Arpin Regulates Migration Persistence by Interacting with Both Tankyrases and the Arp2/3 Complex
Abstract
1. Introduction
2. Results
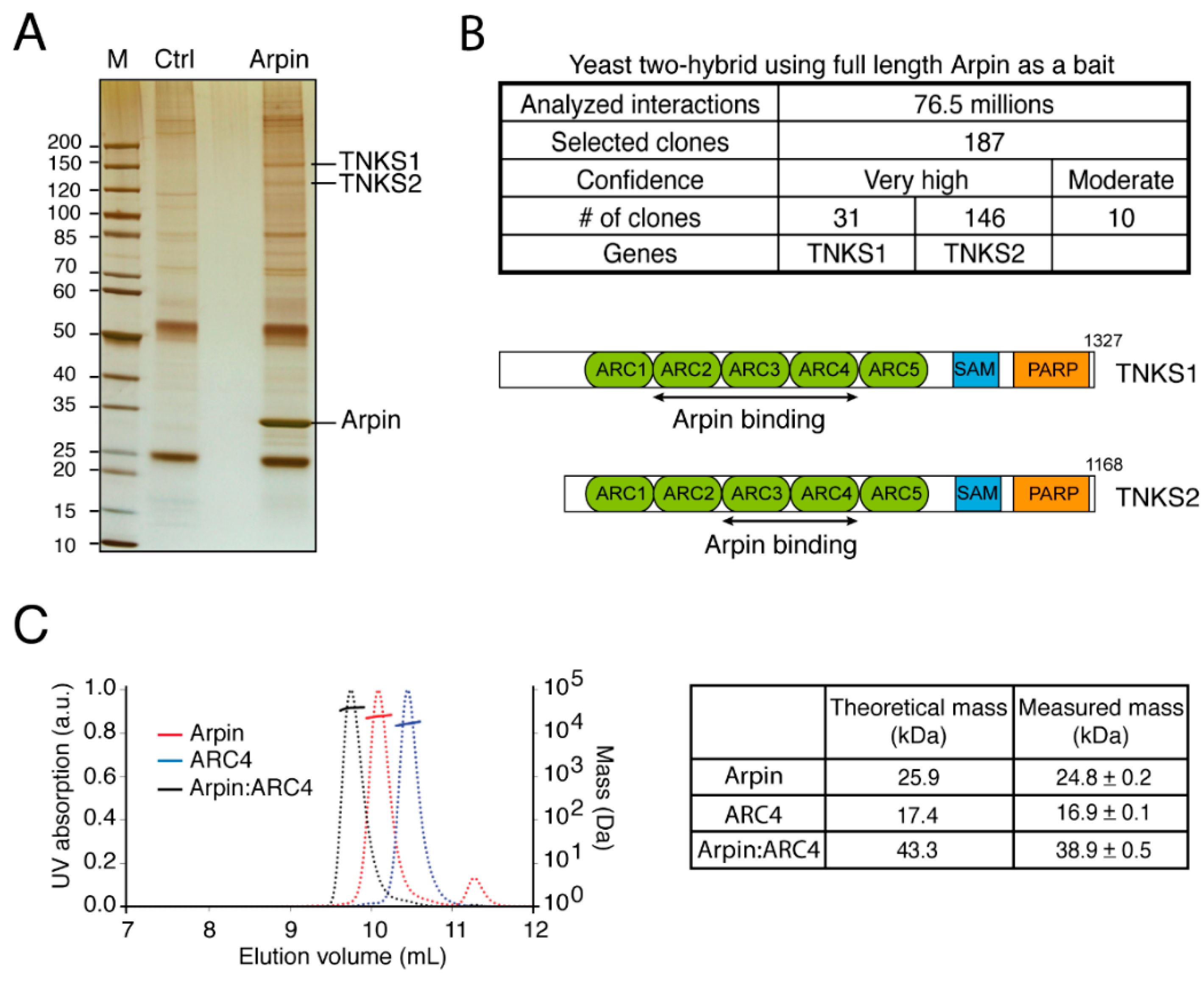
2.1. Arpin Binds to Tankyrase 1 and 2
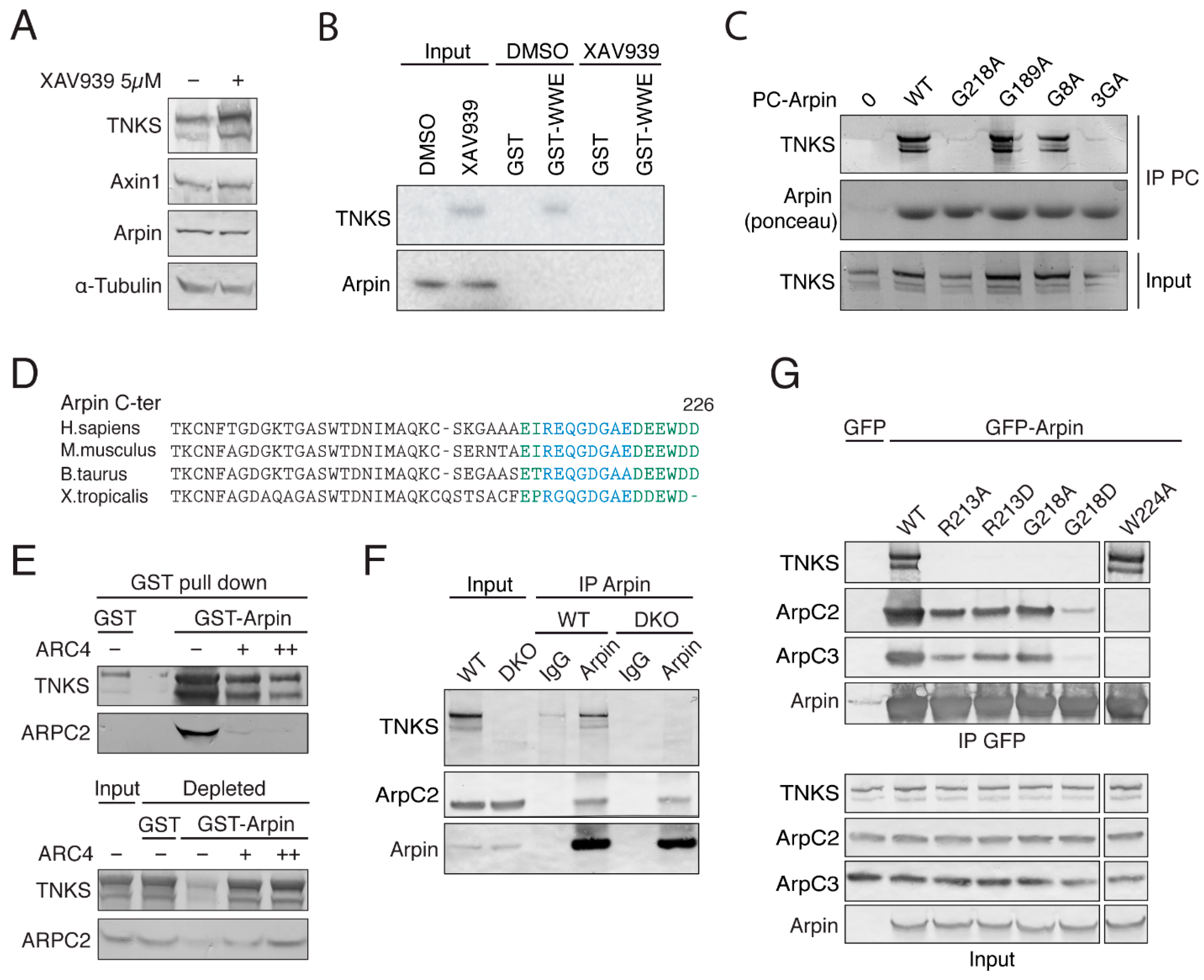
2.2. TNKS Do Not Regulate Arpin Levels
2.3. Arpin Binds to TNKS via Its C-Terminal Acidic Tail
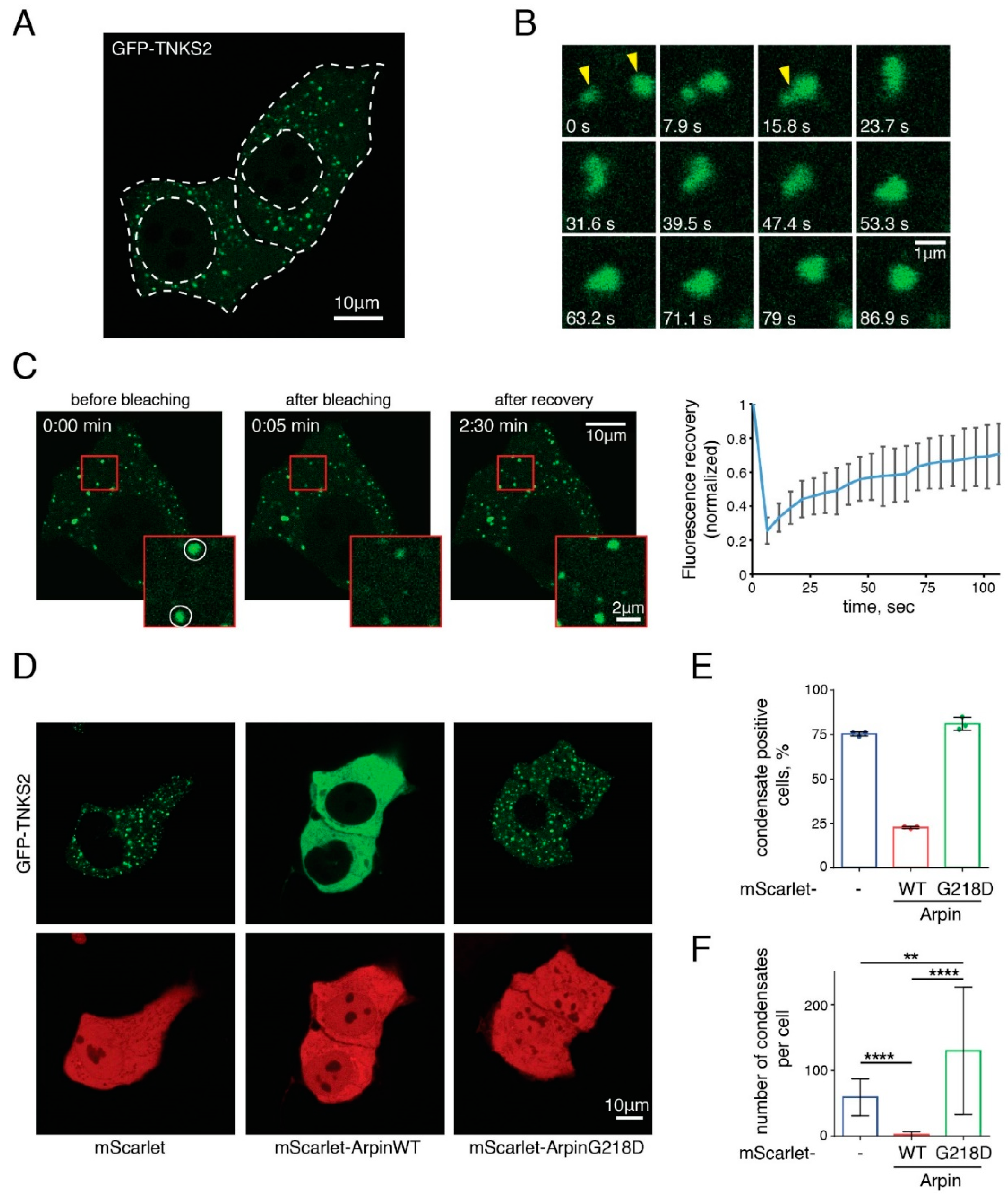
2.4. Arpin Controls the Ability of TNKS to Form Biomolecular Condensates
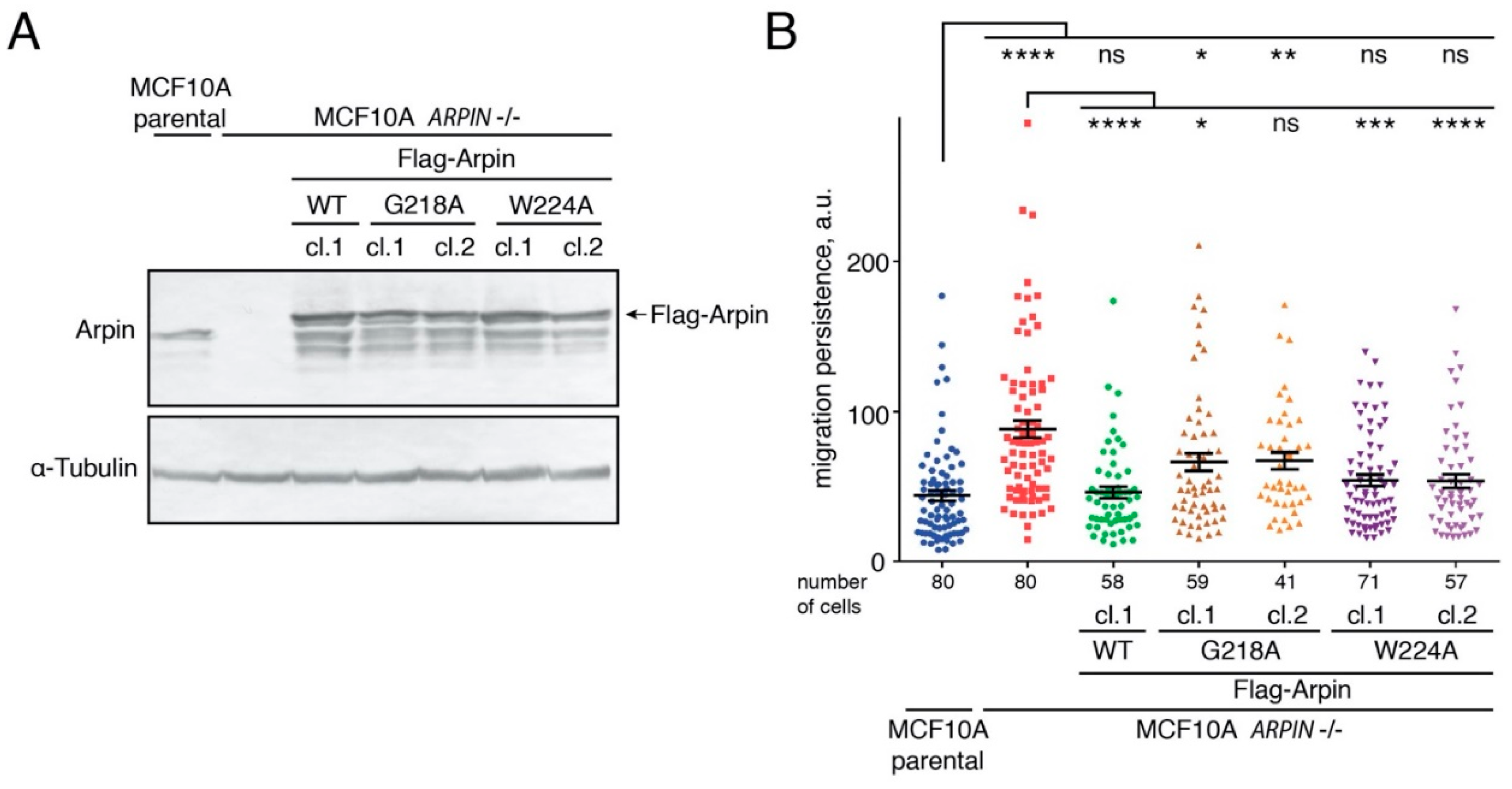
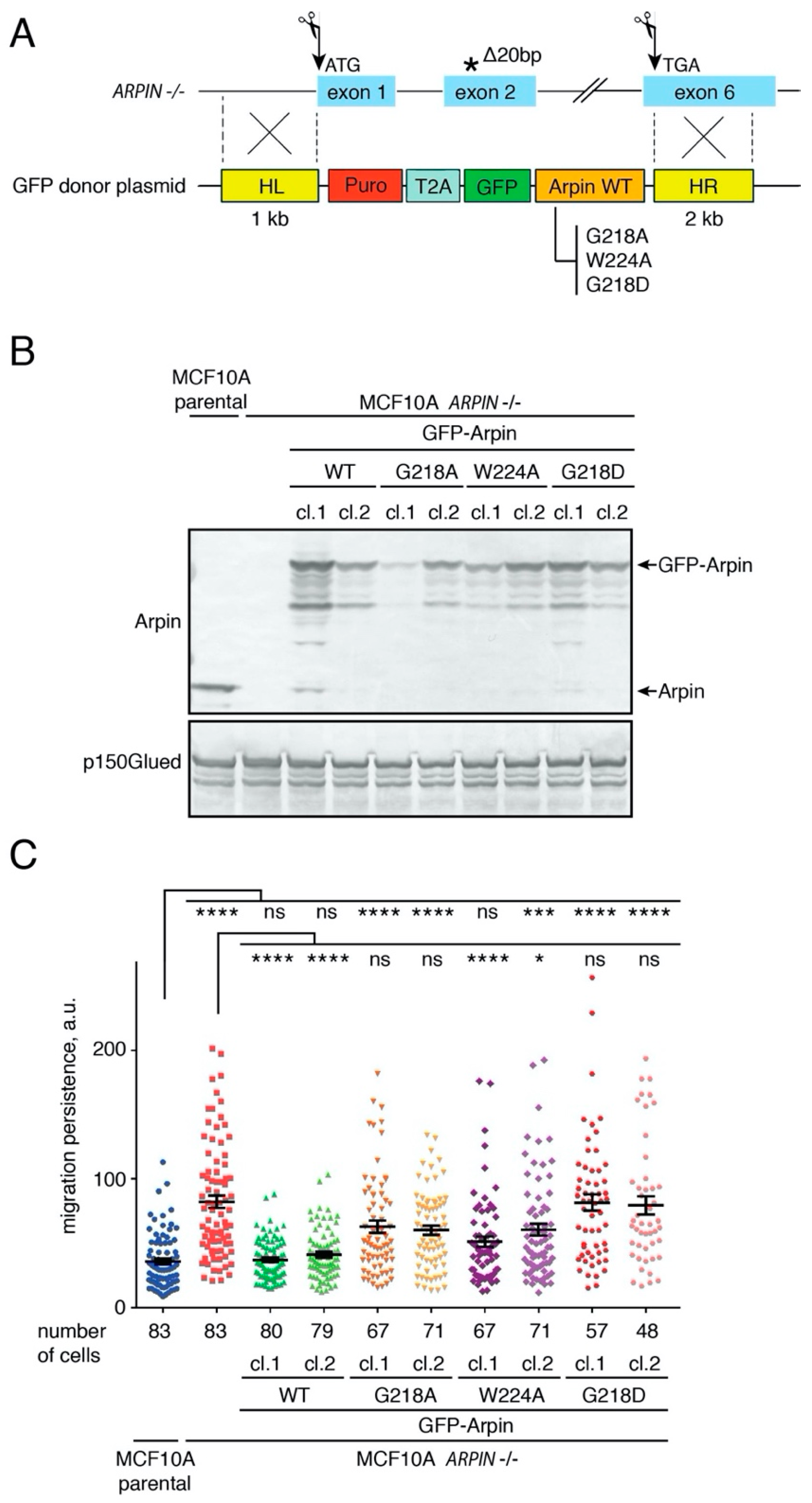
2.5. The Arpin–TNKS Interaction Participates in the Regulation of Cell Migration
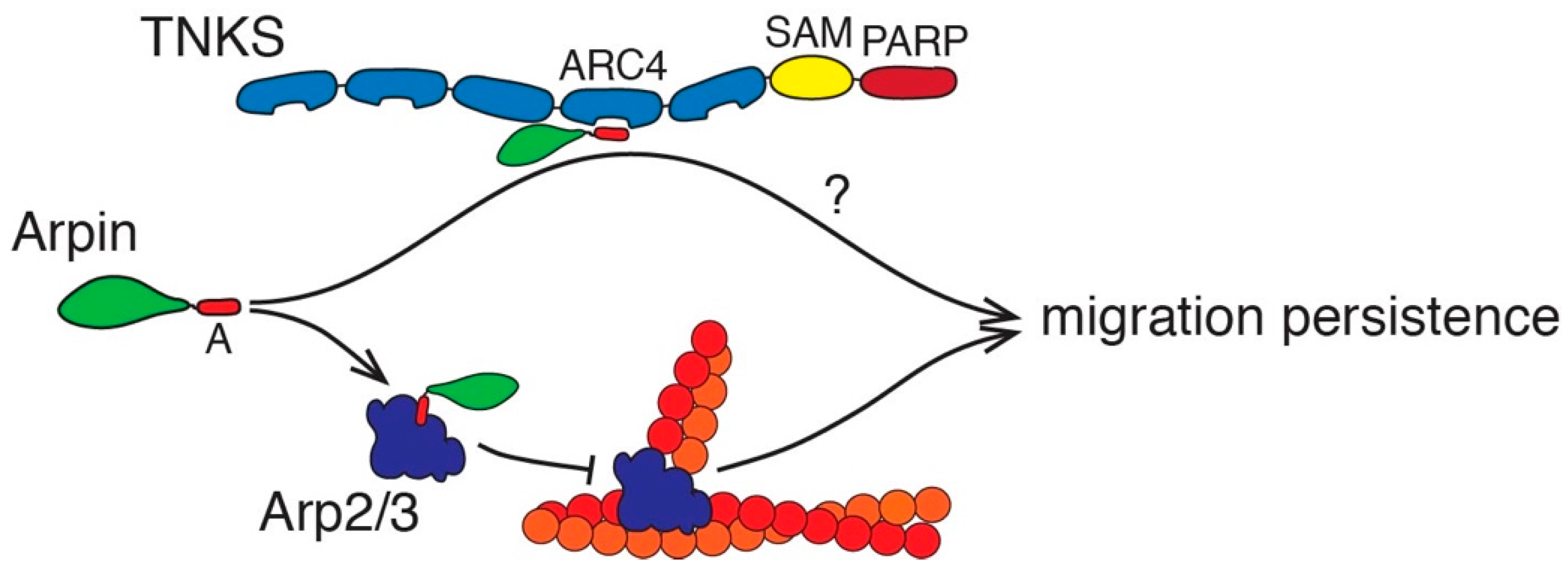
3. Discussion
4. Materials and Methods
4.1. Plasmids, gRNAs, and Transfection
4.2. Cell Culture and Drugs
4.3. Immunoprecipitation and GST Pulldown
4.4. SEC-MALS
4.5. Protein Purification and Analysis of Arpin Expression
4.6. SDS-PAGE, Western Blots, and Antibodies
4.7. Immunofluorescence
4.8. Live Confocal Imaging
4.9. Live Imaging and Analysis of Cell Migration and Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ARC | Ankyrin Repeat Cluster |
| DSB | Double-Stranded Break |
| FRAP | Fluorescence Recovery After Photobleaching |
| KI | Knock-in |
| KO | Knockout |
| HDR | Homology-Directed Repair |
| MEF | Murine Embryonic Fibroblast |
| NPF | Nucleation-Promoting Factor |
| PARP | Poly ADP Ribose Polymerase |
| PC | Protein C |
| SAM | Sterile Alpha Motif |
| SEC-MALS | Size Exclusion Chromatography–Multi-Angle Light Scattering |
| TNKS | Tankyrases |
| WT | Wild type |
References
- Ridley, A.J. Life at the Leading Edge. Cell 2011, 145, 1012–1022. [Google Scholar] [CrossRef]
- Steffen, A.; Koestler, S.A.; Rottner, K. Requirements for and consequences of rac-dependent protrusion. Eur. J. Cell Biol. 2014, 93, 184–193. [Google Scholar] [CrossRef] [PubMed]
- Molinie, N.; Gautreau, A. The Arp2/3 regulatory system and its deregulation in cancer. Physiol. Rev. 2017, 98, 215–238. [Google Scholar] [CrossRef]
- Dang, I.; Gorelik, R.; Sousa-Blin, C.; Derivery, E.; Guérin, C.; Linkner, J.; Nemethova, M.; Dumortier, J.G.; Giger, F.A.; Chipysheva, T.A.; et al. Inhibitory signalling to the Arp2/3 complex steers cell migration. Nature 2013, 503, 281–284. [Google Scholar] [CrossRef]
- Fetics, S.; Thureau, A.; Campanacci, V.; Aumont-Nicaise, M.; Dang, I.; Gautreau, A.; Pérez, J.; Cherfils, J. Hybrid structural analysis of the Arp2/3 regulator arpin identifies its acidic tail as a primary binding epitope. Structure 2016, 24, 252–260. [Google Scholar] [CrossRef]
- Krause, M.; Gautreau, A. Steering cell migration: Lamellipodium dynamics and the regulation of directional persistence. Nat. Rev. Mol. Cell Biol. 2014, 15, 577–590. [Google Scholar] [CrossRef]
- Dimchev, V.; Lahmann, I.; Koestler, S.A.; Kage, F.; Dimchev, G.; Steffen, A.; Stradal, T.E.B.; Vauti, F.; Arnold, H.-H.; Rottner, K. Induced Arp2/3 complex depletion increases FMNL2/3 formin expression and filopodia formation. Front. Cell Dev. Biol. 2021, 9. [Google Scholar] [CrossRef] [PubMed]
- Gorelik, R.; Gautreau, A. The Arp2/3 inhibitory protein arpin induces cell turning by pausing cell migration. Cytoskeleton 2015, 72, 362–371. [Google Scholar] [CrossRef]
- Hsiao, S.J.; Smith, S. Tankyrase function at telomeres, spindle poles, and beyond. Biochimie 2008, 90, 83–92. [Google Scholar] [CrossRef]
- Guettler, S.; LaRose, J.; Petsalaki, E.; Gish, G.; Scotter, A.; Pawson, T.; Rottapel, R.; Sicheri, F. Structural Basis and sequence rules for substrate recognition by tankyrase explain the basis for cherubism disease. Cell 2011, 147, 1340–1354. [Google Scholar] [CrossRef]
- Smith, S.; Giriat, I.; Schmitt, A.; de Lange, T. Tankyrase, a Poly(ADP-Ribose) polymerase at human telomeres. Science 1998, 282, 1484–1487. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.; Coughlin, M.; Mitchison, T.J. Tankyrase-1 polymerization of poly(ADP-Ribose) is required for spindle structure and function. Nat. Cell Biol. 2005, 7, 1133–1139. [Google Scholar] [CrossRef]
- Chang, W.; Dynek, J.N.; Smith, S. NuMA is a major acceptor of poly(ADP-Ribosyl)ation by tankyrase 1 in mitosis. Biochem. J. 2005, 391, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Riffell, J.L.; Lord, C.J.; Ashworth, A. Tankyrase-targeted therapeutics: Expanding opportunities in the PARP family. Nat. Rev. Drug Discov. 2012, 11, 923–936. [Google Scholar] [CrossRef]
- Kim, M.K.; Dudognon, C.; Smith, S. Tankyrase 1 regulates centrosome function by controlling CPAP stability. EMBO Rep. 2012, 13, 724–732. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.-M.A.; Mishina, Y.M.; Liu, S.; Cheung, A.; Stegmeier, F.; Michaud, G.A.; Charlat, O.; Wiellette, E.; Zhang, Y.; Wiessner, S.; et al. Tankyrase inhibition stabilizes axin and antagonizes wnt signalling. Nature 2009, 461, 614–620. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Li, N.; Li, X.; Tran, M.K.; Han, X.; Chen, J. Tankyrase inhibitors target YAP by stabilizing angiomotin family proteins. Cell Rep. 2015, 13, 524–532. [Google Scholar] [CrossRef] [PubMed]
- Eisemann, T.; McCauley, M.; Langelier, M.-F.; Gupta, K.; Roy, S.; Van Duyne, G.D.; Pascal, J.M. Tankyrase-1 ankyrin repeats form an adaptable binding platform for targets of ADP-ribose modification. Structure 2016, 24, 1679–1692. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, S.; Mickanin, C.; Feng, Y.; Charlat, O.; Michaud, G.A.; Schirle, M.; Shi, X.; Hild, M.; Bauer, A.; et al. RNF146 is a poly(ADP-Ribose)-directed E3 ligase that regulates axin degradation and wnt signalling. Nat. Cell Biol. 2011, 13, 623–629. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, A.; Yang, Y.; Ueberheide, B.; Smith, S. Whole Proteome analysis of human tankyrase knockout cells reveals targets of tankyrase-mediated degradation. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef]
- Li, X.; Han, H.; Zhou, M.-T.; Yang, B.; Ta, A.P.; Li, N.; Chen, J.; Wang, W. Proteomic Analysis of the human tankyrase protein interaction network reveals its role in pexophagy. Cell Rep. 2017, 20, 737–749. [Google Scholar] [CrossRef] [PubMed]
- Bae, J.; Donigian, J.R.; Hsueh, A.J.W. Tankyrase 1 interacts with MCL-1 proteins and inhibits their regulation of apoptosis. J. Biol. Chem. 2003, 278, 5195–5204. [Google Scholar] [CrossRef]
- Bisht, K.K.; Dudognon, C.; Chang, W.G.; Sokol, E.S.; Ramirez, A.; Smith, S. GDP-mannose-4,6-Dehydratase is a cytosolic partner of tankyrase 1 that inhibits its poly(ADP-Ribose) polymerase activity. Mol. Cell. Biol. 2012, 32, 3044–3053. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kuusela, S.; Wang, H.; Wasik, A.A.; Suleiman, H.; Lehtonen, S. Tankyrase inhibition aggravates kidney injury in the absence of CD2AP. Cell Death Dis. 2016, 7, e2302. [Google Scholar] [CrossRef]
- Perdreau-Dahl, H.; Progida, C.; Barfeld, S.J.; Guldsten, H.; Thiede, B.; Arntzen, M.; Bakke, O.; Mills, I.G.; Krauss, S.; Morth, J.P. Sjögren syndrome/scleroderma autoantigen 1 is a direct tankyrase binding partner in cancer cells. Commun. Biol. 2020, 3, 1–11. [Google Scholar] [CrossRef]
- Pollard, T.D. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu. Rev. Biophys. Biomol. Struct. 2007, 36, 451–477. [Google Scholar] [CrossRef]
- Mariotti, L.; Templeton, C.M.; Ranes, M.; Paracuellos, P.; Cronin, N.; Beuron, F.; Morris, E.; Guettler, S. Tankyrase requires SAM domain-dependent polymerization to support Wnt-β-Catenin signaling. Mol. Cell 2016, 63, 498–513. [Google Scholar] [CrossRef] [PubMed]
- Riccio, A.A.; McCauley, M.; Langelier, M.-F.; Pascal, J.M. Tankyrase sterile α motif domain polymerization is required for its role in wnt signaling. Structure 2016, 24, 1573–1581. [Google Scholar] [CrossRef]
- De Rycker, M.; Price, C.M. Tankyrase Polymerization is controlled by its sterile alpha motif and poly(ADP-Ribose) polymerase domains. Mol. Cell. Biol. 2004, 24, 9802–9812. [Google Scholar] [CrossRef]
- Alberti, S.; Gladfelter, A.; Mittag, T. Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell 2019, 176, 419–434. [Google Scholar] [CrossRef]
- Morrone, S.; Cheng, Z.; Moon, R.T.; Cong, F.; Xu, W. Crystal structure of a tankyrase-axin complex and its implications for axin turnover and tankyrase substrate recruitment. Proc. Natl. Acad. Sci. USA 2012, 109, 1500–1505. [Google Scholar] [CrossRef]
- Molinie, N.; Rubtsova, S.N.; Fokin, A.; Visweshwaran, S.P.; Rocques, N.; Polesskaya, A.; Schnitzler, A.; Vacher, S.; Denisov, E.V.; Tashireva, L.A.; et al. Cortical branched actin determines cell cycle progression. Cell Res. 2019, 29, 432–445. [Google Scholar] [CrossRef] [PubMed]
- Gorelik, R.; Gautreau, A. Quantitative and unbiased analysis of directional persistence in cell migration. Nat. Protoc. 2014, 9, 1931–1943. [Google Scholar] [CrossRef] [PubMed]
- Bao, R.; Christova, T.; Song, S.; Angers, S.; Yan, X.; Attisano, L. Inhibition of tankyrases induces axin stabilization and blocks wnt signalling in breast cancer cells. PLoS ONE 2012, 7, e48670. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.-W.; Wang, F.; Wei, Q.; Zhao, Y.-F.; Liu, M.; Li, X.; Tang, H. MiR-20a promotes migration and invasion by regulating TNKS2 in human cervical cancer cells. FEBS Lett. 2012, 586, 897–904. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Hou, W.; Bai, S.; Fan, J.; Tong, H.; Xu, H. XAV939 Inhibits the stemness and migration of neuroblastoma cancer stem cells via repression of tankyrase 1. Int. J. Oncol. 2014, 45, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Lupo, B.; Vialard, J.; Sassi, F.; Angibaud, P.; Puliafito, A.; Pupo, E.; Lanzetti, L.; Comoglio, P.M.; Bertotti, A.; Trusolino, L. Tankyrase inhibition impairs directional migration and invasion of lung cancer cells by affecting microtubule dynamics and polarity signals. BMC Biol. 2016, 14, 5. [Google Scholar] [CrossRef]
- Li, C.; Zheng, X.; Han, Y.; Lv, Y.; Lan, F.; Zhao, J. XAV939 inhibits the proliferation and migration of lung adenocarcinoma A549 cells through the WNT pathway. Oncol. Lett. 2018, 15, 8973–8982. [Google Scholar] [CrossRef]
- Ha, G.-H.; Kim, D.-Y.; Breuer, E.-K.; Kim, C.K. Combination treatment of polo-like kinase 1 and tankyrase-1 inhibitors enhances anticancer effect in triple-negative breast cancer cells. Anticancer Res. 2018, 38, 1303–1310. [Google Scholar]
- Yang, H.-Y.; Shen, J.-X.; Wang, Y.; Liu, Y.; Shen, D.-Y.; Quan, S. Tankyrase promotes aerobic glycolysis and proliferation of ovarian cancer through activation of Wnt/β-Catenin signaling. BioMed. Res. Int. 2019, 2019, 2686340. [Google Scholar] [CrossRef]
- Huang, J.; Qu, Q.; Guo, Y.; Xiang, Y.; Feng, D. Tankyrases/β-Catenin signaling pathway as an anti-proliferation and anti-metastatic target in hepatocarcinoma cell lines. J. Cancer 2020, 11, 432–440. [Google Scholar] [CrossRef]
- Ohishi, T.; Yoshida, H.; Katori, M.; Migita, T.; Muramatsu, Y.; Miyake, M.; Ishikawa, Y.; Saiura, A.; Iemura, S.; Natsume, T.; et al. Tankyrase-binding protein TNKS1BP1 Regulates actin cytoskeleton rearrangement and cancer cell invasion. Cancer Res. 2017, 77, 2328–2338. [Google Scholar] [CrossRef]
- Li, P.; Banjade, S.; Cheng, H.-C.; Kim, S.; Chen, B.; Guo, L.; Llaguno, M.; Hollingsworth, J.V.; King, D.S.; Banani, S.F.; et al. Phase transitions in the assembly of multivalent signalling proteins. Nature 2012, 483, 336–340. [Google Scholar] [CrossRef] [PubMed]
- Diamante, A.; Chaturbedy, P.K.; Rowling, P.J.E.; Kumita, J.R.; Eapen, R.S.; McLaughlin, S.H.; de la Roche, M.; Perez-Riba, A.; Itzhaki, L.S. Engineering Mono- and multi-valent inhibitors on a modular scaffold. Chem. Sci. 2021, 12, 880–895. [Google Scholar] [CrossRef] [PubMed]
- Lomakina, M.E.; Lallemand, F.; Vacher, S.; Molinie, N.; Dang, I.; Cacheux, W.; Chipysheva, T.A.; Ermilova, V.D.; de Koning, L.; Dubois, T.; et al. Arpin downregulation in breast cancer is associated with poor prognosis. Br. J. Cancer 2016, 114, 545–553. [Google Scholar] [CrossRef]
- Liu, X.; Zhao, B.; Wang, H.; Wang, Y.; Niu, M.; Sun, M.; Zhao, Y.; Yao, R.; Qu, Z. Aberrant expression of arpin in human breast cancer and its clinical significance. J. Cell. Mol. Med. 2016, 20, 450–458. [Google Scholar] [CrossRef][Green Version]
- Li, T.; Zheng, H.-M.; Deng, N.-M.; Jiang, Y.-J.; Wang, J.; Zhang, D.-L. Clinicopathological and Prognostic significance of aberrant arpin expression in gastric cancer. World J. Gastroenterol. 2017, 23, 1450–1457. [Google Scholar] [CrossRef]
- Zhang, S.-R.; Li, H.; Wang, W.-Q.; Jin, W.; Xu, J.-Z.; Xu, H.-X.; Wu, C.-T.; Gao, H.-L.; Li, S.; Li, T.-J.; et al. Arpin downregulation is associated with poor prognosis in pancreatic ductal adenocarcinoma. Eur. J. Surg. Oncol. 2019, 45, 769–775. [Google Scholar] [CrossRef] [PubMed]
- Derivery, E.; Gautreau, A. Assaying WAVE and WASH Complex Constitutive Activities toward the Arp2/3 Complex. In Methods in Enzymology; Conn, P.M., Ed.; Academic Press: Cambridge, MA, USA, 2010; Volume 484, Chapter 33; pp. 677–695. [Google Scholar]
- Sladitschek, H.L.; Neveu, P.A. MXS-chaining: A highly efficient cloning platform for imaging and flow cytometry approaches in mammalian systems. PLoS ONE 2015, 10, e0124958. [Google Scholar] [CrossRef] [PubMed]
- Polesskaya, A.; Boutillon, A.; Wang, Y.; Lavielle, M.; Vacher, S.; Schnitzler, A.; Molinie, N.; Rocques, N.; Fokin, A.; Bièche, I.; et al. CYFIP2 containing WAVE complexes inhibit cell migration. Biorxiv 2020. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Simanov, G.; Dang, I.; Fokin, A.I.; Oguievetskaia, K.; Campanacci, V.; Cherfils, J.; Gautreau, A.M. Arpin Regulates Migration Persistence by Interacting with Both Tankyrases and the Arp2/3 Complex. Int. J. Mol. Sci. 2021, 22, 4115. https://doi.org/10.3390/ijms22084115
Simanov G, Dang I, Fokin AI, Oguievetskaia K, Campanacci V, Cherfils J, Gautreau AM. Arpin Regulates Migration Persistence by Interacting with Both Tankyrases and the Arp2/3 Complex. International Journal of Molecular Sciences. 2021; 22(8):4115. https://doi.org/10.3390/ijms22084115
Chicago/Turabian StyleSimanov, Gleb, Irene Dang, Artem I. Fokin, Ksenia Oguievetskaia, Valérie Campanacci, Jacqueline Cherfils, and Alexis M. Gautreau. 2021. "Arpin Regulates Migration Persistence by Interacting with Both Tankyrases and the Arp2/3 Complex" International Journal of Molecular Sciences 22, no. 8: 4115. https://doi.org/10.3390/ijms22084115
APA StyleSimanov, G., Dang, I., Fokin, A. I., Oguievetskaia, K., Campanacci, V., Cherfils, J., & Gautreau, A. M. (2021). Arpin Regulates Migration Persistence by Interacting with Both Tankyrases and the Arp2/3 Complex. International Journal of Molecular Sciences, 22(8), 4115. https://doi.org/10.3390/ijms22084115





