Abstract
The sigma-1 receptor (S1R) is a 223 amino acid-long transmembrane endoplasmic reticulum (ER) protein. The S1R modulates the activity of multiple effector proteins, but its signaling functions are poorly understood. S1R is associated with cholesterol, and in our recent studies we demonstrated that S1R association with cholesterol induces the formation of S1R clusters. We propose that these S1R-cholesterol interactions enable the formation of cholesterol-enriched microdomains in the ER membrane. We hypothesize that a number of secreted and signaling proteins are recruited and retained in these microdomains. This hypothesis is consistent with the results of an unbiased screen for S1R-interacting partners, which we performed using the engineered ascorbate peroxidase 2 (APEX2) technology. We further propose that S1R agonists enable the disassembly of these cholesterol-enriched microdomains and the release of accumulated proteins such as ion channels, signaling receptors, and trophic factors from the ER. This hypothesis may explain the pleotropic signaling functions of the S1R, consistent with previously observed effects of S1R agonists in various experimental systems.
1. Introduction
The sigma-1 receptor (S1R) is a 223 amino acid-long transmembrane protein residing in the endoplasmic reticulum (ER) [1,2,3]. S1R attracts significant attention as a potential drug target for treating neurological disorders [2,4,5,6] and cancers [6].
S1R is expressed at high levels in the CNS and specifically in the cortex, basal ganglia, and motor neurons of the spinal cord and brainstem [7,8,9,10]. The S1R is a chaperone protein that is enriched at the ER/mitochondria-associated membrane (MAM), where it plays an important role in the regulation of multiple cellular mechanisms and is key to maintaining neuronal function and health. This is further supported by human genetic studies, showing that complete loss of function (LOF) mutations in the S1R are associated with a juvenile form of amyotrophic lateral sclerosis/frontotemporal dementia (ALS/FTD), while partial LOF mutations cause late onset ALS [11,12,13,14]. Thus, there is a gene dosage relationship between S1R activity and the age of onset of ALS with the complete loss of S1R associated with the earliest age of onset. Additional LOF mutations in the S1R cause distal hereditary motor neuropathies (dHMN) [15,16,17,18,19]. Furthermore, S1R expression levels are reduced in sporadic ALS [20], Parkinson’s disease (PD), and Alzheimer’s disease (AD) patients [21,22]. In preclinical models, the genetic ablation of S1R in mice exacerbated pathology and phenotypic presentation of several neurological disorders [23,24,25]. These results suggest that S1R plays an important role in healthy neuronal physiology.
The first prototypic S1R agonist, SKF-10047, was identified in animal behavioral assays, which led to the proposed existence of sigma opioid receptors [26]. However, SKF-10047 binding to sigma binding sites was not blocked by naloxone, an opioid receptor antagonist, and displayed a different stereospecificity [27,28]. Subsequent cloning of sigma binding sites confirmed that they share no homology with opioid G protein-coupled receptors (GPCRs), as well as sharing little homology to any other mammalian protein [29,30]. Sequence analysis revealed homology with the fungi C7–C8 sterol isomerase. While the S1R does not have isomerase activity, it contains two sterol-like binding domains as part of its ligand-binding site [29]. Recent biochemical and structural analysis indicated that the S1R is a single transmembrane domain protein with a short cytoplasmic tail and a large luminal ligand-binding domain [31,32]. It is suggested that the S1R acts as a molecular chaperone, which can stabilize the native conformation of multiple client proteins in stress conditions [1,33,34]. The S1R can be activated with highly selective synthetic ligands with nanomolar affinity [34,35,36]. The identity of an endogenous ligand is under investigation, with endogenous steroids (pregnenolone, dehydroepiandrosterone sulfate (DHEA), progesterone) being the most likely candidate [37,38], and N,N-dimethyltryptamine [39], sphingolipids [40], and more recently, choline also investigated [41].
Despite its importance in physiology and disease, the biological function of S1R is poorly understood [3]. This protein is involved in many biological processes and signaling pathways including maintenance of calcium homeostasis [42,43,44,45], protein folding [42], stress-response [1,42,46,47], autophagy [48,49], and the regulation of cellular excitability [50,51,52]. The S1R modulates the activity of ion channels via protein–protein interaction [52,53]. The S1R mode of action is not coupled to any known signaling cascade and is more consistent with its role as a modulatory or adaptor protein, or, using a term first coined by Hayashi and Su, a “molecular chaperone” [34,42,54].
Several S1R-interacting partners have been identified and multiple recent reviews comprehensively summarized these S1R interactors and the S1R-induced modulation of their activities [2,3]. Apart from that, S1R is known to interact and mediate the clustering of cholesterol and ceramides in the ER, as shown in cell-based assays [55,56,57,58]. We recently demonstrated that S1R is associated with cholesterol-enriched clusters in the membranes using in vitro reconstitution approach [59].
In this review we propose a hypothesis that the biological functions of the S1R are mediated by its ability to form ER signaling cholesterol-enriched lipid microdomains, analogous to the lipid rafts in the plasma membrane [60].
2. Intracellular Localization of the S1R
S1R primarily resides in the ER membrane where it forms microdomains [42,59,61,62]. Its localization is in contrast to the uniform distribution pattern of ER markers, such as the Sec61b protein. A significant proportion of S1R is localized to MAMs, an ER sub-compartment closely associated with the mitochondria [42,59], in proximity to lipid droplets [63], and at the ER-plasma membrane (PM) junctions [59,64]. It is likely that S1R are localized to additional inter-organelle contact sites, but this has not been systematically investigated.
MAMs are distinct from the rest of the ER as they contain enzymes involved in lipid synthesis, calcium signaling, cholesterol metabolism, and the ER stress-response pathways [65,66,67,68,69]. A detailed protein composition of MAMs was initially characterized by biochemical purifications [70,71] and more recently established using sophisticated proximity labeling approaches [72,73,74].
While the precise lipid composition of the MAMs has not yet been elucidated, recent evidence suggests that cholesterol and ceramide content is significantly higher in MAMs compared to the rest of the ER [57,75,76]. Therefore, MAMs can be thought of as specialized ER signaling domains characterized by unique protein and lipid compositions, similar to PM lipid rafts [60].
While PM lipid and protein heterogeneity was visualized using the giant plasma membrane-derived vesicle technique [77,78,79], only recently was a similar method to yield endomembrane-derived giant unilamellar vesicle (GUV)-like vesicles developed [80]. Using this approach, it was observed that certain, but not all ER contact sites (such as ER-mitochondria, ER-PM, and ER-lipid droplets) showed separation of the glycosylphosphatidylinositol (GPI) ER-targeted marker with a strong affinity for lipid-ordered phase [80]. Similar lipid and protein compartmentalization were recently observed at the inter-organelle contact sites in yeast [81], providing additional experimental evidence for microscopic lipid heterogeneity in the ER.
Binding immunoglobulin protein/glucose-regulated protein 78 (BiP/GRP78) was identified as a major S1R binding partner using pull-down experiments [42]. The S1R interacts with BiP in a calcium- and agonist-dependent manner. At high Ca2+ concentrations or in the absence of an agonist, the S1R forms a complex with BiP, keeping it in an inactive state. Therefore, BiP interaction contributes to S1R retention in the ER.
Under conditions of calcium depletion, or agonist activation, the S1R dissociates from BiP [42]. Agonist stimulation leads to redistribution of S1R from clusters to the ER, plasma membrane, and extracellular space [55,61]. In flotation assays, activation by an agonist causes the S1R to translocate from detergent-resistant to detergent-soluble fractions [55,61,63].
S1R activation releases the inhibitory interaction with BiP and allows for the S1R to interact with various partners inside and outside of the MAMs, including the inositol-1,4,5-triphosphate receptor type 3 (InsP3R3) [34,42]. Our recent results [59] and previous studies [57] suggest that a direct, high-affinity association of the S1R with cholesterol and ceramides may also contribute to S1R targeting by MAMs.
3. Interaction of S1R with ER Membranes
S1R was shown to interact with cholesterol in vitro [55], suggesting that S1R association with cholesterol plays an important role in MAM targeting of the S1R [57] and in modulation of PM cholesterol levels [58].
Recently, we demonstrated that cholesterol promotes the formation of S1R domains in a model lipid bilayer system [59]. Using GUVs with reconstituted fluorescent-labeled S1R, we observed that cholesterol was sufficient to cause clustering of recombinant S1R in the absence of any other proteins [59]. This study further identified a novel cholesterol-binding site within the transmembrane (TM) domain of the S1R. Additionally, point mutations in the TM domain, which weaken the interaction of the S1R with cholesterol, result in the impaired redistribution of S1R into the entire ER network [59].
In vitro, S1R clustering was observed in a narrow range between 2.5% and 5.0% mol cholesterol [59], comparable with the cholesterol-dependence of sterol regulatory element binding protein-2: sterol regulatory element-binding protein cleavage-activating protein complex (SREBP-2:Scap) [82]. Our data suggests that S1R actively participates in the assembly of micrometer-size cholesterol-enriched microdomains. On the basis of these observations, we propose that S1Rs promote the formation and stabilization of MAM microdomains, and potentially other ER contact sites [59]. Consistent with this idea, the genetic deletion of S1R impairs MAM stability and results in a reduced number of contacts, as observed by electron microscopy (EM) and biochemical fractionation [43].
The S1R has an unusually long, single transmembrane domain [59]. Local “measurements” of bilayer thickness with transmembrane sensors showed that bilayers surrounding S1R domains are thicker [59]. In the crystal structure, the S1R molecules are organized as trimers, with their C-terminal ligand-binding domain partially embedded in the membrane [31,83]. These amphipathic helices at the C-terminus of the S1R are rich in aromatic residues and, thus, can play a role in additional stabilization of the bilayer structure, a phenomenon previously observed for other amphipathic helices [84,85].
Thus, we reasoned that the local membrane thickness is increased in S1R-formed ER microdomains [59], which likely plays a role in the sorting of ER membrane proteins to these domains [79,81].
4. The S1R Interactome
To understand the functional significance of S1R-formed microdomains in the ER, we performed an unbiased screen aimed at identifying proteins located in proximity to S1R in cells. Analysis of such a “S1R interactome” further clarifies the composition and biological function of these microdomains.
Our experiments utilized a Tet-inducible vector expressing S1R fused to the peroxidase APEX2 in a proximity labeling technique, which captures weak and transient interactions [86].
For that purpose, we generated a plasmid encoding the S1R and fused to a genetically engineered APEX2 peroxidase, under the Tet-On tetracycline-inducible promoter (S1R-APEX2). Proteins in close proximity of S1R-APEX2 were biotinylated (see Materials and Methods Section for details) and the biotinylated proteins were pulled down using streptavidin-agarose. The eluted proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by mass spectrometry. Control non-induced cells were treated exactly the same way.
Using this approach, we identified several hundred proteins enriched in samples from S1R-APEX2-induced HeLa cells (Figure 1A, Supplementary Table S1). To analyze these hits, we first utilized the “cellular component” gene ontology (GO) term and determined that the majority of these proteins were ER membrane or luminal proteins (Table 1), consistent with the luminal localization of the S1R C-terminus [32,87]. Other enriched proteins were components of the extracellular matrix (ECM) or the plasma membrane (PM), and a small fraction were identified as Golgi and lysosomal proteins (Table 1). Since no S1R-APEX2 staining was detected on the plasma membrane by electron microscopy analysis ([32,87] and our unpublished observations), we hypothesized that the PM proteins and ECM components corresponded to a newly synthesized pool of proteins, that have not yet exited from the ER membrane compartment (full lists of ECM and cell surface proteins are provided in Supplementary Tables S1 and S2, respectively).
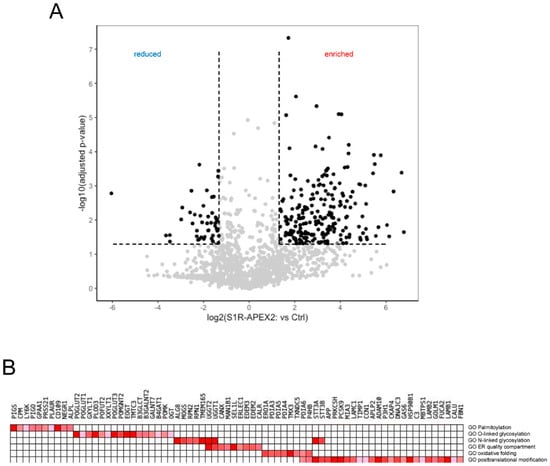
Figure 1.
Identification of sigma-1 receptor (S1R) interactome. (A) The volcano plot of proteins identified in the S1R-APEX2 proteomic screen: x-axis—log2 fold change in S1R-APEX2-expressing HeLa cell line vs. non-induced control cells; y-axis—log10 adjusted p-value; (B) Gene-set enrichment analysis (GSEA) identified S1R clusters as sites for protein folding and post-translational modifications; the hits identified in the screen were compared to manually curated lists using GSEA.

Table 1.
Cellular component gene oncology (GO) classification of top hits from S1R-APEX2 screen.
Hits from the S1R-APEX2 screen were then analyzed on the basis of their “biological process” GO terms. The top GO terms identified in this analysis were protein folding, oxidation-reduction processes, extracellular matrix organization, response to ER stress, protein modification, and protein glycosylation (Table 2).

Table 2.
Biological process GO classification of top hits from S1R-APEX2 screen.
More specific analysis was performed using a gene-set enrichment analysis (GSEA) [88] of these hits. GSE analysis (Figure 1B) revealed that the identified proteins (referred here by their UniProt names) were involved in: (1) the formation and shuffling of disulfide bonds (multiple protein disulfide isomerases protein disulfide isomerases (PDIs) PDIA3/4/5/6, thioredoxin-related transmembrane protein 3 (TMX3) and disulfide regenerating enzyme ERO1-like protein alpha (ERO1A)); (2) N- and O-linked glycosylation (including enzymes involved in multiple steps of the co-translational attachment of the dolichol-phosphate oligosaccharyl precursor (RPN1/2, STT3A/B), initial trimming of mannose chains (mannosyl-oligosaccharide glucosidase (MOGS); neutral alpha-glucosidase AB (GANAB)), and quality control and refolding of sugar chains (UDP-glucose:glycoprotein glucosyltransferase 1/2 (UGGG1/2) and GANAB); (3) the attachment of a glycosylphosphatidylinositol (GPI) anchor (PIGS, PIGO, GPAA1); and (4) in ER quality control including lectins, calnexin (CALX)/calreticulin (CALR), and machinery involved in targeting of misfolded proteins to the retrotranslocation complex (ER degradation-enhancing alpha-mannosidase-like proteins (EDEM2/3), ERLEC1, OS9 and protein SEL1 homolog (SEL1L)). No significant components of the ER ubiquitin-ligase complex were identified, except for SEL1L.
5. The Functional Role of S1R Microdomains: A Hypothesis
The results obtained in our studies with a reconstituted S1R [59] and in the S1R-APEX2 screen (Figure 1) led us to propose a novel hypothesis regarding the biological function ofS1R in cells. We propose that S1R organizes cholesterol-enriched microdomains in the ER (Figure 2). We reason that these microdomains are analogous to lipid rafts in the plasma membrane [60] and that, similar to lipid rafts, these microdomains have unique lipid and protein compositions when compared to the rest of the ER membrane. We also reason that these microdomains are thicker than the rest of the ER [59]. Our data [59] suggest that these microdomains are preferentially formed at ER membrane contact sites such as MAMs. We propose that these microdomains serve as a platform for the post-translational modification (PTM) and maturation of proteins in the ER, on a pre-secretory stage. These platforms may also play a role in protein folding-modification processes that take place in the early stages of protein synthesis, before properly folded proteins can be sorted to post-ER organelles.
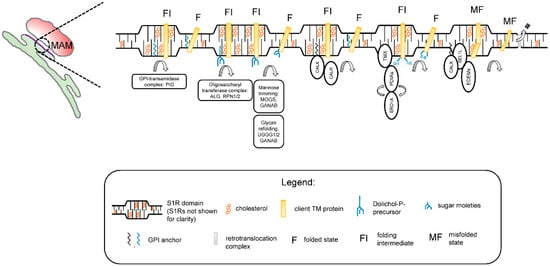
Figure 2.
Proposed function of S1R microdomains in the endoplasmic reticulum (ER). S1R microdomains are formed in the ER membranes and enriched in MAMs. S1R microdomains are characterized by higher cholesterol content and thicker bilayer structure when compared to the rest of the ER membrane. In S1R domains, protein folding-modification machinery is compartmentalized and allows a protein to proceed from a folding intermediate (FI) state to a fully folded (F) state. Client proteins (shown here as a single transmembrane helix for simplicity), which can include ER resident proteins, components of extracellular matrix, and PM channels/receptors, are recruited to these clusters. Folding machinery include enzymes involved in the following processes: GPI transamidase complex (PIG) catalyzing the transfer of a GPI moiety to nascent protein chains; initial glycosylation steps (components of oligosacharyl transferase complex alpha-1,3/1,6-mannosyltransferase (ALG), recognition particle 1/2 (RPN1/2)), initial mannose trimming steps (MOGS, GANAB), and glycan refolding (UGGG1/2, GANAB); ER lectins involved in protein quality control and refolding (calnexin (CALX)/calreticulin (CALR)); oxidative folding enzymes (TMX and protein disulfide isomerases); and finally, quality control lectins guiding unfolded proteins for recognition by the ER-associated protein degradation pathway (ERAD). Agonists affect the S1R oligomerization state and result in partial disassembly of S1R domains, leading to release of proteins from S1R clusters.
No major ER exit site markers were identified in the S1R-APEX2 screen (Figure 1A, Supplementary Table S1), suggesting they are localized outside of the S1R-formed microdomains. According to our hypothesis, PM proteins and proteins destined for secretion to the extracellular space are temporarily “trapped” in rigid and cholesterol-rich S1R-ER microdomains. While located in the S1R microdomains, these proteins can be processed through folding-modification cycles with the help of the enzymatic machinery residing in these domains. Strategic placement of S1R microdomains at MAMs ensures constant ATP supply for high-energy-dependent oxidative folding and protein modification reactions.
In agreement with this hypothesis, the experimental evidence indicates that membrane-bound calnexin and TMX are recruited to MAMs through palmitoylation, a known raft targeting mechanism [89,90]. Components of GPI machinery are localized to ER detergent-resistant membranes [91]. ER detergent-resistant membranes play an important role in assembly and secretion of viral particles [92,93]. The timeframe of protein maturation in the ER is known to be tightly controlled by mannose-trimming enzymes [94]. This timing can be especially important for transmembrane proteins (such as ion channels and receptors) that contain multiple transmembrane domains. Local thinning of the ER membrane was recently proposed to be required for the retrotranslocation of proteins though the ER-associated degradation (ERAD) mechanism [95,96]. Therefore, client proteins and enzymatic machinery localized to the “thick” S1R microdomains [59] are largely protected from a premature proteasomal degradation by ERAD.
6. The S1R as a Therapeutic Target for the Treatment of Neurodegenerative Diseases
The S1R is a well-established target for the treatment of neurodegenerative disorders, and it plays a key role in neurodegenerative diseases. Several S1R mutations have been identified to be associated with ALS and frontotemporal dementia (FTD) [97]. Two complete loss of function (LOF) mutations cause a juvenile form of ALS [12,43]. However, missense mutations that partially impair protein function are associated with an adult form of ALS, showing a dose response between the function of S1R and the age of onset of ALS [98]. Additional LOF mutations in S1R cause hereditary motor neuropathies [15,18]. Furthermore, some variants of the S1R gene are associated with increased risk for Alzheimer’s disease (AD) [99]. In addition, S1R expression levels are reduced in sporadic ALS [20], Parkinson’s disease (PD), and Alzheimer’s disease (AD) patients [21,100].
Further support for the role of S1R in neurodegenerative diseases comes from animal models. In preclinical models, genetic ablation of S1R (S1R−/−) in mice exacerbates pathology and phenotypic presentation of several neurological disorders. For example, S1R−/− mice display impairments in motor function and degeneration of motor neurons at 5 months of age [101]. AD mice (APPsweInd), which lack S1R, show enhanced behavioral and cognitive impairments, as well as a significant reduction in the levels of the brain-derived neurotrophic factor (BDNF) compared to APPsweInd mice expressing the S1R [102]. In the ALS SOD1G93A mouse model that also lacks S1R expression (SOD1G93A/S1R KO), disease progression is accelerated, as revealed by earlier weight loss and by a ~32% decrease in survival time relative to SOD1G93A mice with normal S1R expression [101]. These results suggest that the S1R plays an important role in healthy neuronal physiology.
S1R activation by agonists has demonstrated neuroprotective effects in multiple cellular and animal models of neurodegeneration (reviewed in [5,45,103,104]). Extensive evidence suggests that the mechanisms responsible for the neuroprotective effects of S1R agonists include the stabilization of Ca2+ signaling [42,44,105,106], an increase in the secretion of BDNF and the potentiation of BDNF-tropomyosin-related kinase B (TrkB) signaling [107,108,109,110,111,112], the stimulation of cyclic AMP-responsive element-binding protein (CREB)-mediated transcription [110], changes in the activity of the plasma membrane ion channels and receptors [113,114,115,116], the potentiation of the N-Methyl-D-aspartate (NMDA) receptor response [113,117,118,119], and an improvement in mitochondrial function [43,120,121,122].
For example, we and others have shown that the highly selective and potent S1R agonist pridopidine restores the dysregulated ER Ca2+ signaling and enhances spine density in Huntington’s disease (HD) and Alzheimer’s disease (AD) neurons [44,45,123]. Furthermore, S1R activation by pridopidine enhances synaptic plasticity in HD cortical neurons [123] and exhibits a robust neuroprotective effect against mutant huntingtin-induced cell death in mice’s primary HD neurons and in HD patient-derived induced-pluripotent stem cells (iPSCs) [124]. Pridopidine has been found to upregulate BDNF secretion, potentiate BDNF-TrkB signaling, and enhance BDNF axonal transport in several different models of neurodegenerative diseases including HD and ALS [109,110,125]. Pridopidine has shown protective effects on several mitochondrial functions in various human and mouse models of HD. In primary HD neurons, pridopidine enhances mitochondria-ER tethering and restores mitochondrial function as measured by increased ATP production, respiration, and mitochondrial membrane potential [126]. All these effects are exquisitely mediated by the activation of the S1R, as either a genetic deletion of the S1R or a pharmacological inhibition using an S1R antagonist, completely abolishes pridopidine’s neuroprotective effects, as shown in the studies mentioned above [126].
Recent clinical studies have shown the potential efficacy of the selective S1R agonist pridopidine in HD patients, demonstrating maintenance or slowing the decline of the patient’s functional capacity [127,128]. The non-selective S1R/Muscarinic (M1R) agonist blarcamesine shows a potential beneficial effect in AD [129]. Clinical pivotal studies with pridopidine are currently ongoing for HD and ALS (NCT04556656, NCT04297683). Blarcamesine is currently being evaluated for AD, Rett syndrome, and PD dementia patients (NCT04314934, NCT04304482, NCT04575259). Results of completed clinical trials of S1R agonists in variety of disorders have been comprehensively summarized in recent reviews [4,130].
How can the activation of S1R exert such pleotropic and variable effects on cellular signaling? We propose that the agonist activation of S1R results in the remodeling of S1R microdomains (Figure 2). In our experiments [59] and in published studies [55,61], the activation of S1R has resulted in the disassembly of the S1R oligomers. We propose that agonists cause partial disassembly and remodeling of S1R microdomains in the ER, leading to a rapid release of mature proteins that are trapped in these microdomains.
Consistent with this hypothesis, increases in the levels of PM proteins are often observed following S1R stimulation with an agonist. For example, the S1R agonist SKF-10047 increased the plasma membrane fraction of the GluN1, GluN2A, and GluN2b NMDAR subunits [118]. Cocaine increased the PM fraction of Kv1.2 [50,51]. Cell surface expression of programmed cell death 1 ligand 1 (PD-L1) was increased after the agonist stimulation, and lower levels of PD-L1 were observed in S1R knockdown (KD) cells [131]. On the other hand, S1R knockdown (KD) decreased the stability and levels of the mature human ether-à-go-go-related gene (hERG), as well as hERG currents [132]. A shorter protein half-life was observed for InsP3R3 in MAMs in S1R KD cells [42], and the turnover of p35, which is dependent on protein myrostyilation, was lower in S1R deleted cells [133]. In contrast, however, the stability of UDP-galactose:ceramide galactosyltransferase (UGT8) was increased in S1R deleted cells [134].
The same idea applies not only to PM proteins, but also to secreted proteins. BDNF release increased after treatment with the S1R agonists pridopidine and cutamesine in B104 cells and in astrocyte cultures [111,135]. Levels of secreted BDNF and glial cell-derived neurotrophic factor (GDNF) were increased in substantia nigra after pridopidine and sigma receptor agonist PRE-084 treatment in the experimental Parkinsonism model [112,136]. Moreover, it was shown that BDNF processing is modulated by S1R at the post-translational stage [135], in line with our hypothesis.
On the basis of these results, we propose that there is a “reserve pool” of PM and secreted proteins which accumulate in cholesterol-rich ER microdomains. S1R agonists facilitate the remodeling of these microdomains and the rapid release of PM and secreted proteins, resulting in a robust response to stress and in neuroprotective effects.
Additional neuroprotective effects of S1R activation may also be related to the modulation of ER Ca2+ signaling, in particular inositol trisphosphate receptor (InsP3R)-mediated Ca2+ signaling [42,44,137] and ER-mitochondrial Ca2+ transfer [42,43]. The possible explanations for the observed potentiation of InsP3-induced Ca2+ release [137,138,139] can include the direct effects of cholesterol on the activity of the InsP3Rs, the modulation of ER Ca2+ content through cholesterol regulation of sacro/endoplasmic reticulum Ca2+ (SERCA) pump [140,141,142], the removal of the ankyrin inhibition of InsP3Rs [137], and the redistribution of InsP3Rs from MAMs to the peripheral ER [42,43].
This proposed model may explain the pleiotropic effects of S1R agonists and provide appropriate context for the development of S1R-targeting therapeutic agents.
7. Materials and Methods
7.1. Construct Design and Molecular Cloning
For cloning the S1R-APEX2 fusion gene, APEX2 (https://www.addgene.org/92158, accessed on 1 March 2019) and human S1R (NM_005866) genes were amplified by PCR using the following primers: S1R-EcoRI-F 5′ TAAATGAATTCATGCAGTGGGCCGTGGGCCGG, S1R-NotI-R 5′ GATGCGGCCGCAGGGTCCTGGCCAAAGAGGTAGGT, APEX2-NotI-F 5′ ATCGCGGCCGCCACCATGGACTACAAG, APEX2-BamHI-Rev 5′ ATTGGATCCTTAGGCATCAGCAAACCCAAGCTC. The NotI site was introduced to the APEX2 5′ primer and to the S1R 3′ primer. PCR products were ligated using T4 ligase (NEB, Ipswich, MA, USA) and amplified using the outer primers S1R-EcoRI-F and APEX2-BamHI-Rev to produce the fusion gene S1R-APEX2. The resulting S1R-APEX2 gene was cloned into pTRE-3g expression vector (Takara, Kusatsu, Japan) using EcoRI and BamHI cloning sites.
7.2. Generation of Stable HeLa Cell Lines
For the generation of stable lines, pTRE-3g-S1R-APEX2 plasmid was transfected in HeLa Tet-On inducible cell line (kindly provided by Phillip Thomas’s lab, UTSW) together with a linear puromycin-resistant marker (Takara, Kusatsu, Japan). Stable doxycycline-responsive monoclonal lines were isolated and expanded. The induction of the S1R-APEX2 fusion protein was confirmed by Western blot analysis using anti-APEX2 horseradish peroxidase-conjugate (ab192968, 1:1000, Abcam, Cambridge, UK) and anti-S1R (B-5, 1:300, Santa-Cruz, CA, USA) antibodies.
7.3. Proximity Biotinylation Experiments
For the APEX2-based proximity-labeling experiments, we followed a procedure described in [143]. Briefly, for each experiment, the S1R-APEX2 HeLa cells were cultured on six 10 cm2 dishes. S1R-APEX2 production was induced by the addition of 1 μM doxycycline to the medium. After 72 h post-induction, the cells were incubated in 500 μM biotin-phenol (Iris Biotech, Marktredwitz, Germany) in complete medium at 37 °C for 1 h. Then, proteins were labeled by the addition of 1 mM H2O2 for 1 min and quenched with 10 mM sodium ascorbate, 5 mM Trolox, and 10 mM sodium azide in phosphate buffered saline (PBS). Cells were lysed in radioimmunoprecipitation assay buffer (RIPA) buffer (50 mM Tris-HCl pH = 7.4, 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% Triton X-100, 1× complete protease inhibitor cocktail) for 15 min at 4 °C on a rocker shaker. After centrifugation at 14,000× g for 10 min, 1 mL of lysate was mixed with 50 μL of streptavidin-agarose (Pierce) and incubated at 4 °C for 4 h on a rotary shaker. Resin was washed twice with 1 mL of RIPA buffer, once with 1 M KCl, once with 0.1 M Na2CO3, once with 2 M urea in 25 mM Tris-HCl pH = 8.0, and twice with RIPA buffer. Proteins were eluted by boiling beads in 50 μL of 2× SDS Laemmli loading buffer plus 2 mM biotin. Protein biotinylation was confirmed by Western blot analysis using streptavidin-HRP (7403, 1:20,000, Abcam, Cambridge, UK) antibody. Experiments were performed in duplicates with non-induced cells serving as the control.
7.4. Protein Identification by LC-Tandem Mass Spectrometry and Data Analysis
Eluates were loaded on a 12% gel (BioRad, Hercules, CA, USA). Gels were stained with Coomassie blue. Stained 1-cm bands were cut out of gels, sliced into 1-mm cubes and transferred to 1.5 mL Eppendorf tubes for submission. Raw mass spectrometry data was pre-proceeded and provided by UT Southwestern Proteomics Core. The statistically significant protein hits fold enrichment (calculated by diving a sum of spectral index values in doxycycline-induced samples vs. control samples) and p-values were calculated for each identified protein with at least three peptide sequences. Hits were selected by applying the following criteria: fold change >2.5 and p-value < 0.05. For gene ontology analyses, UniProt IDs were converted to gene names and analyzed by DAVID bioinformatic resources v6.8 (Laboratory of Human Retrovirology and Immunoinformatics, Frederick National Laboratory) [144]. Gene-set enrichment analyses (GSEA) were performed using GSEA v.4.1.0 (UC San Diego/Broad Institute) [88] software using the indicated molecular signature databases according to the instructions (https://www.gsea-msigdb.org/gsea, accessed on 1 March 2020). Data was visualized in R using the ROTS [145] package and GSEA software [88].
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ijms22084082/s1, Table S1: List of protein hits identified in the S1R-APEX2 proteomics screen. Table S2: Extracellular matrix components. Table S3: Cell surface proteins and receptors.
Author Contributions
Conceptualization, V.Z. and I.B.; methodology, V.Z.; formal analysis, V.Z. and I.B.; investigation, V.Z.; resources, I.B. and M.R.H.; data curation, V.Z. and I.B.; writing—original draft preparation, V.Z. and I.B.; writing—review and editing, V.Z., M.G., M.R.H. and I.B.; visualization, V.Z. and I.B.; supervision, I.B. and M.R.H.; project administration, I.B. and M.R.H.; funding acquisition, I.B. and M.R.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Russian Science Foundation grant 19-15-00184 (I.B.) and by the National Institutes of Health grants R01NS056224 (I.B.) and R01AG055577 (I.B.). I.B. holds the Carl J. and Hortense M. Thomsen Chair in Alzheimer’s Disease Research.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Results of S1R-APEX2 screen are available as Table S1.
Acknowledgments
We are thankful to Meewhi Kim and other members of Bezprozvanny laboratory for useful discussions.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Hayashi, T. The Sigma-1 Receptor in Cellular Stress Signaling. Front. Neurosci. 2019, 13, 733. [Google Scholar] [CrossRef]
- Ryskamp, D.A.; Korban, S.; Zhemkov, V.; Kraskovskaya, N.; Bezprozvanny, I. Neuronal Sigma-1 Receptors: Signaling Functions and Protective Roles in Neurodegenerative Diseases. Front. Neurosci. 2019, 13, 862. [Google Scholar] [CrossRef]
- Schmidt, H.R.; Kruse, A.C. The Molecular Function of sigma Receptors: Past, Present, and Future. Trends Pharmacol. Sci. 2019, 40, 636–654. [Google Scholar] [CrossRef]
- Maurice, T. Bi-phasic dose response in the preclinical and clinical developments of sigma-1 receptor ligands for the treatment of neurodegenerative disorders. Expert Opin. Drug Discov. 2021, 16, 373–389. [Google Scholar] [CrossRef]
- Herrando-Grabulosa, M.; Gaja-Capdevila, N.; Vela, J.M.; Navarro, X. Sigma 1 receptor as a therapeutic target for amyotrophic lateral sclerosis. Br. J. Pharmacol. 2020, 178, 1336–1352. [Google Scholar] [CrossRef]
- Kim, F.J.; Maher, C.M. Sigma1 Pharmacology in the Context of Cancer. Handb. Exp. Pharmacol. 2017, 244, 237–308. [Google Scholar] [PubMed]
- Jbilo, O.; Vidal, H.; Paul, R.; De Nys, N.; Bensaid, M.; Silve, S.; Carayon, P.; Davi, D.; Galiegue, S.; Bourrie, B.; et al. Purification and characterization of the human SR 31747A-binding protein. A nuclear membrane protein related to yeast sterol isomerase. J. Biol. Chem. 1997, 272, 27107–27115. [Google Scholar] [CrossRef] [PubMed]
- Alonso, G.; Phan, V.; Guillemain, I.; Saunier, M.; Legrand, A.; Anoal, M.; Maurice, T. Immunocytochemical localization of the sigma(1) receptor in the adult rat central nervous system. Neuroscience 2000, 97, 155–170. [Google Scholar] [CrossRef]
- Kitaichi, K.; Chabot, J.G.; Moebius, F.F.; Flandorfer, A.; Glossmann, H.; Quirion, R. Expression of the purported sigma(1) (sigma(1)) receptor in the mammalian brain and its possible relevance in deficits induced by antagonism of the NMDA receptor complex as revealed using an antisense strategy. J. Chem. Neuroanat. 2000, 20, 375–387. [Google Scholar] [CrossRef]
- Mavlyutov, T.A.; Guo, L.W.; Epstein, M.L.; Ruoho, A.E. Role of the Sigma-1 receptor in Amyotrophic Lateral Sclerosis (ALS). J. Pharmacol. Sci. 2015, 127, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Honma, M.; Sakuraba, M.; Koizumi, T.; Takashima, Y.; Sakamoto, H.; Hayashi, M. Non-homologous end-joining for repairing I-SceI-induced DNA double strand breaks in human cells. DNA Repair 2007, 6, 781–788. [Google Scholar] [CrossRef] [PubMed]
- Al-Saif, A.; Al-Mohanna, F.; Bohlega, S. A mutation in sigma-1 receptor causes juvenile amyotrophic lateral sclerosis. Ann. Neurol. 2011, 70, 913–919. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Kwon, M.J.; Choi, W.J.; Oh, K.W.; Oh, S.I.; Ki, C.S.; Kim, S.H. Mutations in UBQLN2 and SIGMAR1 genes are rare in Korean patients with amyotrophic lateral sclerosis. Neurobiol. Aging 2014, 35, 1957.e7–1957.e8. [Google Scholar] [CrossRef] [PubMed]
- Ullah, M.I.; Ahmad, A.; Raza, S.I.; Amar, A.; Ali, A.; Bhatti, A.; John, P.; Mohyuddin, A.; Ahmad, W.; Hassan, M.J. In silico analysis of SIGMAR1 variant (rs4879809) segregating in a consanguineous Pakistani family showing amyotrophic lateral sclerosis without frontotemporal lobar dementia. Neurogenetics 2015, 16, 299–306. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Hu, Z.; Liu, L.; Xie, Y.; Zhan, Y.; Zi, X.; Wang, J.; Wu, L.; Xia, K.; Tang, B.; et al. A SIGMAR1 splice-site mutation causes distal hereditary motor neuropathy. Neurology 2015, 84, 2430–2437. [Google Scholar] [CrossRef]
- Almendra, L.; Laranjeira, F.; Fernandez-Marmiesse, A.; Negrao, L. SIGMAR1 gene mutation causing Distal Hereditary Motor Neuropathy in a Portuguese family. Acta Myol. 2018, 37, 2–4. [Google Scholar] [PubMed]
- Ververis, A.; Dajani, R.; Koutsou, P.; Aloqaily, A.; Nelson-Williams, C.; Loring, E.; Arafat, A.; Mubaidin, A.F.; Horany, K.; Bader, M.B.; et al. Distal hereditary motor neuronopathy of the Jerash type is caused by a novel SIGMAR1 c.500A>T missense mutation. J. Med. Genet. 2020, 57, 178–186. [Google Scholar] [CrossRef] [PubMed]
- Gregianin, E.; Pallafacchina, G.; Zanin, S.; Crippa, V.; Rusmini, P.; Poletti, A.; Fang, M.; Li, Z.; Diano, L.; Petrucci, A.; et al. Loss-of-function mutations in the SIGMAR1 gene cause distal hereditary motor neuropathy by impairing ER-mitochondria tethering and Ca2+ signalling. Hum. Mol. Genet. 2016, 25, 3741–3753. [Google Scholar] [CrossRef]
- Horga, A.; Tomaselli, P.J.; Gonzalez, M.A.; Laura, M.; Muntoni, F.; Manzur, A.Y.; Hanna, M.G.; Blake, J.C.; Houlden, H.; Zuchner, S.; et al. SIGMAR1 mutation associated with autosomal recessive Silver-like syndrome. Neurology 2016, 87, 1607–1612. [Google Scholar] [CrossRef]
- Prause, J.; Goswami, A.; Katona, I.; Roos, A.; Schnizler, M.; Bushuven, E.; Dreier, A.; Buchkremer, S.; Johann, S.; Beyer, C.; et al. Altered localization, abnormal modification and loss of function of Sigma receptor-1 in amyotrophic lateral sclerosis. Hum. Mol. Genet. 2013, 22, 1581–1600. [Google Scholar] [CrossRef] [PubMed]
- Jansen, K.L.; Faull, R.L.; Storey, P.; Leslie, R.A. Loss of sigma binding sites in the CA1 area of the anterior hippocampus in Alzheimer’s disease correlates with CA1 pyramidal cell loss. Brain Res. 1993, 623, 299–302. [Google Scholar] [CrossRef]
- Mishina, M.; Ohyama, M.; Ishii, K.; Kitamura, S.; Kimura, Y.; Oda, K.; Kawamura, K.; Sasaki, T.; Kobayashi, S.; Katayama, Y.; et al. Low density of sigma1 receptors in early Alzheimer’s disease. Ann. Nucl. Med. 2008, 22, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Mavlyutov, T.A.; Epstein, M.L.; Verbny, Y.I.; Huerta, M.S.; Zaitoun, I.; Ziskind-Conhaim, L.; Ruoho, A.E. Lack of sigma-1 receptor exacerbates ALS progression in mice. Neuroscience 2013, 240, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.; Wang, L.; Zhang, T.; Zhang, B.; Chen, L. Sigma-1 receptor knockout increases alpha-synuclein aggregation and phosphorylation with loss of dopaminergic neurons in substantia nigra. Neurobiol. Aging 2017, 59, 171–183. [Google Scholar] [CrossRef]
- Wang, J.; Saul, A.; Cui, X.; Roon, P.; Smith, S.B. Absence of Sigma 1 Receptor Accelerates Photoreceptor Cell Death in a Murine Model of Retinitis Pigmentosa. Investig. Ophthalmol. Vis. Sci. 2017, 58, 4545–4558. [Google Scholar] [CrossRef]
- Gilbert, P.E.; Martin, W.R. The effects of morphine and nalorphine-like drugs in the nondependent, morphine-dependent and cyclazocine-dependent chronic spinal dog. J. Pharmacol. Exp. Ther. 1976, 198, 66–82. [Google Scholar]
- Su, T.P. Evidence for sigma opioid receptor: Binding of [3H]SKF-10047 to etorphine-inaccessible sites in guinea-pig brain. J. Pharmacol. Exp. Ther. 1982, 223, 284–290. [Google Scholar]
- Hayashi, T.; Su, T. The sigma receptor: Evolution of the concept in neuropsychopharmacology. Curr. Neuropharmacol. 2005, 3, 267–280. [Google Scholar] [CrossRef] [PubMed]
- Hanner, M.; Moebius, F.F.; Flandorfer, A.; Knaus, H.G.; Striessnig, J.; Kempner, E.; Glossmann, H. Purification, molecular cloning, and expression of the mammalian sigma1-binding site. Proc. Natl. Acad. Sci. USA 1996, 93, 8072–8077. [Google Scholar] [CrossRef]
- Kekuda, R.; Prasad, P.D.; Fei, Y.J.; Leibach, F.H.; Ganapathy, V. Cloning and functional expression of the human type 1 sigma receptor (hSigmaR1). Biochem. Biophys. Res. Commun. 1996, 229, 553–558. [Google Scholar] [CrossRef]
- Schmidt, H.R.; Zheng, S.; Gurpinar, E.; Koehl, A.; Manglik, A.; Kruse, A.C. Crystal structure of the human sigma1 receptor. Nature 2016, 532, 527–530. [Google Scholar] [CrossRef]
- Mavylutov, T.; Chen, X.; Guo, L.; Yang, J. APEX2- tagging of Sigma 1-receptor indicates subcellular protein topology with cytosolic N-terminus and ER luminal C-terminus. Protein Cell 2018, 9, 733–737. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.; Lucke-Wold, B.P.; Mookerjee, S.; Kaushal, N.; Matsumoto, R.R. Sigma-1 Receptors and Neurodegenerative Diseases: Towards a Hypothesis of Sigma-1 Receptors as Amplifiers of Neurodegeneration and Neuroprotection. Adv. Exp. Med. Biol. 2017, 964, 133–152. [Google Scholar]
- Su, T.P.; Hayashi, T.; Maurice, T.; Buch, S.; Ruoho, A.E. The sigma-1 receptor chaperone as an inter-organelle signaling modulator. Trends Pharmacol. Sci. 2010, 31, 557–566. [Google Scholar] [CrossRef]
- Maurice, T.; Su, T.P. The pharmacology of sigma-1 receptors. Pharmacol. Ther. 2009, 124, 195–206. [Google Scholar] [CrossRef]
- Chu, U.B.; Ruoho, A.E. Biochemical Pharmacology of the Sigma-1 Receptor. Mol. Pharmacol. 2016, 89, 142–153. [Google Scholar] [CrossRef]
- Maurice, T.; Su, T.P.; Privat, A. Sigma1 (sigma 1) receptor agonists and neurosteroids attenuate B25-35-amyloid peptide-induced amnesia in mice through a common mechanism. Neuroscience 1998, 83, 413–428. [Google Scholar] [CrossRef]
- Maurice, T. Neurosteroids and sigma1 receptors, biochemical and behavioral relevance. Pharmacopsychiatry 2004, 37 (Suppl. 3), S171–S182. [Google Scholar] [CrossRef]
- Fontanilla, D.; Johannessen, M.; Hajipour, A.R.; Cozzi, N.V.; Jackson, M.B.; Ruoho, A.E. The hallucinogen N,N-dimethyltryptamine (DMT) is an endogenous sigma-1 receptor regulator. Science 2009, 323, 934–937. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, S.; Chu, U.B.; Mavlyutov, T.A.; Pal, A.; Pyne, S.; Ruoho, A.E. The sigma1 receptor interacts with N-alkyl amines and endogenous sphingolipids. Eur. J. Pharmacol. 2009, 609, 19–26. [Google Scholar] [CrossRef]
- Brailoiu, E.; Chakraborty, S.; Brailoiu, G.C.; Zhao, P.; Barr, J.L.; Ilies, M.A.; Unterwald, E.M.; Abood, M.E.; Taylor, C.W. Choline Is an Intracellular Messenger Linking Extracellular Stimuli to IP3-Evoked Ca2+ Signals through Sigma-1 Receptors. Cell Rep. 2019, 26, 330–337.e4. [Google Scholar] [CrossRef]
- Hayashi, T.; Su, T.P. Sigma-1 receptor chaperones at the ER-mitochondrion interface regulate Ca2+ signaling and cell survival. Cell 2007, 131, 596–610. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, S.; Ilieva, H.; Tamada, H.; Nomura, H.; Komine, O.; Endo, F.; Jin, S.; Mancias, P.; Kiyama, H.; Yamanaka, K. Mitochondria-associated membrane collapse is a common pathomechanism in SIGMAR1- and SOD1-linked ALS. EMBO Mol. Med. 2016, 8, 1421–1437. [Google Scholar] [CrossRef] [PubMed]
- Ryskamp, D.; Wu, J.; Geva, M.; Kusko, R.; Grossman, I.; Hayden, M.; Bezprozvanny, I. The sigma-1 receptor mediates the beneficial effects of pridopidine in a mouse model of Huntington disease. Neurobiol. Dis. 2017, 97 Pt A, 46–59. [Google Scholar] [CrossRef]
- Ryskamp, D.; Wu, L.; Wu, J.; Kim, D.; Rammes, G.; Geva, M.; Hayden, M.; Bezprozvanny, I. Pridopidine stabilizes mushroom spines in mouse models of Alzheimer’s disease by acting on the sigma-1 receptor. Neurobiol. Dis. 2019, 124, 489–504. [Google Scholar] [CrossRef]
- Mori, T.; Hayashi, T.; Hayashi, E.; Su, T.P. Sigma-1 receptor chaperone at the ER-mitochondrion interface mediates the mitochondrion-ER-nucleus signaling for cellular survival. PLoS ONE 2013, 8, e76941. [Google Scholar] [CrossRef] [PubMed]
- Rosen, D.A.; Seki, S.M.; Fernandez-Castaneda, A.; Beiter, R.M.; Eccles, J.D.; Woodfolk, J.A.; Gaultier, A. Modulation of the sigma-1 receptor-IRE1 pathway is beneficial in preclinical models of inflammation and sepsis. Sci. Transl. Med. 2019, 11, eaau5266. [Google Scholar] [CrossRef]
- Vollrath, J.T.; Sechi, A.; Dreser, A.; Katona, I.; Wiemuth, D.; Vervoorts, J.; Dohmen, M.; Chandrasekar, A.; Prause, J.; Brauers, E.; et al. Loss of function of the ALS protein SigR1 leads to ER pathology associated with defective autophagy and lipid raft disturbances. Cell Death Dis. 2014, 5, e1290. [Google Scholar] [CrossRef]
- Yang, H.; Shen, H.; Li, J.; Guo, L.W. SIGMAR1/Sigma-1 receptor ablation impairs autophagosome clearance. Autophagy 2019, 15, 1539–1557. [Google Scholar] [CrossRef]
- Delint-Ramirez, I.; Garcia-Oscos, F.; Segev, A.; Kourrich, S. Cocaine engages a non-canonical, dopamine-independent, mechanism that controls neuronal excitability in the nucleus accumbens. Mol. Psychiatry 2020, 25, 680–691. [Google Scholar] [CrossRef]
- Kourrich, S.; Hayashi, T.; Chuang, J.Y.; Tsai, S.Y.; Su, T.P.; Bonci, A. Dynamic interaction between sigma-1 receptor and Kv1.2 shapes neuronal and behavioral responses to cocaine. Cell 2013, 152, 236–247. [Google Scholar] [CrossRef]
- Kourrich, S. Sigma-1 Receptor and Neuronal Excitability. Handb. Exp. Pharmacol. 2017, 244, 109–130. [Google Scholar] [PubMed]
- Su, T.P.; Su, T.C.; Nakamura, Y.; Tsai, S.Y. The Sigma-1 Receptor as a Pluripotent Modulator in Living Systems. Trends Pharmacol. Sci. 2016, 37, 262–278. [Google Scholar] [CrossRef] [PubMed]
- Tsai, S.Y.; Hayashi, T.; Mori, T.; Su, T.P. Sigma-1 receptor chaperones and diseases. Cent. Nerv. Syst. Agents Med. Chem. 2009, 9, 184–189. [Google Scholar] [CrossRef] [PubMed]
- Palmer, C.P.; Mahen, R.; Schnell, E.; Djamgoz, M.B.; Aydar, E. Sigma-1 receptors bind cholesterol and remodel lipid rafts in breast cancer cell lines. Cancer Res. 2007, 67, 11166–11175. [Google Scholar] [CrossRef]
- Hayashi, T.; Su, T.P. Cholesterol at the endoplasmic reticulum: Roles of the sigma-1 receptor chaperone and implications thereof in human diseases. Cholest. Bind. Cholest. Transp. Proteins 2010, 51, 381–398. [Google Scholar]
- Hayashi, T.; Fujimoto, M. Detergent-resistant microdomains determine the localization of sigma-1 receptors to the endoplasmic reticulum-mitochondria junction. Mol. Pharmacol. 2010, 77, 517–528. [Google Scholar] [CrossRef]
- Takebayashi, M.; Hayashi, T.; Su, T.P. Sigma-1 receptors potentiate epidermal growth factor signaling towards neuritogenesis in PC12 cells: Potential relation to lipid raft reconstitution. Synapse 2004, 53, 90–103. [Google Scholar] [CrossRef]
- Zhemkov, V.; Ditlev, J.A.; Lee, W.-R.; Liou, J.; Rosen, M.K.; Bezprozvanny, I. The role of sigma-1 receptor in organization of endoplasmic reticulum signaling microdomains. bioRxiv 2020. [Google Scholar] [CrossRef]
- Levental, I.; Levental, K.R.; Heberle, F.A. Lipid Rafts: Controversies Resolved, Mysteries Remain. Trends Cell Biol. 2020, 30, 341–353. [Google Scholar] [CrossRef]
- Hayashi, T.; Su, T.P. Intracellular dynamics of sigma-1 receptors (sigma(1) binding sites) in NG108-15 cells. J. Pharmacol. Exp. Ther. 2003, 306, 726–733. [Google Scholar] [CrossRef]
- Hayashi, T.; Su, T.P. Sigma-1 receptors at galactosylceramide-enriched lipid microdomains regulate oligodendrocyte differentiation. Proc. Natl. Acad. Sci. USA 2004, 101, 14949–14954. [Google Scholar] [CrossRef]
- Hayashi, T.; Su, T.P. Sigma-1 receptors (sigma(1) binding sites) form raft-like microdomains and target lipid droplets on the endoplasmic reticulum: Roles in endoplasmic reticulum lipid compartmentalization and export. J. Pharmacol. Exp. Ther. 2003, 306, 718–725. [Google Scholar] [CrossRef] [PubMed]
- Srivats, S.; Balasuriya, D.; Pasche, M.; Vistal, G.; Edwardson, J.M.; Taylor, C.W.; Murrell-Lagnado, R.D. Sigma1 receptors inhibit store-operated Ca2+ entry by attenuating coupling of STIM1 to Orai1. J. Cell Biol. 2016, 213, 65–79. [Google Scholar] [CrossRef]
- Csordas, G.; Weaver, D.; Hajnoczky, G. Endoplasmic Reticulum-Mitochondrial Contactology: Structure and Signaling Functions. Trends Cell Biol. 2018, 28, 523–540. [Google Scholar] [CrossRef] [PubMed]
- Prinz, W.A.; Toulmay, A.; Balla, T. The functional universe of membrane contact sites. Nat. Rev. Mol. Cell Biol. 2020, 21, 7–24. [Google Scholar] [CrossRef] [PubMed]
- Hajnoczky, G.; Csordas, G.; Yi, M. Old players in a new role: Mitochondria-associated membranes, VDAC, and ryanodine receptors as contributors to calcium signal propagation from endoplasmic reticulum to the mitochondria. Cell Calcium 2002, 32, 363–377. [Google Scholar] [CrossRef]
- Vance, J.E. MAM (mitochondria-associated membranes) in mammalian cells: Lipids and beyond. Biochim. Biophys. Acta 2014, 1841, 595–609. [Google Scholar] [CrossRef] [PubMed]
- Garofalo, T.; Matarrese, P.; Manganelli, V.; Marconi, M.; Tinari, A.; Gambardella, L.; Faggioni, A.; Misasi, R.; Sorice, M.; Malorni, W. Evidence for the involvement of lipid rafts localized at the ER-mitochondria associated membranes in autophagosome formation. Autophagy 2016, 12, 917–935. [Google Scholar] [CrossRef]
- Poston, C.N.; Krishnan, S.C.; Bazemore-Walker, C.R. In-depth proteomic analysis of mammalian mitochondria-associated membranes (MAM). J. Proteomics 2013, 79, 219–230. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.H.; Shen, S.; Wang, J.J.; He, Z.; Poon, A.; Li, J.; Qu, J.; Zhang, S.X. Comparative Proteomic Analysis of the Mitochondria-associated ER Membrane (MAM) in a Long-term Type 2 Diabetic Rodent Model. Sci. Rep. 2017, 7, 2062. [Google Scholar] [CrossRef]
- Kwak, C.; Shin, S.; Park, J.S.; Jung, M.; Nhung, T.T.M.; Kang, M.G.; Lee, C.; Kwon, T.H.; Park, S.K.; Mun, J.Y.; et al. Contact-ID, a tool for profiling organelle contact sites, reveals regulatory proteins of mitochondrial-associated membrane formation. Proc. Natl. Acad. Sci. USA 2020, 117, 12109–12120. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.F.; Branon, T.C.; Rajeev, S.; Svinkina, T.; Udeshi, N.D.; Thoudam, T.; Kwak, C.; Rhee, H.W.; Lee, I.K.; Carr, S.A.; et al. Split-TurboID enables contact-dependent proximity labeling in cells. Proc. Natl. Acad. Sci. USA 2020, 117, 12143–12154. [Google Scholar] [CrossRef]
- Hung, V.; Lam, S.S.; Udeshi, N.D.; Svinkina, T.; Guzman, G.; Mootha, V.K.; Carr, S.A.; Ting, A.Y. Proteomic mapping of cytosol-facing outer mitochondrial and ER membranes in living human cells by proximity biotinylation. Elife 2017, 6, e24463. [Google Scholar] [CrossRef]
- Fujimoto, M.; Hayashi, T.; Su, T.P. The role of cholesterol in the association of endoplasmic reticulum membranes with mitochondria. Biochem. Biophys. Res. Commun. 2012, 417, 635–639. [Google Scholar] [CrossRef]
- Area-Gomez, E.; Del Carmen Lara Castillo, M.; Tambini, M.D.; Guardia-Laguarta, C.; de Groof, A.J.; Madra, M.; Ikenouchi, J.; Umeda, M.; Bird, T.D.; Sturley, S.L.; et al. Upregulated function of mitochondria-associated ER membranes in Alzheimer disease. EMBO J. 2012, 31, 4106–4123. [Google Scholar] [CrossRef] [PubMed]
- Baumgart, T.; Hammond, A.T.; Sengupta, P.; Hess, S.T.; Holowka, D.A.; Baird, B.A.; Webb, W.W. Large-scale fluid/fluid phase separation of proteins and lipids in giant plasma membrane vesicles. Proc. Natl. Acad. Sci. USA 2007, 104, 3165–3170. [Google Scholar] [CrossRef]
- Sezgin, E.; Levental, I.; Mayor, S.; Eggeling, C. The mystery of membrane organization: Composition, regulation and roles of lipid rafts. Nat. Rev. Mol. Cell Biol. 2017, 18, 361–374. [Google Scholar] [CrossRef]
- Lorent, J.H.; Diaz-Rohrer, B.; Lin, X.; Spring, K.; Gorfe, A.A.; Levental, K.R.; Levental, I. Structural determinants and functional consequences of protein affinity for membrane rafts. Nat. Commun. 2017, 8, 1219. [Google Scholar] [CrossRef]
- King, C.; Sengupta, P.; Seo, A.Y.; Lippincott-Schwartz, J. ER membranes exhibit phase behavior at sites of organelle contact. Proc. Natl. Acad. Sci. USA 2020, 117, 7225–7235. [Google Scholar] [CrossRef] [PubMed]
- Prasad, R.; Sliwa-Gonzalez, A.; Barral, Y. Mapping bilayer thickness in the ER membrane. Sci. Adv. 2020, 6, eaba5130. [Google Scholar] [CrossRef] [PubMed]
- Radhakrishnan, A.; Goldstein, J.L.; McDonald, J.G.; Brown, M.S. Switch-like control of SREBP-2 transport triggered by small changes in ER cholesterol: A delicate balance. Cell Metab. 2008, 8, 512–521. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, H.R.; Betz, R.M.; Dror, R.O.; Kruse, A.C. Structural basis for sigma1 receptor ligand recognition. Nat. Struct. Mol. Biol. 2018, 25, 981–987. [Google Scholar] [CrossRef]
- Gimenez-Andres, M.; Copic, A.; Antonny, B. The Many Faces of Amphipathic Helices. Biomolecules 2018, 8, 45. [Google Scholar] [CrossRef] [PubMed]
- Olsinova, M.; Jurkiewicz, P.; Kishko, I.; Sykora, J.; Sabo, J.; Hof, M.; Cwiklik, L.; Cebecauer, M. Roughness of Transmembrane Helices Reduces Lipid Membrane Dynamics. iScience 2018, 10, 87–97. [Google Scholar] [CrossRef]
- Rhee, H.W.; Zou, P.; Udeshi, N.D.; Martell, J.D.; Mootha, V.K.; Carr, S.A.; Ting, A.Y. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science 2013, 339, 1328–1331. [Google Scholar] [CrossRef]
- Mavlyutov, T.A.; Yang, H.; Epstein, M.L.; Ruoho, A.E.; Yang, J.; Guo, L.W. APEX2-enhanced electron microscopy distinguishes sigma-1 receptor localization in the nucleoplasmic reticulum. Oncotarget 2017, 8, 51317–51330. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Lynes, E.M.; Bui, M.; Yap, M.C.; Benson, M.D.; Schneider, B.; Ellgaard, L.; Berthiaume, L.G.; Simmen, T. Palmitoylated TMX and calnexin target to the mitochondria-associated membrane. EMBO J. 2012, 31, 457–470. [Google Scholar] [CrossRef]
- Lynes, E.M.; Raturi, A.; Shenkman, M.; Ortiz Sandoval, C.; Yap, M.C.; Wu, J.; Janowicz, A.; Myhill, N.; Benson, M.D.; Campbell, R.E.; et al. Palmitoylation is the switch that assigns calnexin to quality control or ER Ca2+ signaling. J. Cell Sci. 2013, 126 Pt 17, 3893–3903. [Google Scholar] [CrossRef]
- Pielsticker, L.K.; Mann, K.J.; Lin, W.L.; Sevlever, D. Raft-like membrane domains contain enzymatic activities involved in the synthesis of mammalian glycosylphosphatidylinositol anchor intermediates. Biochem. Biophys. Res. Commun. 2005, 330, 163–171. [Google Scholar] [CrossRef]
- Boyer, A.; Dreneau, J.; Dumans, A.; Burlaud-Gaillard, J.; Bull-Maurer, A.; Roingeard, P.; Meunier, J.C. Endoplasmic Reticulum Detergent-Resistant Membranes Accommodate Hepatitis C Virus Proteins for Viral Assembly. Cells 2019, 8, 487. [Google Scholar] [CrossRef]
- Friesland, M.; Mingorance, L.; Chung, J.; Chisari, F.V.; Gastaminza, P. Sigma-1 receptor regulates early steps of viral RNA replication at the onset of hepatitis C virus infection. J. Virol. 2013, 87, 6377–6390. [Google Scholar] [CrossRef]
- Shenkman, M.; Lederkremer, G.Z. Compartmentalization and Selective Tagging for Disposal of Misfolded Glycoproteins. Trends Biochem. Sci. 2019, 44, 827–836. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Siggel, M.; Ovchinnikov, S.; Mi, W.; Svetlov, V.; Nudler, E.; Liao, M.; Hummer, G.; Rapoport, T.A. Structural basis of ER-associated protein degradation mediated by the Hrd1 ubiquitin ligase complex. Science 2020, 368, eaaz2449. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Rapoport, T.A. Translocation of Proteins through a Distorted Lipid Bilayer. Trends Cell Biol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Luty, A.A.; Kwok, J.B.; Dobson-Stone, C.; Loy, C.T.; Coupland, K.G.; Karlstrom, H.; Sobow, T.; Tchorzewska, J.; Maruszak, A.; Barcikowska, M.; et al. Sigma nonopioid intracellular receptor 1 mutations cause frontotemporal lobar degeneration-motor neuron disease. Ann. Neurol. 2010, 68, 639–649. [Google Scholar] [CrossRef] [PubMed]
- Izumi, Y.; Morino, H.; Miyamoto, R.; Matsuda, Y.; Ohsawa, R.; Kurashige, T.; Shimatani, Y.; Kaji, R.; Kawakami, H. Compound heterozygote mutations in the SIGMAR1 gene in an oldest-old patient with amyotrophic lateral sclerosis. Geriatr. Gerontol. Int. 2018, 18, 1519–1520. [Google Scholar] [CrossRef]
- Brimson, J.M.; Akula, K.K.; Abbas, H.; Ferry, D.R.; Kulkarni, S.K.; Russell, S.T.; Tisdale, M.J.; Tencomnao, T.; Safrany, S.T. Simple ammonium salts acting on sigma-1 receptors yield potential treatments for cancer and depression. Sci. Rep. 2020, 10, 9251. [Google Scholar] [CrossRef] [PubMed]
- Maurice, T.; Goguadze, N. Sigma-1 (sigma1) Receptor in Memory and Neurodegenerative Diseases. Handb. Exp. Pharmacol. 2017, 244, 81–108. [Google Scholar]
- Uchida, N.; Ujike, H.; Tanaka, Y.; Sakai, A.; Yamamoto, M.; Fujisawa, Y.; Kanzaki, A.; Kuroda, S. A variant of the sigma receptor type-1 gene is a protective factor for Alzheimer disease. Am. J. Geriatr. Psychiatry 2005, 13, 1062–1066. [Google Scholar] [CrossRef]
- Mishina, M.; Ishiwata, K.; Ishii, K.; Kitamura, S.; Kimura, Y.; Kawamura, K.; Oda, K.; Sasaki, T.; Sakayori, O.; Hamamoto, M.; et al. Function of sigma1 receptors in Parkinson’s disease. Acta Neurol. Scand. 2005, 112, 103–107. [Google Scholar] [CrossRef] [PubMed]
- Mavlyutov, T.A.; Epstein, M.L.; Andersen, K.A.; Ziskind-Conhaim, L.; Ruoho, A.E. The sigma-1 receptor is enriched in postsynaptic sites of C-terminals in mouse motoneurons. An anatomical and behavioral study. Neuroscience 2010, 167, 247–255. [Google Scholar] [CrossRef]
- Maurice, T.; Strehaiano, M.; Duhr, F.; Chevallier, N. Amyloid toxicity is enhanced after pharmacological or genetic invalidation of the sigma1 receptor. Behav. Brain Res. 2018, 339, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Monnet, F.P. Sigma-1 receptor as regulator of neuronal intracellular Ca2+: Clinical and therapeutic relevance. Biol. Cell 2005, 97, 873–883. [Google Scholar] [CrossRef] [PubMed]
- Tchedre, K.T.; Yorio, T. sigma-1 receptors protect RGC-5 cells from apoptosis by regulating intracellular calcium, Bax levels, and caspase-3 activation. Investig. Ophthalmol. Vis. Sci. 2008, 49, 2577–2588. [Google Scholar] [CrossRef]
- Kikuchi-Utsumi, K.; Nakaki, T. Chronic treatment with a selective ligand for the sigma-1 receptor chaperone, SA4503, up-regulates BDNF protein levels in the rat hippocampus. Neurosci. Lett. 2008, 440, 19–22. [Google Scholar] [CrossRef]
- Peviani, M.; Salvaneschi, E.; Bontempi, L.; Petese, A.; Manzo, A.; Rossi, D.; Salmona, M.; Collina, S.; Bigini, P.; Curti, D. Neuroprotective effects of the Sigma-1 receptor (S1R) agonist PRE-084, in a mouse model of motor neuron disease not linked to SOD1 mutation. Neurobiol. Dis. 2014, 62, 218–232. [Google Scholar] [CrossRef] [PubMed]
- Geva, M.; Kusko, R.; Soares, H.; Fowler, K.D.; Birnberg, T.; Barash, S.; Wagner, A.M.; Fine, T.; Lysaght, A.; Weiner, B.; et al. Pridopidine activates neuroprotective pathways impaired in Huntington Disease. Hum. Mol. Genet. 2016, 25, 3975–3987. [Google Scholar] [CrossRef]
- Kusko, R.; Dreymann, J.; Ross, J.; Cha, Y.; Escalante-Chong, R.; Garcia-Miralles, M.; Tan, L.J.; Burczynski, M.E.; Zeskind, B.; Laifenfeld, D.; et al. Large-scale transcriptomic analysis reveals that pridopidine reverses aberrant gene expression and activates neuroprotective pathways in the YAC128 HD mouse. Mol. Neurodegener. 2018, 13, 25. [Google Scholar] [CrossRef]
- Malik, M.; Rangel-Barajas, C.; Sumien, N.; Su, C.; Singh, M.; Chen, Z.; Huang, R.Q.; Meunier, J.; Maurice, T.; Mach, R.H.; et al. The effects of sigma (sigma1) receptor-selective ligands on muscarinic receptor antagonist-induced cognitive deficits in mice. Br. J. Pharmacol. 2015, 172, 2519–2531. [Google Scholar] [CrossRef]
- Francardo, V.; Bez, F.; Wieloch, T.; Nissbrandt, H.; Ruscher, K.; Cenci, M.A. Pharmacological stimulation of sigma-1 receptors has neurorestorative effects in experimental parkinsonism. Brain 2014, 137 Pt 7, 1998–2014. [Google Scholar] [CrossRef]
- Klette, K.L.; Lin, Y.; Clapp, L.E.; DeCoster, M.A.; Moreton, J.E.; Tortella, F.C. Neuroprotective sigma ligands attenuate NMDA and trans-ACPD-induced calcium signaling in rat primary neurons. Brain Res. 1997, 756, 231–240. [Google Scholar] [CrossRef]
- Zhang, X.J.; Liu, L.L.; Jiang, S.X.; Zhong, Y.M.; Yang, X.L. Activation of the zeta receptor 1 suppresses NMDA responses in rat retinal ganglion cells. Neuroscience 2011, 177, 12–22. [Google Scholar] [CrossRef]
- Kourrich, S.; Su, T.P.; Fujimoto, M.; Bonci, A. The sigma-1 receptor: Roles in neuronal plasticity and disease. Trends Neurosci. 2012, 35, 762–771. [Google Scholar] [CrossRef]
- Kimura, Y.; Fujita, Y.; Shibata, K.; Mori, M.; Yamashita, T. Sigma-1 receptor enhances neurite elongation of cerebellar granule neurons via TrkB signaling. PLoS ONE 2013, 8, e75760. [Google Scholar] [CrossRef] [PubMed]
- Martina, M.; Turcotte, M.E.; Halman, S.; Bergeron, R. The sigma-1 receptor modulates NMDA receptor synaptic transmission and plasticity via SK channels in rat hippocampus. J. Physiol. 2007, 578 Pt 1, 143–157. [Google Scholar] [CrossRef]
- Pabba, M.; Wong, A.Y.; Ahlskog, N.; Hristova, E.; Biscaro, D.; Nassrallah, W.; Ngsee, J.K.; Snyder, M.; Beique, J.C.; Bergeron, R. NMDA receptors are upregulated and trafficked to the plasma membrane after sigma-1 receptor activation in the rat hippocampus. J. Neurosci. 2014, 34, 11325–11338. [Google Scholar] [CrossRef]
- Hayashi, T.; Kagaya, A.; Takebayashi, M.; Shimizu, M.; Uchitomi, Y.; Motohashi, N.; Yamawaki, S. Modulation by sigma ligands of intracellular free Ca++ mobilization by N-methyl-D-aspartate in primary culture of rat frontal cortical neurons. J. Pharmacol. Exp. Ther. 1995, 275, 207–214. [Google Scholar]
- Meunier, J.; Hayashi, T. Sigma-1 receptors regulate Bcl-2 expression by reactive oxygen species-dependent transcriptional regulation of nuclear factor kappaB. J. Pharmacol. Exp. Ther. 2010, 332, 388–397. [Google Scholar] [CrossRef] [PubMed]
- Tsai, S.Y.; Hayashi, T.; Harvey, B.K.; Wang, Y.; Wu, W.W.; Shen, R.F.; Zhang, Y.; Becker, K.G.; Hoffer, B.J.; Su, T.P. Sigma-1 receptors regulate hippocampal dendritic spine formation via a free radical-sensitive mechanism involving Rac1xGTP pathway. Proc. Natl. Acad. Sci. USA 2009, 106, 22468–22473. [Google Scholar] [CrossRef]
- Weng, T.Y.; Tsai, S.A.; Su, T.P. Roles of sigma-1 receptors on mitochondrial functions relevant to neurodegenerative diseases. J. Biomed. Sci. 2017, 24, 74. [Google Scholar] [CrossRef]
- Smith-Dijak, A.I.; Nassrallah, W.B.; Zhang, L.Y.J.; Geva, M.; Hayden, M.R.; Raymond, L.A. Impairment and Restoration of Homeostatic Plasticity in Cultured Cortical Neurons From a Mouse Model of Huntington Disease. Front. Cell. Neurosci. 2019, 13, 209. [Google Scholar] [CrossRef]
- Eddings, C.R.; Arbez, N.; Akimov, S.; Geva, M.; Hayden, M.R.; Ross, C.A. Pridopidine protects neurons from mutant-huntingtin toxicity via the sigma-1 receptor. Neurobiol. Dis. 2019, 129, 118–129. [Google Scholar] [CrossRef] [PubMed]
- Ionescu, A.; Gradus, T.; Altman, T.; Maimon, R.; Saraf Avraham, N.; Geva, M.; Hayden, M.; Perlson, E. Targeting the Sigma-1 Receptor via Pridopidine Ameliorates Central Features of ALS Pathology in a SOD1(G93A) Model. Cell Death Dis. 2019, 10, 210. [Google Scholar] [CrossRef] [PubMed]
- Naia, L.; Ly, P.; Mota, S.I.; Lopes, C.; Maranga, C.; Gershoni-Emek, N.; Geva, M.; Hayden, M.R.; Rego, A.C. The Sigma-1 receptor mediates pridopidine rescue of mitochondrial function in Huntington Disease models. Neurotherapeutics 2021, in press. [Google Scholar] [CrossRef]
- McGarry, A.; Leinonen, M.; Kieburtz, K.; Geva, M.; Olanow, C.W.; Hayden, M. Effects of Pridopidine on Functional Capacity in Early-Stage Participants from the PRIDE-HD Study. J. Huntingt. Dis. 2020, 9, 371–380. [Google Scholar] [CrossRef] [PubMed]
- McGarry, A.; Auinger, P.; Kieburtz, K.; Geva, M.; Mehra, M.; Abler, V.; Grachev, I.D.; Gordon, M.F.; Savola, J.M.; Gandhi, S.; et al. Additional Safety and Exploratory Efficacy Data at 48 and 60 Months from Open-HART, an Open-Label Extension Study of Pridopidine in Huntington Disease. J. Huntingt. Dis. 2020, 9, 173–184. [Google Scholar] [CrossRef] [PubMed]
- Hampel, H.; Afshar, M.; Parmentier, F.; Williams, C.; Etcheto, A.; Goodsaid, F.; Missling, C. Longitudinal 148-Week Extension Study for ANAVEX® 2-73 Phase 2a Alzheimer’s Disease Demonstrates Maintained Activities of Daily Living Score (ADCS-ADL) and Reduced Cognitive Decline (MMSE) for Patient Cohort on Higher Drug Concentration and Confirms Role of Patient Selection Biomarkers. J. Prev. Alzheimers Dis. 2018, 5, S43. [Google Scholar]
- Ye, N.; Qin, W.; Tian, S.; Xu, Q.; Wold, E.A.; Zhou, J.; Zhen, X.C. Small Molecules Selectively Targeting Sigma-1 Receptor for the Treatment of Neurological Diseases. J. Med. Chem. 2020, 63, 15187–15217. [Google Scholar] [CrossRef] [PubMed]
- Maher, C.M.; Thomas, J.D.; Haas, D.A.; Longen, C.G.; Oyer, H.M.; Tong, J.Y.; Kim, F.J. Small-Molecule Sigma1 Modulator Induces Autophagic Degradation of PD-L1. Mol. Cancer Res. 2018, 16, 243–255. [Google Scholar] [CrossRef] [PubMed]
- Crottes, D.; Martial, S.; Rapetti-Mauss, R.; Pisani, D.F.; Loriol, C.; Pellissier, B.; Martin, P.; Chevet, E.; Borgese, F.; Soriani, O. Sig1R protein regulates hERG channel expression through a post-translational mechanism in leukemic cells. J. Biol. Chem. 2011, 286, 27947–27958. [Google Scholar] [CrossRef] [PubMed]
- Tsai, S.Y.; Pokrass, M.J.; Klauer, N.R.; Nohara, H.; Su, T.P. Sigma-1 receptor regulates Tau phosphorylation and axon extension by shaping p35 turnover via myristic acid. Proc. Natl. Acad. Sci. USA 2015, 112, 6742–6747. [Google Scholar] [CrossRef]
- Hayashi, T.; Hayashi, E.; Fujimoto, M.; Sprong, H.; Su, T.P. The lifetime of UDP-galactose:ceramide galactosyltransferase is controlled by a distinct endoplasmic reticulum-associated degradation (ERAD) regulated by sigma-1 receptor chaperones. J. Biol. Chem. 2012, 287, 43156–43169. [Google Scholar] [CrossRef]
- Fujimoto, M.; Hayashi, T.; Urfer, R.; Mita, S.; Su, T.P. Sigma-1 receptor chaperones regulate the secretion of brain-derived neurotrophic factor. Synapse 2012, 66, 630–639. [Google Scholar] [CrossRef]
- Francardo, V.; Geva, M.; Bez, F.; Denis, Q.; Steiner, L.; Hayden, M.R.; Cenci, M.A. Pridopidine Induces Functional Neurorestoration Via the Sigma-1 Receptor in a Mouse Model of Parkinson’s Disease. Neurotherapeutics 2019, 16, 465–479. [Google Scholar] [CrossRef]
- Hayashi, T.; Su, T.P. Regulating ankyrin dynamics: Roles of sigma-1 receptors. Proc. Natl. Acad. Sci. USA 2001, 98, 491–496. [Google Scholar] [CrossRef] [PubMed]
- Walker, J.M.; Bowen, W.D.; Walker, F.O.; Matsumoto, R.R.; De Costa, B.; Rice, K.C. Sigma receptors: Biology and function. Pharmacol. Rev. 1990, 42, 355–402. [Google Scholar] [PubMed]
- Skuza, G.; Sadaj, W.; Kabzinski, M.; Cassano, G.; Gasparre, G.; Abate, C.; Berardi, F. The effects of new sigma (sigma) receptor ligands, PB190 and PB212, in the models predictive of antidepressant activity. Pharmacol. Rep. 2014, 66, 320–324. [Google Scholar] [CrossRef]
- Gustavsson, M.; Traaseth, N.J.; Veglia, G. Activating and deactivating roles of lipid bilayers on the Ca2+-ATPase/phospholamban complex. Biochemistry 2011, 50, 10367–10374. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ge, M.; Ciani, L.; Kuriakose, G.; Westover, E.J.; Dura, M.; Covey, D.F.; Freed, J.H.; Maxfield, F.R.; Lytton, J.; et al. Enrichment of endoplasmic reticulum with cholesterol inhibits sarcoplasmic-endoplasmic reticulum calcium ATPase-2b activity in parallel with increased order of membrane lipids: Implications for depletion of endoplasmic reticulum calcium stores and apoptosis in cholesterol-loaded macrophages. J. Biol. Chem. 2004, 279, 37030–37039. [Google Scholar]
- Madden, T.D.; Chapman, D.; Quinn, P.J. Cholesterol modulates activity of calcium-dependent ATPase of the sarcoplasmic reticulum. Nature 1979, 279, 538–541. [Google Scholar] [CrossRef] [PubMed]
- Hung, V.; Udeshi, N.D.; Lam, S.S.; Loh, K.H.; Cox, K.J.; Pedram, K.; Carr, S.A.; Ting, A.Y. Spatially resolved proteomic mapping in living cells with the engineered peroxidase APEX2. Nat. Protoc. 2016, 11, 456–475. [Google Scholar] [CrossRef] [PubMed]
- Jiao, X.; Sherman, B.T.; Huang, D.W.; Stephens, R.; Baseler, M.W.; Lane, H.C.; Lempicki, R.A. DAVID-WS: A stateful web service to facilitate gene/protein list analysis. Bioinformatics 2012, 28, 1805–1806. [Google Scholar] [CrossRef] [PubMed]
- Suomi, T.; Seyednasrollah, F.; Jaakkola, M.K.; Faux, T.; Elo, L.L. ROTS: An R package for reproducibility-optimized statistical testing. PLoS Comput. Biol. 2017, 13, e1005562. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).