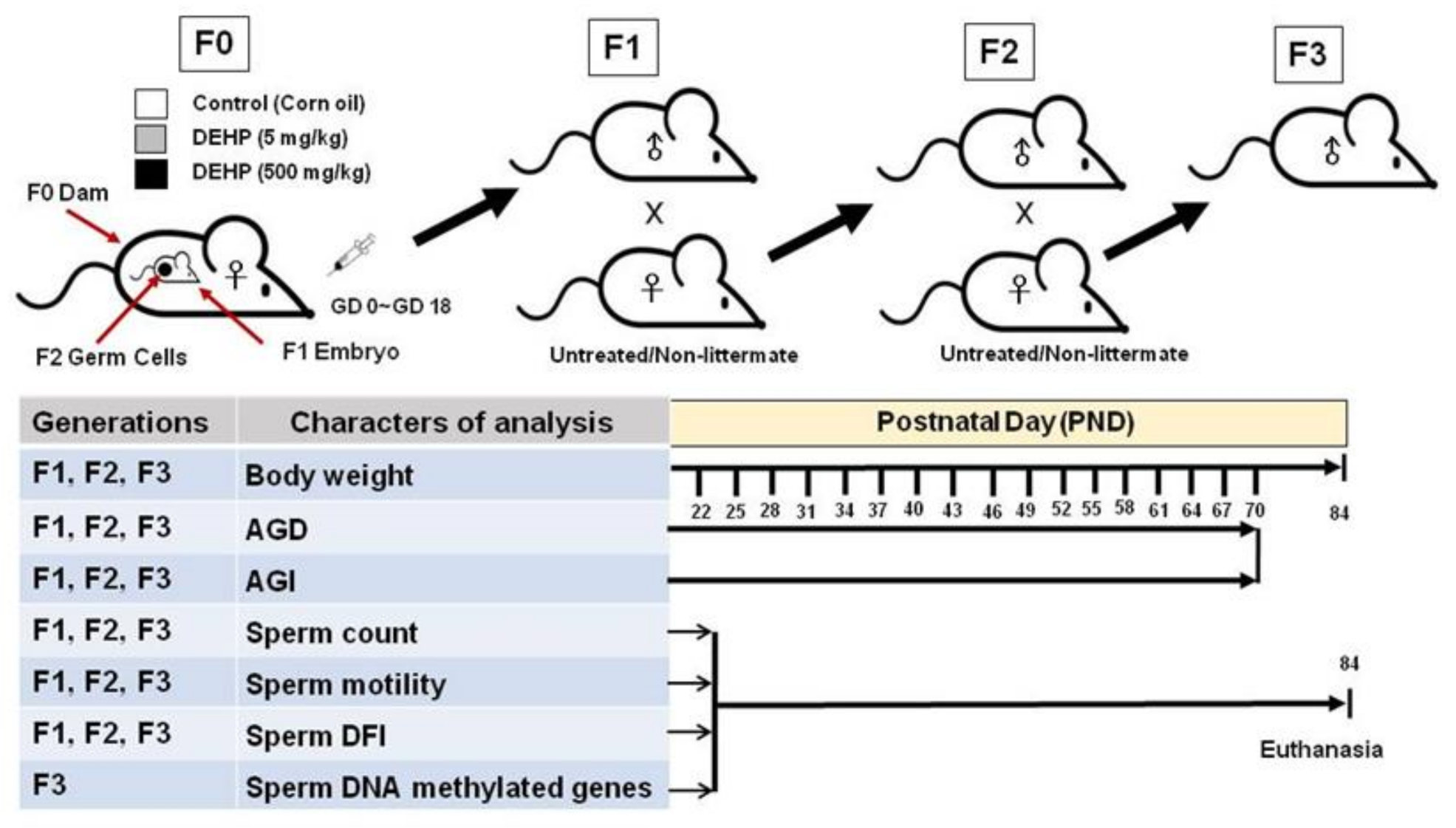
4.1. Study Design
4.2. Animals and Treatment
4.3. Breeding the F1, F2, and F3 Generations
4.4. Body Weight, AGD, and AGI
4.5. Sperm Count and Motility Analysis
4.6. Sperm Chromatin Structure Assay
4.7. Sperm DNA Extraction
4.8. Methyl-CpG Binding Domain Sequencing
4.9. Analysis of Hypermethylation and Hypomethylation in Sperm DNA
4.10. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AGD | anogenital distance |
| AGI | anogenital index |
| ATG16L1 | autophagy Atg16L1 |
| BP | biological processes |
| CPSF2 | cleavage and polyadenylation specific factor 2 |
| EDC | endocrine disrupting chemical |
| DEHP | di(2-ethylhexyl) phthalate |
| DFI | DNA fragmentation index |
| FAM13A | family with sequence similarity 13, member A |
| FAM222A | family with sequence similarity 222, member A |
| FCM | flow cytometry |
| GAPDH | glyceraldehyde-3-phosphate Dehydrogenase |
| GD | gestation day |
| GO | gene ontology |
| MBD-seq | methyl-CpG binding domain sequencing |
| MeDIP | methylated DNA immunoprecipitation |
| PND | postnatal day |
| PVC | polyvinylchloride |
| ROS | reactive oxygen species |
| RPKM | reads per kilobase per million |
| SCSA | sperm chromatin structure assay |
| SD | Sprague-Dawley |
References
- Tanaka, T. Reproductive and neurobehavioural effects of bis(2-ethylhexyl) phthalate (DEHP) in a cross-mating toxicity study of mice. Food Chem. Toxicol. 2005, 43, 581–589. [Google Scholar] [CrossRef] [PubMed]
- Mendiola, J.; Meeker, J.D.; Jorgensen, N.; Andersson, A.M.; Liu, F.; Calafat, A.M.; Redmon, J.B.; Drobnis, E.Z.; Sparks, A.E.; Wang, C.; et al. Urinary concentrations of di(2-ethylhexyl) phthalate metabolites and serum reproductive hormones: Pooled analysis of fertile and infertile men. J. Androl. 2012, 33, 488–498. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.C.; Kuo, Y.T.; Guo, Y.L.; Chen, J.R.; Tsai, S.S.; Chao, H.R.; Teng, Y.N.; Pan, M.H. The adverse effects of low-dose exposure to di(2-ethylhexyl) phthalate during adolescence on sperm function in adult rats. Environ. Toxicol. 2016, 31, 706–712. [Google Scholar] [CrossRef] [PubMed]
- Silva, M.J.; Reidy, J.A.; Herbert, A.R.; Preau, J.L.; Needham, L.L.; Calafat, A.M. Detection of phthalate metabolites in human amniotic fluid. Bull. Environ. Contam. Toxicol. 2004, 72, 1226–1231. [Google Scholar] [CrossRef]
- Swan, S.H. Environmental phthalate exposure in relation to reproductive outcomes and other health endpoints in humans. Environ. Res. 2008, 108, 177–184. [Google Scholar] [CrossRef]
- Philippat, C.; Mortamais, M.; Chevrier, C.; Petit, C.; Calafat, A.M.; Ye, X.; Silva, M.J.; Brambilla, C.; Pin, I.; Charles, M.A.; et al. Exposure to phthalates and phenols during pregnancy and offspring size at birth. Environ. Health Perspect. 2012, 120, 464–470. [Google Scholar] [CrossRef]
- Suzuki, Y.; Yoshinaga, J.; Mizumoto, Y.; Serizawa, S.; Shiraishi, H. Foetal exposure to phthalate esters and anogenital distance in male newborns. Int. J. Androl. 2012, 35, 236–244. [Google Scholar] [CrossRef]
- Jensen, M.S.; Anand-Ivell, R.; Norgaard-Pedersen, B.; Jonsson, B.A.; Bonde, J.P.; Hougaard, D.M.; Cohen, A.; Lindh, C.H.; Ivell, R.; Toft, G. Amniotic fluid phthalate levels and male fetal gonad function. Epidemiology 2015, 26, 91–99. [Google Scholar] [CrossRef]
- Kita, D.H.; Meyer, K.B.; Venturelli, A.C.; Adams, R.; Machado, D.L.; Morais, R.N.; Swan, S.H.; Gennings, C.; Martino-Andrade, A.J. Manipulation of pre and postnatal androgen environments and anogenital distance in rats. Toxicology 2016, 368, 152–161. [Google Scholar] [CrossRef]
- Thankamony, A.; Pasterski, V.; Ong, K.K.; Acerini, C.L.; Hughes, I.A. Anogenital distance as a marker of androgen exposure in humans. Andrology 2016, 4, 616–625. [Google Scholar] [CrossRef]
- Gallavan, R.H., Jr.; Holson, J.F.; Stump, D.G.; Knapp, J.F.; Reynolds, V.L. Interpreting the toxicologic significance of alterations in anogenital distance: Potential for confounding effects of progeny body weights. Reprod. Toxicol. 1999, 13, 383–390. [Google Scholar] [CrossRef]
- Stenz, L.; Escoffier, J.; Rahban, R.; Nef, S.; Paoloni-Giacobino, A. Testicular dysgenesis syndrome and long-lasting epigenetic silencing of mouse sperm genes involved in the reproductive system after prenatal exposure to DEHP. PLoS ONE 2017, 12, e0170441. [Google Scholar] [CrossRef]
- Kay, V.R.; Bloom, M.S.; Foster, W.G. Reproductive and developmental effects of phthalate diesters in males. Crit. Rev. Toxicol. 2014, 44, 467–498. [Google Scholar] [CrossRef]
- Huang, P.C.; Kuo, P.L.; Chou, Y.Y.; Lin, S.J.; Lee, C.C. Association between prenatal exposure to phthalates and the health of newborns. Environ. Int. 2009, 35, 14–20. [Google Scholar] [CrossRef]
- Reamon-Buettner, S.M.; Borlak, J. A new paradigm in toxicology and teratology: Altering gene activity in the absence of DNA sequence variation. Reprod. Toxicol. 2007, 24, 20–30. [Google Scholar] [CrossRef]
- Van Otterdijk, S.D.; Michels, K.B. Transgenerational epigenetic inheritance in mammals: How good is the evidence? FASEB J. 2016, 30, 2457–2465. [Google Scholar] [CrossRef]
- Chung, F.F.-L.; Herceg, Z. The promises and Challenges of toxico-epigenomics: Environmental chemicals and their impacts on the epigenome. Environ. Health Perspect. 2020, 128, 15001. [Google Scholar] [CrossRef]
- Ruiz-Hernandez, A.; Kuo, C.C.; Rentero-Garrido, P.; Tang, W.Y.; Redon, J.; Ordovas, J.M.; Navas-Acien, A.; Tellez-Plaza, M. Environmental chemicals and DNA methylation in adults: A systematic review of the epidemiologic evidence. Clin. Epigenet. 2015, 7, 55. [Google Scholar] [CrossRef]
- Singh, S.; Li, S.S.L. Epigenetic effects of environmental chemicals bisphenol A and phthalates. Int. J. Mol. Sci. 2012, 13, 10143–10153. [Google Scholar] [CrossRef]
- Van Cauwenbergh, O.; Di Serafino, A.; Tytgat, J.; Soubry, A. Transgenerational epigenetic effects from male exposure to endocrine-disrupting compounds: A systematic review on research in mammals. Clin. Epigenet. 2020, 12, 65. [Google Scholar] [CrossRef]
- Prados, J.; Stenz, L.; Somm, E.; Stouder, C.; Dayer, A.; Paoloni-Giacobino, A. Prenatal exposure to DEHP affects spermatogenesis and sperm DNA methylation in a strain-dependent manner. PLoS ONE 2015, 10, e0132136. [Google Scholar] [CrossRef]
- Chen, C.H.; Jiang, S.S.; Chang, I.S.; Wen, H.J.; Sun, C.W.; Wang, S.L. Association between fetal exposure to phthalate endocrine disruptor and genome-wide DNA methylation at birth. Environ. Res. 2018, 162, 261–270. [Google Scholar] [CrossRef]
- Manikkam, M.; Guerrero-Bosagna, C.; Tracey, R.; Haque, M.M.; Skinner, M.K. Transgenerational actions of environmental compounds on reproductive disease and identification of epigenetic biomarkers of ancestral exposures. PLoS ONE 2012, 7, e31901. [Google Scholar] [CrossRef]
- Manikkam, M.; Tracey, R.; Guerrero-Bosagna, C.; Skinner, M.K. Plastics derived endocrine disruptors (BPA, DEHP and DBP) induce epigenetic transgenerational inheritance of obesity, reproductive disease and sperm epimutations. PLoS ONE 2013, 8, e55387. [Google Scholar] [CrossRef]
- Chen, J.; Wu, S.; Wen, S.; Shen, L.; Peng, J.; Yan, C.; Cao, X.; Zhou, Y.; Long, C.; Lin, T.; et al. The Mechanism of Environmental Endocrine Disruptors (DEHP) Induces Epigenetic Transgenerational Inheritance of Cryptorchidism. PLoS ONE 2015, 10, e0126403. [Google Scholar] [CrossRef]
- Vandenberg, L.N.; Colborn, T.; Hayes, T.B.; Heindel, J.J.; Jacobs, D.R., Jr.; Lee, D.H.; Shioda, T.; Soto, A.M.; vom Saal, F.S.; Welshons, W.V.; et al. Hormones and endocrine-disrupting chemicals: Low-dose effects and nonmonotonic dose responses. Endocr. Rev. 2012, 33, 378–455. [Google Scholar] [CrossRef]
- Agarwal, D.K.; Lawrence, W.H.; Turner, J.E.; Autian, J. Effects of parenteral di-(2-ethylhexyl) phthalate (DEHP) on gonadal biochemistry, pathology, and reproductive performance of mice. J. Toxicol. Environ. Health 1989, 26, 39–59. [Google Scholar] [CrossRef]
- Barakat, R.; Lin, P.-C.P.; Rattan, S.; Brehm, E.; Canisso, I.F.; Abosalum, M.E.; Flaws, J.A.; Hess, R.; Ko, C. Prenatal exposure to DEHP induces premature reproductive senescence in male mice. Toxicol. Sci. 2017, 156, 96–108. [Google Scholar] [CrossRef] [PubMed]
- Doyle, T.J.; Bowman, J.L.; Windell, V.L.; McLean, D.J.; Kim, K.H. Transgenerational effects of di-(2-ethylhexyl) phthalate on testicular germ cell associations and spermatogonial stem cells in mice. Boil. Reprod. 2013, 88, 112. [Google Scholar] [CrossRef] [PubMed]
- Yuan, B.; Wu, W.; Chen, M.; Gu, H.; Tang, Q.; Guo, D.; Chen, T.; Chen, Y.; Lu, C.; Song, L.; et al. From the cover: Metabolomics reveals a role of betaine in prenatal DBP exposure-induced epigenetic transgenerational failure of spermatogenesis in rats. Toxicol. Sci. 2017, 158, 356–366. [Google Scholar] [CrossRef] [PubMed]
- Satoh, J.H.; Kino, Y.; Yanaizu, M.; Ishida, T.; Saito, Y. Reactive astrocytes express Aggregatin (FAM222A) in the brains of Alzheimer’s disease and Nasu-Hakola disease. Intractable Rare Dis. Res. 2020, 9, 217–221. [Google Scholar] [CrossRef]
- Yan, T.; Liang, J.; Gao, J.; Wang, L.; Fujioka, H.; Zhu, X.; Wang, X. FAM222A encodes a protein which accumulates in plaques in Alzheimer’s disease. Nat. Commun. 2020, 11, 411–427. [Google Scholar] [CrossRef]
- Feiden, S.; Wolfrum, U.; Wegener, G.; Kamp, G. Expression and compartmentalisation of the glycolytic enzymes GAPDH and pyruvate kinase in boar spermatogenesis. Reprod. Fertil. Dev. 2008, 20, 713–723. [Google Scholar] [CrossRef]
- Castillo, J.; Jodar, M.; Oliva, R. The contribution of human sperm proteins to the development and epigenome of the preimplantation embryo. Hum. Reprod. Update 2018, 24, 535–555. [Google Scholar] [CrossRef]
- Nakahira, K.; Haspel, J.A.; Rathinam, V.A.; Lee, S.J.; Dolinay, T.; Lam, H.C.; Englert, J.A.; Rabinovitch, M.; Cernadas, M.; Kim, H.P.; et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat. Immunol. 2011, 12, 222–230. [Google Scholar] [CrossRef]
- Fagerberg, L.; Hallström, B.M.; Oksvold, P.; Kampf, C.; Djureinovic, D.; Odeberg, J.; Habuka, M.; Tahmasebpoor, S.; Danielsson, A.; Edlund, K.; et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell Proteom. 2014, 13, 397–406. [Google Scholar] [CrossRef]
- Huang, L.P.; Lee, C.C.; Hsu, P.C.; Shih, T.S. The association between semen quality in workers and the concentration of di(2-ethylhexyl) phthalate in polyvinyl chloride pellet plant air. Fertil. Steril. 2011, 96, 90–94. [Google Scholar] [CrossRef]
- Huang, L.P.; Lee, C.C.; Fan, J.P.; Kuo, P.H.; Shih, T.S.; Hsu, P.C. Urinary metabolites of di(2-ethylhexyl) phthalate relation to sperm motility, reactive oxygen species generation, and apoptosis in polyvinyl chloride workers. Int. Arch. Occup. Environ. Health 2014, 87, 635–646. [Google Scholar] [CrossRef]
- Chen, H.P.; Pan, M.H.; Chou, Y.Y.; Sung, C.; Lee, K.H.; Leung, C.M.; Hsu, P.C. Effects of di(2-ethylhexyl) phthalate exposure on 1,2-dimethyhydrazine-induced colon tumor promotion in rats. Food Chem. Toxicol. 2017, 103, 157–167. [Google Scholar] [CrossRef]
- Christiansen, S.; Boberg, J.; Axelstad, M.; Dalgaard, M.; Vinggaard, A.M.; Metzdorff, S.B.; Hass, U. Low-dose perinatal exposure to di(2-ethylhexyl) phthalate induces anti-androgenic effects in male rats. Reprod. Toxicol. 2010, 30, 313–321. [Google Scholar] [CrossRef]
- Li, M.; Qiu, L.; Zhang, Y.; Hua, Y.; Tu, S.; He, Y.; Wen, S.; Wang, Q.; Wei, G. Dose-related effect by maternal exposure to di-(2-ethylhexyl) phthalate plasticizer on inducing hypospadiac male rats. Environ. Toxicol. Pharmacol. 2013, 35, 55–60. [Google Scholar] [CrossRef]
- Evenson, D.P.; Wixon, R. Environmental toxicants cause sperm DNA fragmentation as detected by the sperm chromatin structure assay (SCSATM). Toxicol. Appl. Pharmacol. 2005, 207, 532–537. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef]
- Lienhard, M.; Grimm, C.; Morkel, M.; Herwig, R.; Chavez, L. MEDIPS: Genome-wide differential coverage analysis of sequencing data derived from DNA enrichment experiments. Bioinformatics 2014, 30, 284–286. [Google Scholar] [CrossRef]




| Gene Symbol | Gene Name | Chromosome | Relative Fold of RPKM Expression between 5 mg/kg and Control Groups * | Relative Fold of RPKM Expression between 500 mg/kg and 5 mg/kg Groups # |
|---|---|---|---|---|
| FAM222A | Family with sequence similarity 222, member A | Chr12 | 4.44 | 2.32 (10.30) a |
| GAPDH | Glyceraldehyde-3-phosphate Dehydrogenase | Chr4 | 4.23 | 1.86 (7.87) |
| CPSF2 | Cleavage and polyadenylation specific factor 2 | Chr6 | 2.84 | 2.61 (7.41) |
| ESRRA | Estrogen-related receptor alpha | Chr1 | 3.73 | 1.79 (6.68) |
| ANKRD13D | Ankyrin repeat domain 13 family, member D | Chr1 | 4.38 | 1.51 (6.61) |
| TTBK1 | Tau Tubulin Kinase 1 | Chr9 | 3.79 | 1.72 (6.52) |
| TBCCD1 | Tubulin cofactor C TBCC-domain containing 1 | Chr11 | 4.84 | 1.26 (6.10) |
| FAM220A | Family with sequence similarity 220, member A | Chr12 | 4.55 | 1.33 (6.05) |
| CD302 | CD302 antigen | Chr3 | 5.06 | 1.19 (6.02) |
| MTMR7 | Myotubularin-related protein 7 | Chr16 | 2.91 | 2.04 (5.94) |
| Gene Symbol | Gene Name | Chromosome | Relative Fold of RPKM Expression between 5 mg/kg and Control Groups * | Relative Fold of RPKM Expression between 500 mg/kg and 5 mg/kg Groups # |
|---|---|---|---|---|
| ATG16L1 | Autophagy Atg16L1 | Chr9 | 0.47 | 0.50 (0.24) a |
| FAM13A | Family with sequence similarity 13, member A | Chr4 | 0.49 | 0.49 (0.24) |
| ATOH7 | Atonal Homolog 7 | Chr20 | 0.50 | 0.68 (0.34) |
| BCAP29 | B-cell receptor-associated protein 29 | Chr6 | 0.36 | 0.98 (0.35) |
| MRPS18B | Mitochondrial ribosomal protein S18B | Chr20 | 0.42 | 0.85 (0.36) |
| DUSP22 | Dual-specificity phosphatases 22 | Chr17 | 0.39 | 0.92 (0.36) |
| AOX2 | Alcohol oxidase 2 | Chr9 | 0.46 | 0.81 (0.37) |
| RBM | RNA-binding motif | Chr8 | 0.50 | 0.77 (0.39) |
| MED24 | Mediator complex subunit 24 | Chr10 | 0.42 | 0.92 (0.39) |
| DDX19A | DEAD box polypeptide 19A | Chr19 | 0.46 | 0.85 (0.39) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hsu, P.-C.; Jhong, J.-Y.; Huang, L.-P.; Lee, K.-H.; Chen, H.-P.; Guo, Y.-L. Transgenerational Effects of Di(2-Ethylhexyl) Phthalate on Anogenital Distance, Sperm Functions and DNA Methylation in Rat Offspring. Int. J. Mol. Sci. 2021, 22, 4131. https://doi.org/10.3390/ijms22084131
Hsu P-C, Jhong J-Y, Huang L-P, Lee K-H, Chen H-P, Guo Y-L. Transgenerational Effects of Di(2-Ethylhexyl) Phthalate on Anogenital Distance, Sperm Functions and DNA Methylation in Rat Offspring. International Journal of Molecular Sciences. 2021; 22(8):4131. https://doi.org/10.3390/ijms22084131
Chicago/Turabian StyleHsu, Ping-Chi, Jia-Ying Jhong, Li-Ping Huang, Kuo-Hsin Lee, Hsin-Pao Chen, and Yue-Leon Guo. 2021. "Transgenerational Effects of Di(2-Ethylhexyl) Phthalate on Anogenital Distance, Sperm Functions and DNA Methylation in Rat Offspring" International Journal of Molecular Sciences 22, no. 8: 4131. https://doi.org/10.3390/ijms22084131
APA StyleHsu, P.-C., Jhong, J.-Y., Huang, L.-P., Lee, K.-H., Chen, H.-P., & Guo, Y.-L. (2021). Transgenerational Effects of Di(2-Ethylhexyl) Phthalate on Anogenital Distance, Sperm Functions and DNA Methylation in Rat Offspring. International Journal of Molecular Sciences, 22(8), 4131. https://doi.org/10.3390/ijms22084131






