Machine Learning-Based Classification of Lignocellulosic Biomass from Pyrolysis-Molecular Beam Mass Spectrometry Data
Abstract
1. Introduction
2. Results
2.1. Sourcing and Removal of Instrumental Variance
2.2. Statistics, Principal Component Analysis, and Clustering
2.3. Classification Models
2.3.1. Classification of Biomass Prior to Removal of Unwanted Variance (RUV) Correction
2.3.2. Models Constructed from Data after RUV Correction
2.3.3. Classification of Biomass Mixtures
3. Discussion
4. Materials and Methods
4.1. Lignocellulosic Biomass Feedstocks
4.2. Pyrolysis-Molecular Beam Mass Spectrometry (py-MBMS)
4.3. Spectral Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ANN | Artificial neural network |
| DA | Discriminant analysis |
| GLM | Generalized linear model |
| GNB | Gaussian naïve Bayes |
| ML | Machine learning |
| MLP | Multi-layer perceptron |
| PCA | Principal component analysis |
| PCR | Polymerase chain reaction |
| PLS | Partial least squares |
| Py-MBMS | Pyrolysis molecular beam mass spectrometry |
| RF | Random forest |
| RNA | Ribo-nucleic acid |
| RUV | Remove unwanted variation |
| XGB | Extreme gradient boosting |
| MDPI | Multidisciplinary Digital Publishing Institute |
| NIST | National Institute of Standards and Technology |
| DOAJ | Directory of open access journals |
| ANN | Artificial neural network |
| DA | Discriminant analysis |
| GLM | Generalized linear model |
| GNB | Gaussian Naïve Bayes |
| ML | Machine learning |
| MLP | Multi-layer perceptron |
| PCA | Principal component analysis |
References
- Davis, M.F.; Johnson, D.K.; Deutch, S.P.; Agblevor, F.A.; Fennell, J.; Ashley, P. Variability in the Composition of Short Rotation Woody Feedstocks. In Second Biomass Conference of the Americas: Energy, Environment, Agriculture, and Industry Proceedings; National Renewable Energy Laboratory: Golden, CO, USA, 1995; pp. 216–225. [Google Scholar]
- Johnson, D.; Ashley, P.; Deutch, S.; Davis, M.; Fennell, J.; Wiselogel, A. Compositional Variability in Herbaceous Energy Crops. In Second Biomass Conference of the Americas: Energy, Environment, Agriculture, and Industry Proceedings; National Renewable Energy Laboratory: Golden, CO, USA, 1995; pp. 267–277. [Google Scholar]
- Decker, S.R.; Sykes, R.W.; Turner, G.B.; Lupoi, J.S.; Doepkke, C.; Tucker, M.P.; Schuster, L.A.; Mazza, K.; Himmel, M.E.; Davis, M.F.; et al. High-throughput Screening of Recalcitrance Variations in Lignocellulosic Biomass: Total Lignin, Lignin Monomers, and Enzymatic Sugar Release. JoVE 2015, 103, e53163. [Google Scholar] [CrossRef] [PubMed]
- Harman-Ware, A.E.; Davis, M.F.; Peter, G.F.; Wang, Y.; Sykes, R.W. Estimation of terpene content in loblolly pine biomass using a hybrid fast-GC and pyrolysis-molecular beam mass spectrometry method. J. Anal. Appl. Pyrolysis 2017, 124, 343–348. [Google Scholar] [CrossRef]
- Penning, B.W.; Sykes, R.W.; Babcock, N.C.; Dugard, C.K.; Klimek, J.F.; Gamblin, D.; Davis, M.; Filley, T.R.; Mosier, N.S.; Weil, C.F.; et al. Validation of PyMBMS as a High-throughput Screen for Lignin Abundance in Lignocellulosic Biomass of Grasses. Bioenergy Res. 2014, 7, 899–908. [Google Scholar] [CrossRef]
- Sykes, R.; Kodrzycki, B.; Tuskan, G.; Foutz, K.; Davis, M. Within tree variability of lignin composition in Populus. Wood Sci. Technol. 2008, 42, 649–661. [Google Scholar] [CrossRef]
- Sykes, R.; Yung, M.; Novaes, E.; Kirst, M.; Peter, G.; Davis, M. High-Throughput Screening of Plant Cell-Wall Composition Using Pyrolysis Molecular Beam Mass Spectroscopy. In Biofuels: Methods and Protocols; Mielenz, J.R., Ed.; Humana Press: Totowa, NJ, USA, 2009; pp. 169–183. [Google Scholar]
- Evans, R.J.; Milne, T.A. Molecular characterization of the pyrolysis of biomass. Energy Fuels 1987, 1, 123–137. [Google Scholar] [CrossRef]
- Sykes, R.W.; Gjersing, E.L.; Doeppke, C.L.; Davis, M.F. High-Throughput Method for Determining the Sugar Content in Biomass with Pyrolysis Molecular Beam Mass Spectrometry. Bioenergy Res. 2015, 8, 964–972. [Google Scholar] [CrossRef]
- Windig, W.; Meuzelaar, H.L.C.; Shafizadeh, F.; Kelsey, R.G. Biochemical analysis of wood and wood products by pyrolysis-mass spectrometry and multivariate analysis. J. Anal. Appl. Pyrolysis 1984, 6, 233–250. [Google Scholar] [CrossRef]
- Abdel-Maksoud, G.; El-Amin, A.-R. A review on the materials used during the mummification processes in ancient egypt. Mediterr. Archaeol. Archaeom. 2011, 11, 129–150. [Google Scholar]
- Harman-Ware, A.E.; Crocker, M.; Kaur, A.P.; Meier, M.S.; Kato, D.; Lynn, B. Pyrolysis–GC/MS of sinapyl and coniferyl alcohol. J. Anal. Appl. Pyrolysis 2013, 99, 161–169. [Google Scholar] [CrossRef]
- Arias, M.E.; Blánquez, A.; Hernández, M.; Rodríguez, J.; Ball, A.S.; Jiménez-Morillo, N.T.; González-Vila, F.J.; González-Pérez, J.A. Role of a thermostable laccase produced by Streptomyces ipomoeae in the degradation of wheat straw lignin in solid state fermentation. J. Anal. Appl. Pyrolysis 2016, 122, 202–208. [Google Scholar] [CrossRef]
- Safdari, M.-S.; Rahmati, M.; Amini, E.; Howarth, J.E.; Berryhill, J.P.; Dietenberger, M.; Weise, D.R.; Fletcher, T.H. Characterization of pyrolysis products from fast pyrolysis of live and dead vegetation native to the Southern United States. Fuel 2018, 229, 151–166. [Google Scholar] [CrossRef]
- Patwardhan, P.R.; Satrio, J.A.; Brown, R.C.; Shanks, B.H. Influence of inorganic salts on the primary pyrolysis products of cellulose. Bioresour. Technol. 2010, 101, 4646–4655. [Google Scholar] [CrossRef]
- Wang, S.; Dai, G.; Yang, H.; Luo, Z. Lignocellulosic biomass pyrolysis mechanism: A state-of-the-art review. Prog. Energy Combust. Sci. 2017, 62, 33–86. [Google Scholar] [CrossRef]
- Weighill, D.; Jones, P.; Shah, M.; Ranjan, P.; Muchero, W.; Schmutz, J.; Sreedasyam, A.; Macaya-Sanz, D.; Sykes, R.; Zhao, N.; et al. Pleiotropic and Epistatic Network-Based Discovery: Integrated Networks for Target Gene Discovery. Front. Energy Res. 2018, 6, 30. [Google Scholar] [CrossRef]
- Muchero, W.; Guo, J.; DiFazio, S.P.; Chen, J.-G.; Ranjan, P.; Slavov, G.T.; Gunter, L.E.; Jawdy, S.; Bryan, A.C.; Sykes, R.; et al. High-resolution genetic mapping of allelic variants associated with cell wall chemistry in Populus. BMC Genom. 2015, 16, 24. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, J.; Evrendilek, F.; Zhang, X.; Buyukada, M. TG-FTIR and Py-GC/MS analyses of pyrolysis behaviors and products of cattle manure in CO2 and N2 atmospheres: Kinetic, thermodynamic, and machine-learning models. Energy Convers. Manag. 2019, 195, 346–359. [Google Scholar] [CrossRef]
- Goodacre, R.; Neal, M.J.; Kell, D.B. Quantitative Analysis of Multivariate Data Using Artificial Neural Networks: A Tutorial Review and Applications to the Deconvolution of Pyrolysis Mass Spectra. Zent. Für Bakteriol. 1996, 284, 516–539. [Google Scholar] [CrossRef]
- Goodacre, R.; Kell, D.B. Rapid and quantitative analysis and bioprocesses using pyrolysis mass spectrometry and neural networks: Application to indole production. Anal. Chim. Acta 1993, 279, 17–26. [Google Scholar] [CrossRef]
- Goodacre, R.; Neal, M.J.; Kell, D.B. Rapid and Quantitative Analysis of the Pyrolysis Mass Spectra of Complex Binary and Tertiary Mixtures Using Multivariate Calibration and Artificial Neural Networks. Anal. Chem. 1994, 66, 1070–1085. [Google Scholar] [CrossRef]
- Gromski, P.S.; Xu, Y.; Correa, E.; Ellis, D.I.; Turner, M.L.; Goodacre, R. A comparative investigation of modern feature selection and classification approaches for the analysis of mass spectrometry data. Anal. Chim. Acta 2014, 829, 1–8. [Google Scholar] [CrossRef]
- Goodacre, R.; Kell, D.B. Correction of Mass Spectral Drift Using Artificial Neural Networks. Anal. Chem. 1996, 68, 271–280. [Google Scholar] [CrossRef]
- Goodacre, R.; Trew, S.; Wrigley-Jones, C.; Neal, M.J.; Maddock, J.; Ottley, T.W.; Porter, N.; Kell, D.B. Rapid screening for metabolite overproduction in fermentor broths, using pyrolysis mass spectrometry with multivariate calibration and artificial neural networks. Biotechnol. Bioeng. 1994, 44, 1205–1216. [Google Scholar] [CrossRef]
- Workman, J., Jr.; Howard, M. Survey of Chemometrics Methods in Spectroscopy, Spectroscop. Spectroscopy 2020, 35, 9–14. [Google Scholar]
- Risso, D.; Ngai, J.; Speed, T.P.; Dudoit, S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat. Biotechnol. 2014, 32, 896–902. [Google Scholar] [CrossRef]
- Harman-Ware, A.E.; Macaya-Sanz, D.; Abeyratne, C.R.; Doepkke, C.; Haiby, K.; Tuskan, G.A.; Stanton, B.; DiFazio, S.; Davis, M.F. Accurate Determination of Genotypic Variance of Cell Wall Characteristics of a Populus trichocarpa Pedigree Using High-Throughput Pyrolysis-Molecular Beam Mass Spectrometry. Biotechnol. Biofuels 2020, 14, 1–15. [Google Scholar]
- Decker, S.R.; Harman-Ware, A.E.; Happs, R.M.; Wolfrum, E.J.; Tuskan, G.A.; Kainer, D.; Oguntimein, G.B.; Rodriguez, M.; Weighill, D.; Jones, P.; et al. High Throughput Screening Technologies in Biomass Characterization. Front. Energy Res. 2018, 6, 120. [Google Scholar] [CrossRef]
- Agblevor, F.; Evans, R.; Johnson, D.K. Molecular-beam mass-spectrometric analysis of lignocellulosic materials: I. Herbaceous biomass. J. Anal. Appl. Pyrolysis 1994, 30, 125–144. [Google Scholar] [CrossRef]
- Harman-Ware, A.; Crocker, M.; Pace, R.; Placido, A.; Morton, S., III; DeBolt, S. Characterization of Endocarp Biomass and Extracted Lignin Using Pyrolysis and Spectroscopic Methods. Bioenergy Res. 2014, 8, 350–368. [Google Scholar] [CrossRef]
- Brownlee, J. Improve Model Accuracy with Data Pre-Processing; Machine Learning Mastery Pty. Ltd.: San Juan, Puerto Rico, 2014. [Google Scholar]
- Varghese, D. Comparative Study on Classic Machine Learning Algorithms. Available online: https://towardsdatascience.com/comparative-study-on-classic-machine-learning-algorithms-24f9ff6ab222 (accessed on 15 March 2021).
- Dhiraj, K. Top 5 Advantages and Disadvantages of Decision Tree Algorithm. 2019, Volume 2021. Available online: http://Medium.com (accessed on 15 March 2021).
- Rawale, S. Understanding Decision Tree, Algorithm, Drawbacks and Advantages. 2018, Volume 2021. Available online: http://Medium.com (accessed on 15 March 2021).
- Sluiter, A.; Hames, B.; Ruiz, R.; Scarlata, C.; Sluiter, J.; Templeton, D.; Crocker, D. Determination of Structural Carbohydrates and Lignin in Biomass. Lab. Anal. Proc. 2011, 1617, 1–16. [Google Scholar]
- Sluiter, A.; Ruiz, R.; Scarlata, C.; Sluiter, J.; Templeton, D. Determination of extractives in biomass. Lab. Anal. Proced. 2005, 1617. [Google Scholar]
- Hu, Z.; Sykes, R.; Davis, M.F.; Charles Brummer, E.; Ragauskas, A.J. Chemical profiles of switchgrass. Bioresour. Technol. 2010, 101, 3253–3257. [Google Scholar] [CrossRef] [PubMed]
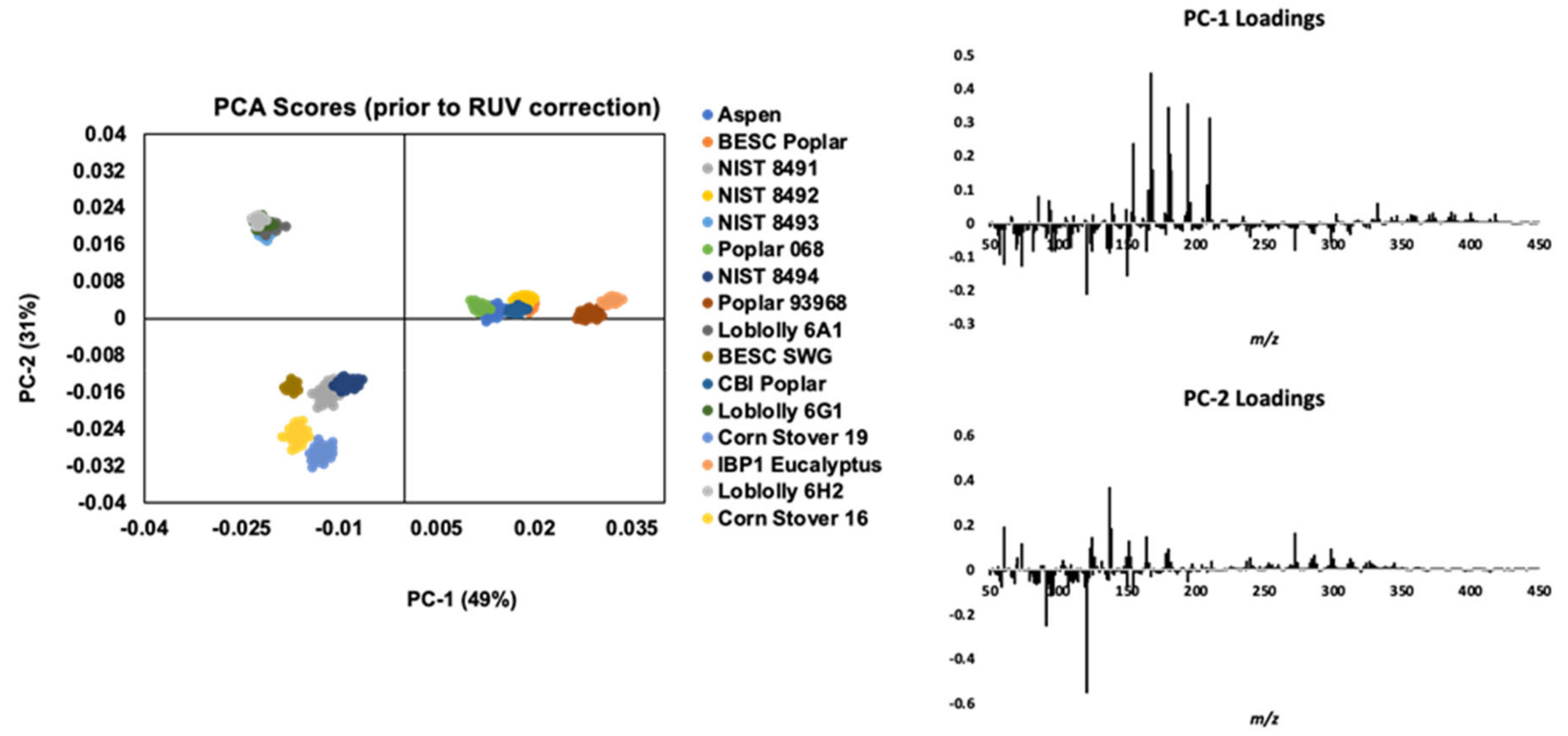

| Number of | Primary Type | Secondary Type | Sample ID |
|---|---|---|---|
| Classes | 3 | 10 | 16 |
| Samples | 816 | 816 | 816 |
| Machine Learning Algorithm | Primary | Secondary | Sample ID | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Random Forest Classifier | Number of Features | Accuracy (CV Score) | Number of Features | Accuracy (CV Score) | Number of Features | Accuracy (CV Score) | |||
| All features | 127 | 1.0 (1.0) | All features | 127 | 1.0 (1.0) | All features | 127 | 0.98 (0.99) | |
| Correlated features removed | 106 | 1.0 (1.0) | Correlated features removed | 106 | 0.99 (0.99) | Correlated features removed | 106 | 0.96 (0.97) | |
| p-value based feature selection | 48 | 1.0 (1.0) | p-value based feature selection | 47 | 1.0 (0.99) | p-value based feature selection | 43 | 0.97 (0.96) | |
| Decision Tree Classifier | Number of Features | Accuracy (CV Score) | Number of Features | Accuracy (CV Score) | Number of Features | Accuracy (CV Score) | |||
| All features | 127 | 1.0 (1.0) | All features | 127 | 0.93 (0.95) | All features | 127 | 0.95 (0.93) | |
| Correlated features removed | 106 | 1.0 (0.99) | Correlated features removed | 106 | 0.94 (0.93) | Correlated features removed | 106 | 0.93 (0.90) | |
| p-value based feature selection | 48 | 1.0 (1.0) | p-value based feature selection | 47 | 0.93 (0.94) | p-value based feature selection | 43 | 0.89 (0.92) | |
| k-NN Classifier | Number of Features | Accuracy (CV Score) | Number of Features | Accuracy (CV Score) | Number of Features | Accuracy (CV Score) | |||
| All features | 127 | 1.0 (1.0) | All features | 127 | 1.0 (1.0) | All features | 127 | 0.97 (0.97) | |
| Correlated features removed | 106 | 1.0 (1.0) | Correlated features removed | 106 | 1.0 (1.0) | Correlated features removed | 106 | 0.98 (0.97) | |
| p-value based feature selection | 48 | 1.0 (1.0) | p-value based feature selection | 47 | 1.0 (1.0) | p-value based feature selection | 43 | 0.96 (0.96) | |
| GNB Classifier | Number of Features | Accuracy (CV Score) | Number of Features | Accuracy (CV Score) | Number of Features | Accuracy (CV Score) | |||
| All features | 127 | 1.0 (1.0) | All features | 127 | 0.96(0.96) | All features | 127 | 0.98 (0.98) | |
| Correlated features removed | 106 | 1.0 (1.0) | Correlated features removed | 106 | 0.95(0.95) | Correlated features removed | 106 | 0.97 (0.97) | |
| p-value based feature selection | 48 | 1.0 (1.0) | p-value based feature selection | 47 | 0.96(0.96) | p-value based feature selection | 43 | 0.97 (0.97) | |
| XGB Classifier | Number of Features | Accuracy (CV Score) | Number of Features | Accuracy (CV Score) | Number of Features | Accuracy (CV Score) | |||
| All features | 127 | 1.0 (1.0) | All features | 127 | 0.99(0.99) | All features | 127 | 0.98 (0.97) | |
| Correlated features removed | 106 | 1.0 (1.0) | Correlated features removed | 106 | 0.99(0.98) | Correlated features removed | 106 | 0.94 (0.95) | |
| p-value based feature selection | 48 | 1.0 (1.0) | p-value based feature selection | 47 | 0.99(0.98) | p-value based feature selection | 43 | 0.95 (0.95) | |
| MLP Classifier | Number of Features | Accuracy (CV Score) | Number of Features | Accuracy (CV Score) | Number of Features | Accuracy (CV Score) | |||
| All features | 127 | 1.0 (1.0) | All features | 127 | 0.99(0.99) | All features | 127 | 0.97 (0.97) | |
| Correlated features removed | 106 | 1.0 (1.0) | Correlated features removed | 106 | 0.99(0.99) | Correlated features removed | 106 | 0.97 (0.96) | |
| p-value based feature selection | 48 | 1.0 (1.0) | p-value based feature selection | 47 | 0.99(0.99) | p-value based feature selection | 43 | 0.95 (0.95) |
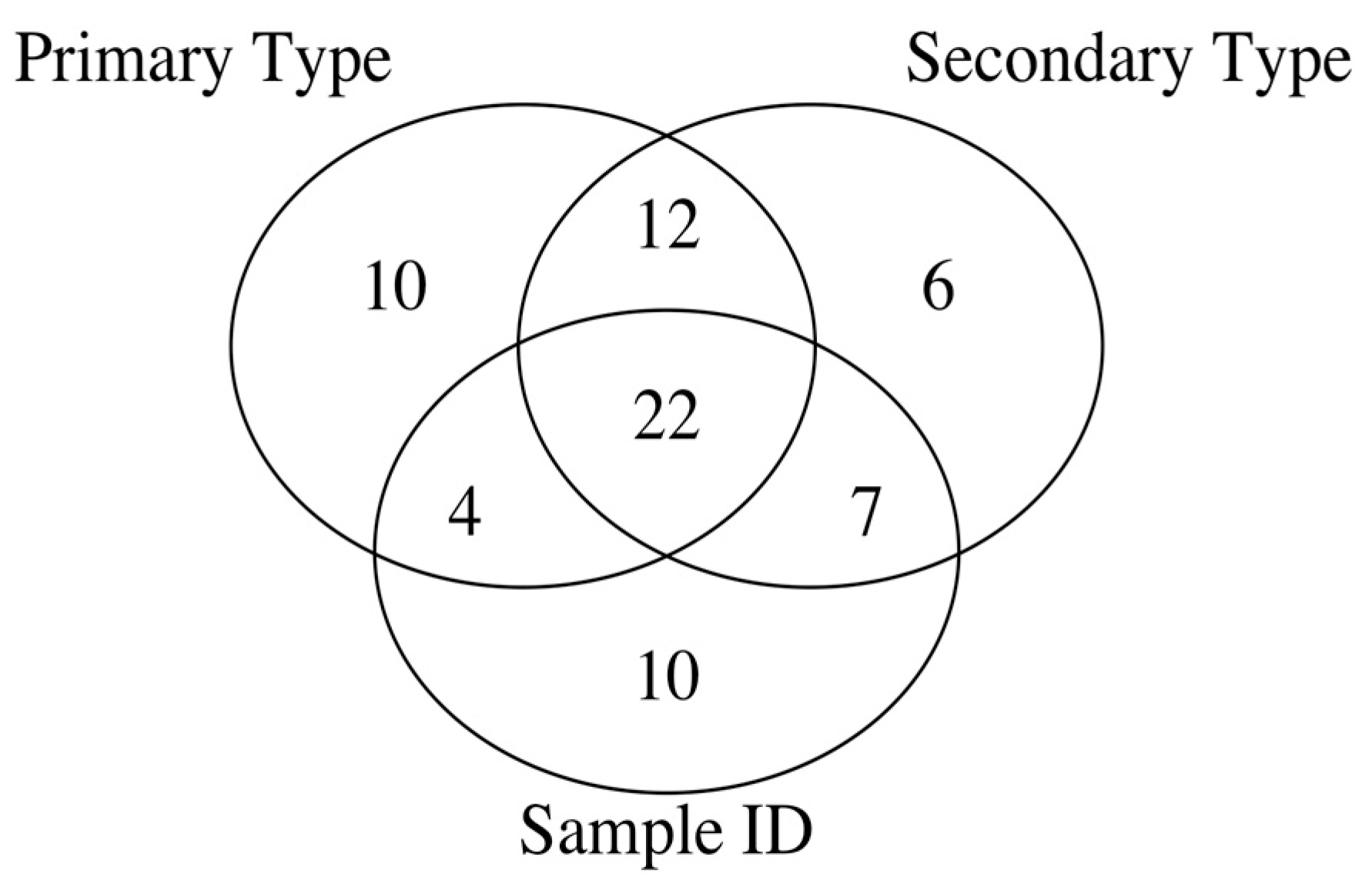
| Primary Type | Secondary Type | Sample ID | Counts | m/z Values |
|---|---|---|---|---|
| True | True | True | 22 | 60, 84, 85, 86, 91, 93, 94, 105, 114, 123, 124, 126, 135, 136, 139, 140, 144, 165, 184, 205, 209, 302 |
| False | True | True | 7 | 55, 58, 64, 115, 119, 125, 131 |
| True | False | True | 4 | 66, 197, 200, 296 |
| False | False | True | 10 | 79, 80, 100, 106, 113, 116, 161, 190, 211, 212 |
| True | True | False | 12 | 74, 92, 99, 103, 107, 110, 121, 129, 137, 148, 149, 166 |
| False | True | False | 6 | 77, 97, 111, 117, 153, 162 |
| True | False | False | 10 | 65, 70, 83, 109, 112, 174, 177, 192, 203, 219 |
| Machine Learning Algorithm | Primary Type | Secondary Type | Sample ID | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Random Forest Classifier | Number of Features | Accuracy (CV Score) | Number of Features | Accuracy (CV Score) | Number of Features | Accuracy (CV Score) | |||
| All features | 127 | 1.0 (1.0) | All features | 127 | 0.98 (1.0) | All features | 127 | 0.96 (0.97) | |
| Correlated features removed | 110 | 1.0 (1.0) | Correlated features removed | 110 | 0.99 (0.99) | Correlated features removed | 110 | 0.95 (0.96) | |
| p-value based feature selection | 46 | 1.0 (1.0) | p-value based feature selection | 49 | 0.99 (0.99) | p-value based feature selection | 35 | 0.95 (0.96) | |
| Decision Tree Classifier | Number of Features | Accuracy (CV Score) | Number of Features | Accuracy (CV Score) | Number of Features | Accuracy (CV Score) | |||
| All features | 127 | 1.0 (1.0) | All features | 127 | 0.98 (0.97) | All features | 127 | 0.91 (0.88) | |
| Correlated features removed | 110 | 1.0 (1.0) | Correlated features removed | 110 | 0.97 (0.96) | Correlated features removed | 110 | 0.90 (0.88) | |
| p-value based feature selection | 46 | 1.0 (1.0) | p-value based feature selection | 49 | 0.98 (0.96) | p-value based feature selection | 35 | 0.93 (0.89) | |
| k-NN Classifier | Number of Features | Accuracy (CV Score) | Number of Features | Accuracy (CV Score) | Number of Features | Accuracy (CV Score) | |||
| All features | 127 | 1.0 (1.0) | All features | 127 | 0.99 (0.99) | All features | 127 | 0.93 (0.93) | |
| Correlated features removed | 110 | 1.0 (1.0) | Correlated features removed | 110 | 0.98 (0.99) | Correlated features removed | 110 | 0.93 (0.93) | |
| p-value based feature selection | 46 | 1.0 (1.0) | p-value based feature selection | 49 | 0.99 (0.98) | p-value based feature selection | 35 | 0.92 (0.92) | |
| GNB Classifier | Number of Features | Accuracy (CV Score) | Number of Features | Accuracy (CV Score) | Number of Features | Accuracy (CV Score) | |||
| All features | 127 | 1.0 (1.0) | All features | 127 | 0.98 (0.98) | All features | 127 | 0.96 (0.96) | |
| Correlated features removed | 110 | 1.0 (1.0) | Correlated features removed | 110 | 0.98 (0.97) | Correlated features removed | 110 | 0.97 (0.96) | |
| p-value based feature selection | 46 | 1.0 (1.0) | p-value based feature selection | 49 | 0.98 (0.98) | p-value based feature selection | 35 | 0.97 (0.95) | |
| XGB Classifier | Number of Features | Accuracy (CV Score) | Number of Features | Accuracy (CV Score) | Number of Features | Accuracy (CV Score) | |||
| All features | 127 | 1.0 (1.0) | All features | 127 | 0.98 (0.98) | All features | 127 | 0.96 (0.97) | |
| Correlated features removed | 110 | 1.0 (1.0) | Correlated features removed | 110 | 0.98 (0.99) | Correlated features removed | 110 | 0.97 (0.97) | |
| p-value based feature selection | 46 | 1.0 (1.0) | p-value based feature selection | 49 | 0.98 (0.98) | p-value based feature selection | 35 | 0.96 (0.97) | |
| MLP Classifier | Number of Features | Accuracy (CV Score) | Number of Features | Accuracy (CV Score) | Number of Features | Accuracy (CV Score) | |||
| All features | 127 | 1.0 (1.0) | All features | 127 | 0.99 (1.0) | All features | 127 | 0.96 (0.95) | |
| Correlated features removed | 110 | 1.0 (1.0) | Correlated features removed | 110 | 1.0 (0.99) | Correlated features removed | 110 | 0.96 (0.96) | |
| p-value based feature selection | 46 | 1.0 | p-value based feature selection | 49 | 0.99 (0.99) | p-value based feature selection | 35 | 0.96 (0.95) | |
| Primary Type | Secondary Type | Sample ID | Counts | m/z Value |
|---|---|---|---|---|
| True | True | True | 15 | 73, 93, 94, 105, 107, 114, 123, 124, 126, 135, 140, 154, 162, 219, 302 |
| False | True | True | 12 | 58, 91, 95, 97, 100, 119, 131, 136, 139, 190, 200, 332 |
| True | False | True | 4 | 60, 79, 165, 203 |
| False | False | True | 4 | 55, 80, 113, 418 |
| True | True | False | 11 | 57, 66, 84, 85, 86, 92, 120, 144, 149, 174, 192 |
| False | True | False | 11 | 68, 103, 106, 117, 122, 129, 130, 148, 153, 209, 211 |
| True | False | False | 16 | 65, 72, 74, 78, 81, 83, 98, 109, 110, 115, 161, 163, 166, 197, 205, 296 |
| Machine Learning Algorithm | Primary Type (Pure + Binary Mixtures) | ||
|---|---|---|---|
| Random Forest Classifier | Number of Features | Accuracy (CV Score) | |
| All features | 127 | 0.97 (0.94) | |
| Correlated features removed | 97 | 0.89 (0.93) | |
| p-value based feature selection | 16 | 0.92 (0.89) | |
| Decision Tree Classifier | Number of Features | Accuracy (CV Score) | |
| All features | 127 | 0.86 (0.91) | |
| Correlated features removed | 97 | 0.83 (0.82) | |
| p-value based feature selection | 16 | 0.72 (0.79) | |
| k-NN Classifier | Number of Features | Accuracy (CV Score) | |
| All features | 127 | 0.92 (0.90) | |
| Correlated features removed | 97 | 0.83 (0.85) | |
| p-value based feature selection | 16 | 0.81 (0.78) | |
| GNB Classifier | Number of Features | Accuracy (CV Score) | |
| All features | 127 | 0.97 (0.94) | |
| Correlated features removed | 97 | 0.97 (0.94) | |
| p-value based feature selection | 16 | 0.92 (0.89) | |
| XGB Classifier | Number of Features | Accuracy (CV Score) | |
| All features | 127 | 0.89 (0.92) | |
| Correlated features removed | 97 | 0.94 (0.90) | |
| p-value based feature selection | 16 | 0.92 (0.87) | |
| MLP Classifier | Number of Features | Accuracy (CV Score) | |
| All features | 127 | 0.97 (0.95) | |
| Correlated features removed | 97 | 0.97 (0.93) | |
| p-value based feature selection | 16 | 0.25 (0.24) | |
| Primary Type (Pure Samples) | Primary Type (Pure Samples + Binary Mixtures) | Counts | m/z Values |
|---|---|---|---|
| True | True | 7 | 154, 167, 168, 181, 194, 208, 210 |
| True | False | 2 | 139, 196 |
| False | True | 4 | 103, 137, 180, 182 |
| Sample ID | Family (Primary Class) | Species (Secondary Class) | Total Ash % | % Ethanol Extractives | % Lignin | % Glucan | % Xylan | % Galactan | % Arabinan | % Mannan |
|---|---|---|---|---|---|---|---|---|---|---|
| BESC SWG | Grass | Switchgrass | 5.23 | 3.68 | 23.90 | 30.97 | 19.18 | 1.79 | 3.33 | 0.00 |
| Corn Stover L | Grass | Corn Stover | 5.09 | 3.37 | 14.78 | 32.49 | 20.21 | 1.67 | 2.40 | 0.00 |
| Corn Stover | Grass | Corn Stover | 5.42 | 3.53 | 15.42 | 34.93 | 18.94 | 1.19 | 1.86 | 0.00 |
| NIST 8491 | Grass | Sugarcane Bagasse | 4.00 | 4.40 | 24.20 | 40.20 | 21.50 | 0.60 | 1.80 | 0.40 |
| NIST 8494 | Grass | Wheat Straw | 10.30 | 13.00 | 18.00 | 37.60 | 21.70 | 0.80 | 2.50 | 0.30 |
| Aspen | Hardwood | Populus tremuloides | 0.72 | 3.22 | 24.05 | 39.08 | 16.43 | 1.24 | 0.25 | 1.98 |
| BESC Poplar | Hardwood | Populus trichocarpa | 0.55 | 1.41 | 26.95 | 46.17 | 14.76 | 0.97 | 0.40 | 2.80 |
| CBI Poplar | Hardwood | Populus trichocarpa | 0.50 | 0.00 | 23.22 | 43.99 | 15.60 | 1.27 | 0.62 | 3.11 |
| IBP1 | Hardwood | Eucalyptus | 0.67 | 1.15 | 29.15 | 39.38 | 13.36 | 1.45 | 0.00 | 2.37 |
| NIST 8492 | Hardwood | Poplulus deltoides | 1.00 | 2.40 | 26.20 | 43.20 | 13.90 | 0.60 | 0.70 | 2.00 |
| Pop 068 | Hardwood | Populus trichocarpa | 0.54 | 8.17 | 20.65 | 35.88 | 13.04 | 0.99 | 0.00 | 3.92 |
| Pop 93968 | Hardwood | Populus trichocarpa | 0.52 | 1.42 | 26.26 | 40.61 | 15.38 | 1.18 | 0.00 | 4.21 |
| Lob 6A1 | Softwood | Loblolly Pine | 0.24 | 1.77 | 33.30 | 37.79 | 6.94 | 2.69 | 0.00 | 13.57 |
| Lob 6G1 | Softwood | Loblolly Pine | 0.24 | 1.81 | 29.13 | 40.75 | 6.03 | 2.16 | 0.00 | 16.00 |
| Lob 6H2 | Softwood | Loblolly Pine | 0.33 | 1.44 | 31.77 | 41.13 | 6.12 | 1.91 | 0.00 | 16.51 |
| NIST 8493 | Softwood | Monterey Pine | 0.30 | 2.70 | 26.60 | 42.90 | 6.20 | 2.40 | 1.50 | 10.90 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nag, A.; Gerritsen, A.; Doeppke, C.; Harman-Ware, A.E. Machine Learning-Based Classification of Lignocellulosic Biomass from Pyrolysis-Molecular Beam Mass Spectrometry Data. Int. J. Mol. Sci. 2021, 22, 4107. https://doi.org/10.3390/ijms22084107
Nag A, Gerritsen A, Doeppke C, Harman-Ware AE. Machine Learning-Based Classification of Lignocellulosic Biomass from Pyrolysis-Molecular Beam Mass Spectrometry Data. International Journal of Molecular Sciences. 2021; 22(8):4107. https://doi.org/10.3390/ijms22084107
Chicago/Turabian StyleNag, Ambarish, Alida Gerritsen, Crissa Doeppke, and Anne E. Harman-Ware. 2021. "Machine Learning-Based Classification of Lignocellulosic Biomass from Pyrolysis-Molecular Beam Mass Spectrometry Data" International Journal of Molecular Sciences 22, no. 8: 4107. https://doi.org/10.3390/ijms22084107
APA StyleNag, A., Gerritsen, A., Doeppke, C., & Harman-Ware, A. E. (2021). Machine Learning-Based Classification of Lignocellulosic Biomass from Pyrolysis-Molecular Beam Mass Spectrometry Data. International Journal of Molecular Sciences, 22(8), 4107. https://doi.org/10.3390/ijms22084107






