Analysis of the Phospholipid Profile of the Collection Strain PAO1 and Clinical Isolates of Pseudomonas aeruginosa in Relation to Their Attachment Capacity
Abstract
1. Introduction
2. Materials and Methods
Isolation, Identification and Growth Conditions of Bacterial Strains
3. Attachment Test
4. Phospholipids Extraction
5. Mass Spectrometry Analyses of Pseudomonas Aeruginosa Phospholipids
5.1. Identification of PL by Shotgun Mass Spectrometry
5.2. Relative Quantification of PLs by Reverse-Phase Liquid Chromatography-Mass Spectrometry (RPLC-MS/MS)
6. Statistical Analysis
7. Results
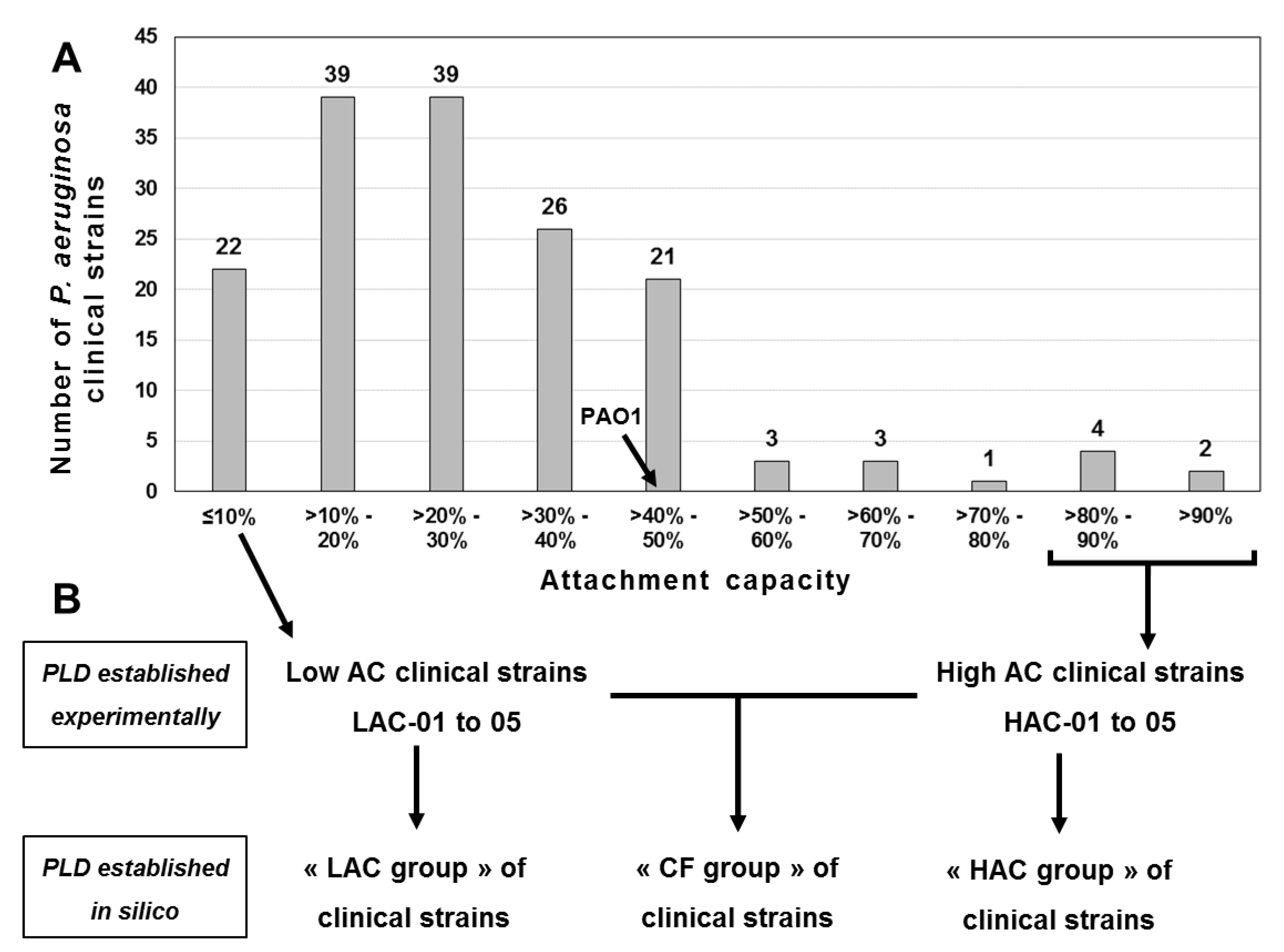
Attachment Capacities and Selection of P. aeruginosa PAO1 and Clinical Strains
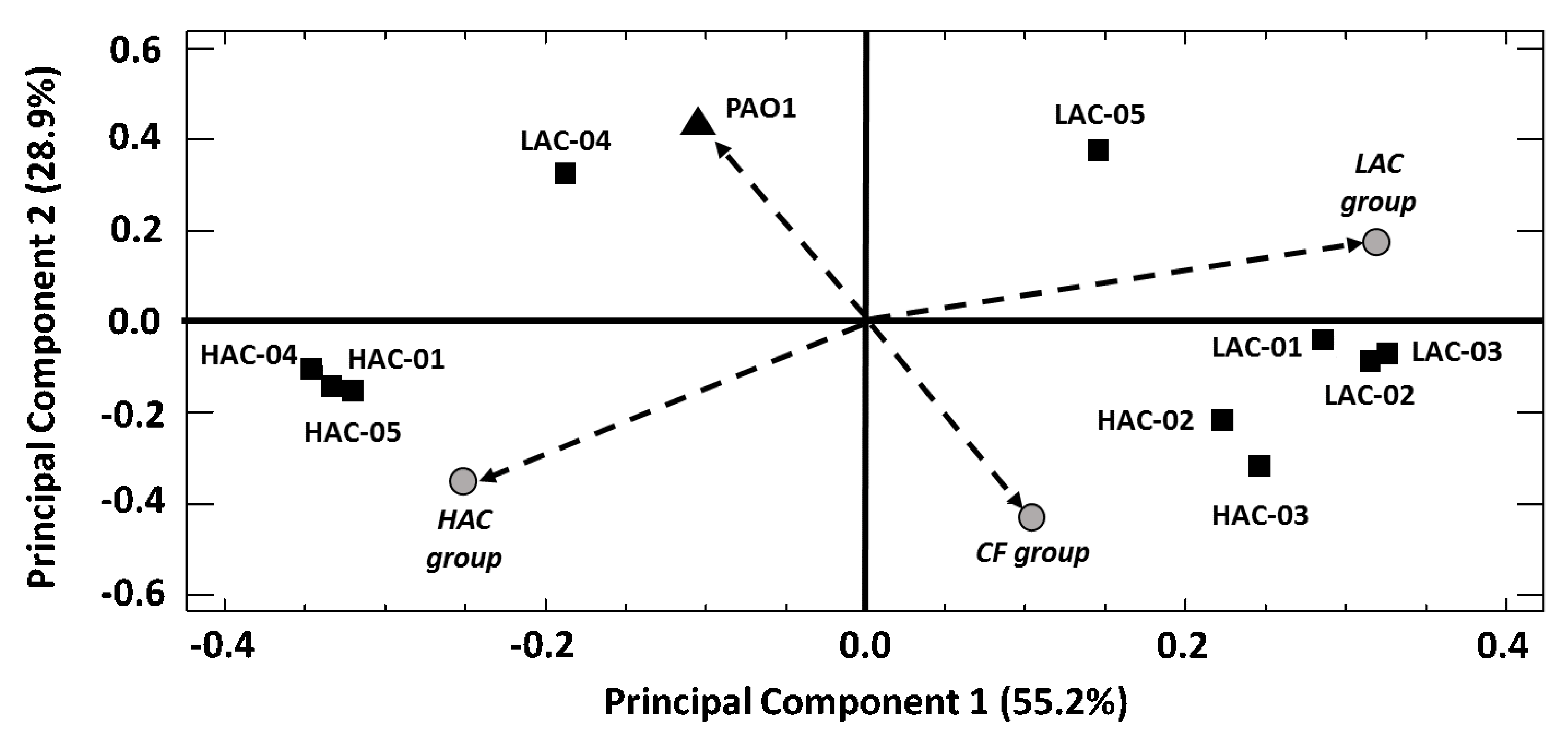
8. Phospholipidome Differences between P. aeruginosa PAO1 and Clinical Strains
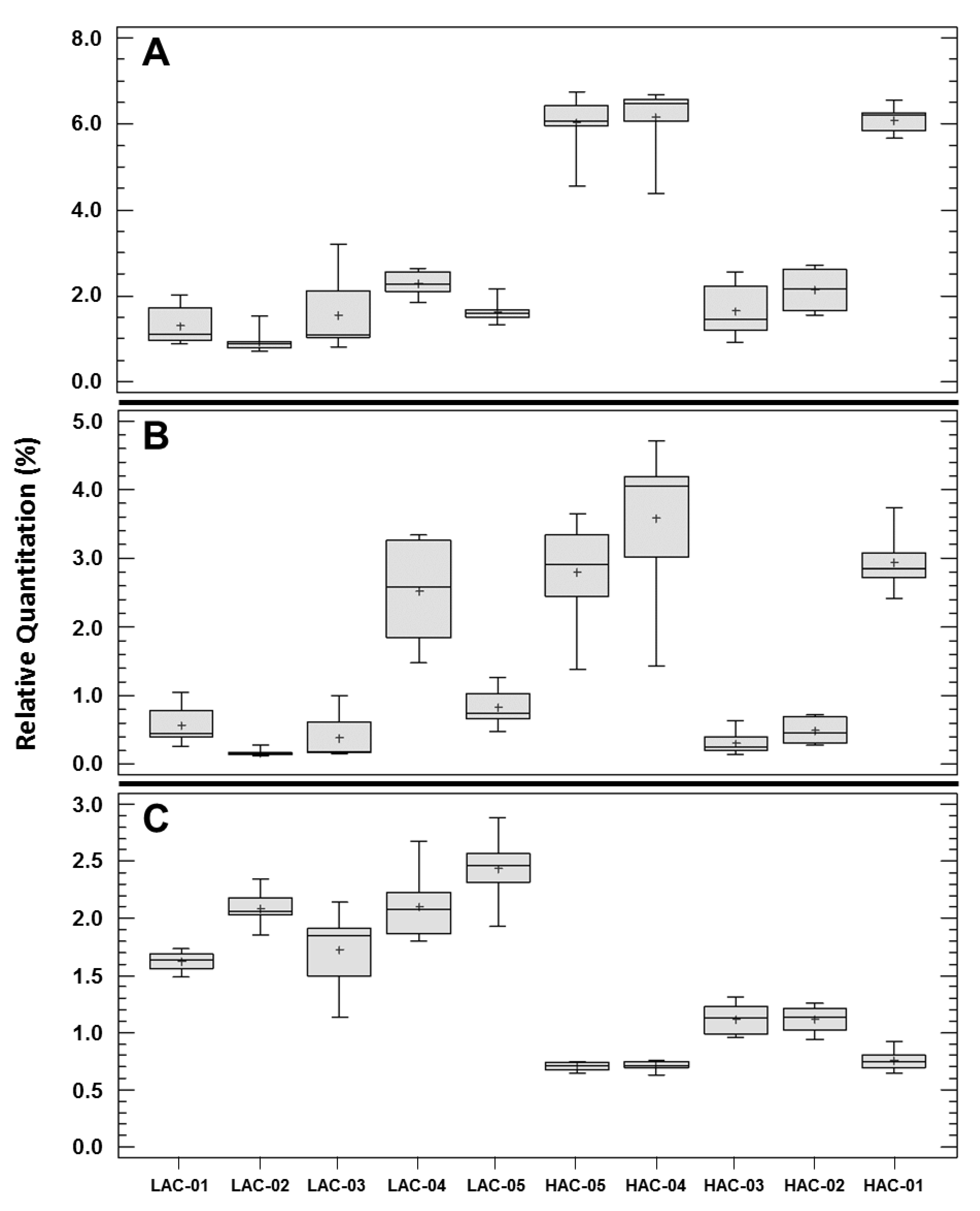
9. Differences among the Phospholipidomes of the P. aeruginosa Clinical Strains
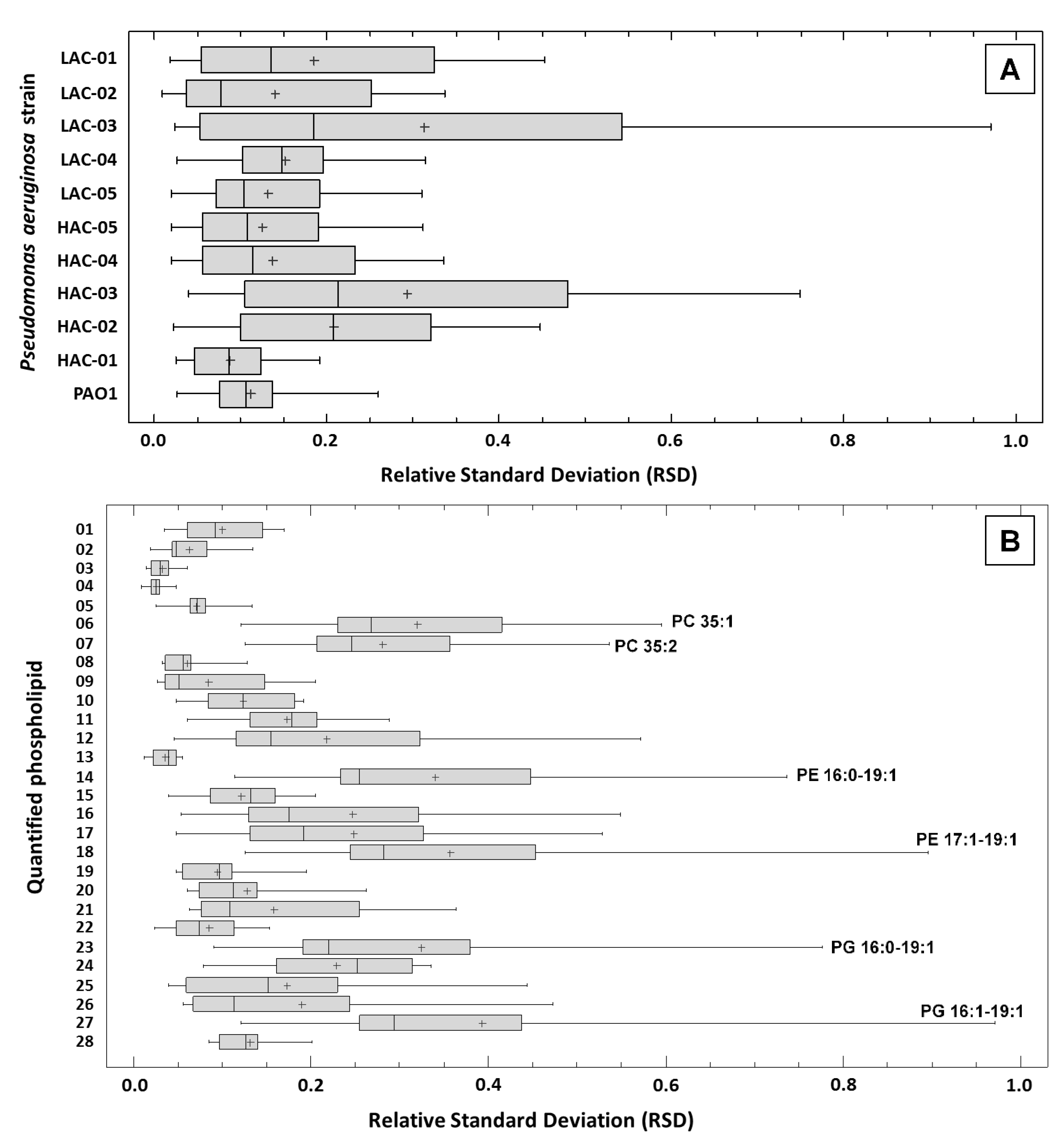
10. Repeatability of Data
11. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kannan, S.; Solomon, A.; Krishnamoorthy, G.; Marudhamuthu, M. Liposome encapsulated surfactant abetted copper nanoparticles alleviates biofilm mediated virulence in pathogenic Pseudomonas aeruginosa and MRSA. Sci. Rep. 2021, 11, 1102. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Lebeaux, D.; Ghigo, J.M.; Beloin, C. Biofilm-related infections: Bridging the gap between clinical management and fundamental aspects of recalcitrance toward antibiotics. Microbiol. Mol. Biol. Rev. 2014, 78, 510–543. [Google Scholar] [CrossRef]
- Moradali, M.F.; Ghods, S.; Rehm, B.H. Pseudomonas aeruginosa lifestyle: A paradigm for adaptation, survival, and persistence. Front. Cell Infect. Microbiol. 2017, 7, 39. [Google Scholar] [CrossRef] [PubMed]
- Muffler, K.; Lakatos, M.; Schlegel, C.; Strieth, D.; Kuhne, S.; Ulber, R. Application of biofilm bioreactors in white biotechnology. Adv. Biochem. Eng. Biotechnol. 2014, 146, 123–161. [Google Scholar] [CrossRef]
- Verderosa, A.D.; Totsika, M.; Fairfull-Smith, K.E. Bacterial biofilm eradication agents: A current review. Front. Chem. 2019, 7, 824. [Google Scholar] [CrossRef] [PubMed]
- Rybtke, M.; Hultqvist, L.D.; Givskov, M.; Tolker-Nielsen, T. Pseudomonas aeruginosa biofilm infections: Community structure, antimicrobial tolerance and immune response. J. Mol. Biol. 2015, 427, 3628–3645. [Google Scholar] [CrossRef] [PubMed]
- Sousa, A.M.; Pereira, M.O. Pseudomonas aeruginosa diversification during infection development in cystic fibrosis lungs—A review. Pathogens 2014, 3, 680–703. [Google Scholar] [CrossRef]
- Nathwani, D.; Raman, G.; Sulham, K.; Gavaghan, M.; Menon, V. Clinical and economic consequences of hospital-acquired resistant and multidrug-resistant Pseudomonas aeruginosa infections: A systematic review and meta-analysis. Antimicrob. Resist. Infect. Control 2014, 3, 32. [Google Scholar] [CrossRef]
- Ricciardelli, A.; Casillo, A.; Vergara, A.; Balasco, N.; Corsaro, M.M.; Tutino, M.L.; Parrilli, E. Environmental conditions shape the biofilm of the Antarctic bacterium Pseudoalteromonas haloplanktis TAC125. Microbiol. Res. 2019, 218, 66–75. [Google Scholar] [CrossRef]
- Lianou, A.; Nychas, G.J.E.; Koutsoumanis, K.P. Strain variability in biofilm formation: A food safety and quality perspective. Food Res. Int. 2020, 137, 109424. [Google Scholar] [CrossRef] [PubMed]
- Seneviratne, C.J.; Suriyanarayanan, T.; Widyarman, A.S.; Lee, L.S.; Lau, M.; Ching, J.; Delaney, C.; Ramage, G. Multi-omics tools for studying microbial biofilms: Current perspectives and future directions. Crit. Rev. Microbiol. 2020, 46, 759–778. [Google Scholar] [CrossRef]
- Coenye, T.; Kjellerup, B.; Stoodley, P.; Bjarnsholt, T. The future of biofilm research—Report on the ‘2019 Biofilm Bash’. Biofilm 2019, 2, 100012. [Google Scholar] [CrossRef] [PubMed]
- Crouzet, M.; Le Senechal, C.; Brözel, V.S.; Costaglioli, P.; Barthe, C.; Bonneu, M.; Garbay, B.; Vilain, S. Exploring early steps in biofilm formation: Set-up of an experimental system for molecular studies. BMC Microbiol. 2014, 14, 253. [Google Scholar] [CrossRef] [PubMed]
- Crouzet, M.; Claverol, S.; Lomenech, A.M.; Le Sénéchal, C.; Costaglioli, P.; Barthe, C.; Garbay, B.; Bonneu, M.; Vilain, S. Pseudomonas aeruginosa cells attached to a surface display a typical proteome early as 20 minutes of incubation. PLoS ONE 2017, 12, e0180341. [Google Scholar] [CrossRef] [PubMed]
- Le Sénéchal, C.; Crouzet, M.; Costaglioli, P.; Barthe, C.; Buré, C.; Vilain, S. Phospholipid content of Pseudomonas aeruginosa PAO1 is modulated by the growth phase rather than the immobilization state. Lipids 2019, 54, 519–529. [Google Scholar] [CrossRef]
- Oluyombo, O.; Penfold, C.N.; Diggle, S.P. Competition in biofilms between cystic fibrosis isolates of Pseudomonas aeruginosa is shaped by R-pyocins. mBio 2019, 10, e01828-18. [Google Scholar] [CrossRef]
- Rossi, E.; La Rosa, R.; Bartell, J.A.; Marvig, R.L.; Haagensen, J.A.J.; Sommer, L.M.; Molin, S.; Johansen, H.K. Pseudomonas aeruginosa adaptation and evolution in patients with cystic fibrosis. Nat. Rev. Microbiol. 2020. [Google Scholar] [CrossRef]
- Buré, C.; Ayciriex, S.; Testet, E.; Schmitter, J.M. A single run LC-MS/MS method for phospholipidomics. Anal. Bioanal. Chem. 2013, 405, 203–213. [Google Scholar] [CrossRef]
- Ejsing, C.S.; Duchoslav, E.; Sampaio, J.; Simons, K.; Bonner, R.; Thiele, C.; Ekroos, K.; Shevchenko, A. Automated identification and quantification of glycerophospholipid molecular species by multiple precursor ion scanning. Anal. Chem. 2006, 78, 6202–6214. [Google Scholar] [CrossRef]
- Kostakioti, M.; Hadjifrangiskou, M.; Hultgren, S.J. Bacterial biofilms: Development, dispersal, and therapeutic strategies in the dawn of the postantibiotic era. Cold Spring Harb. Perspect. Med. 2013, 3, a010306. [Google Scholar] [CrossRef] [PubMed]
- Kearns, D.B.; Losick, R. Cell population heterogeneity during growth of Bacillus subtilis. Genes Dev. 2005, 19, 3083–3094. [Google Scholar] [CrossRef] [PubMed]
- Vissers, T.; Brown, A.T.; Koumakis, N.; Dawson, A.; Hermes, M.; Schwarz-Linek, J.; Schofield, A.B.; French, J.M.; Koutsos, V.; Arlt, J.; et al. Bacteria as living patchy colloids: Phenotypic heterogeneity in surface adhesion. Sci. Adv. 2018, 4, eaao1170. [Google Scholar] [CrossRef]
- Murray, T.S.; Ledizet, M.; Kazmierczak, B.I. Swarming motility, secretion of type 3 effectors and biofilm formation phenotypes exhibited within a large cohort of Pseudomonas aeruginosa clinical isolates. J. Med. Microbiol. 2010, 59 Pt 5, 511–520. [Google Scholar] [CrossRef]
- Milivojevic, D.; Šumonja, N.; Medic, S.; Pavic, A.; Moric, I.; Vasiljevic, B.; Senerovic, L.; Nikodinovic-Runic, J. Biofilm-forming ability and infection potential of Pseudomonas aeruginosa strains isolated from animals and humans. Pathog. Dis. 2018, 76, fty041. [Google Scholar] [CrossRef]
- Ryu, S.Y.; Baek, W.K.; Kim, H.A. Association of biofilm production with colonization among clinical isolates of Acinetobacter baumannii. Korean J. Intern. Med. 2017, 32, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Schaeffer, C.R.; Hoang, T.N.; Sudbeck, C.M.; Alawi, M.; Tolo, I.E.; Robinson, D.A.; Horswill, A.R.; Rohde, H.; Fey, P.D. Versatility of biofilm matrix molecules in Staphylococcus epidermidis clinical isolates and importance of polysaccharide intercellular adhesin expression during high shear stress. mSphere 2016, 1, e00165-16. [Google Scholar] [CrossRef]
- Kalmokoff, M.L.; Austin, J.W.; Wan, X.D.; Sanders, G.; Banerjee, S.; Farber, J.M. Adsorption, attachment and biofilm formation among isolates of Listeria monocytogenes using model conditions. J. Appl. Microbiol. 2001, 91, 725–734. [Google Scholar] [CrossRef]
- McLoon, A.L.; Guttenplan, S.B.; Kearns, D.B.; Kolter, R.; Losick, R. Tracing the domestication of a biofilm-forming bacterium. J. Bacteriol. 2011, 193, 2027–2034. [Google Scholar] [CrossRef]
- Barreto, H.C.; Cordeiro, T.N.; Henriques, A.O.; Gordo, I. Rampant loss of social traits during domestication of a Bacillus subtilis natural isolate. Sci. Rep. 2020, 10, 18886. [Google Scholar] [CrossRef]
- Deligianni, E.; Pattison, S.; Berrar, D.; Ternan, N.G.; Haylock, R.W.; Moore, J.E.; Elborn, S.J.; Dooley, J.S. Pseudomonas aeruginosa cystic fibrosis isolates of similar RAPD genotype exhibit diversity in biofilm forming ability in vitro. BMC Microbiol. 2010, 10, 38. [Google Scholar] [CrossRef] [PubMed]
- Cerca, N.; Pier, G.B.; Vilanova, M.; Oliveira, R.; Azeredo, J. Quantitative analysis of adhesion and biofilm formation on hydrophilic and hydrophobic surfaces of clinical isolates of Staphylococcus epidermidis. Res. Microbiol. 2005, 156, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.Y.; Weibel, D.B. Organization and function of anionic phospholipids in bacteria. Appl. Microbiol. Biotechnol. 2016, 100, 4255–4267. [Google Scholar] [CrossRef]
- Epand, R.F.; Savage, P.B.; Epand, R.M. Bacterial lipid composition and the antimicrobial efficacy of cationic steroid compounds (Ceragenins). Biochim. Biophys. Acta 2007, 1768, 2500–2509. [Google Scholar] [CrossRef] [PubMed]
- Rowlett, V.W.; Mallampalli, V.K.P.S.; Karlstaedt, A.; Dowhan, W.; Taegtmeyer, H.; Margolin, W.; Vitrac, H. Impact of membrane phospholipid alterations in Escherichia coli on cellular function and bacterial stress adaptation. J. Bacteriol. 2017, 199, e00849-16. [Google Scholar] [CrossRef]
- Jiang, J.H.; Bhuiyan, M.S.; Shen, H.H.; Cameron, D.R.; Rupasinghe, T.W.T.; Wu, C.M.; Le Brun, A.P.; Kostoulias, X.; Domene, C.; Fulcher, A.J.; et al. Antibiotic resistance and host immune evasion in Staphylococcus aureus mediated by a metabolic adaptation. Proc. Natl. Acad. Sci. USA 2019, 116, 3722–3727. [Google Scholar] [CrossRef]
- Tashiro, Y.; Inagaki, A.; Shimizu, M.; Ichikawa, S.; Takaya, N.; Nakajima-Kambe, T.; Uchiyama, H.; Nomura, N. Characterization of phospholipids in membrane vesicles derived from Pseudomonas aeruginosa. Biosci. Biotechnol. Biochem. 2011, 75, 605–607. [Google Scholar] [CrossRef]
- Varga, J.J.; Barbier, M.; Mulet, X.; Bielecki, P.; Bartell, J.A.; Owings, J.P.; Martinez-Ramos, I.; Hittle, L.E.; Davis MRJr Damron, F.H.; Liechti, G.W.; et al. Genotypic and phenotypic analyses of a Pseudomonas aeruginosa chronic bronchiectasis isolate reveal differences from cystic fibrosis and laboratory strains. BMC Genom. 2015, 16, 883. [Google Scholar] [CrossRef]
- Clark, S.T.; Guttman, D.S.; Hwang, D.M. Diversification of Pseudomonas aeruginosa within the cystic fibrosis lung and its effects on antibiotic resistance. FEMS Microbiol. Lett. 2018, 365, fny026. [Google Scholar] [CrossRef]
- Deschamps, E.; Schaumann, A.; Schmitz-Afonso, I.; Afonso, C.; Dé, E.; Loutelier-Bourhis, C.; Alexandre, S. Membrane phospholipid composition of Pseudomonas aeruginosa grown in a cystic fibrosis mucus-mimicking medium. Biochim. Biophys. Acta Biomembr. 2021, 1863, 183482. [Google Scholar] [CrossRef]
- Hobby, C.R.; Herndon, J.L.; Morrow, C.A.; Peters, R.E.; Symes, S.J.K.; Giles, D.K. Exogenous fatty acids alter phospholipid composition, membrane permeability, capacity for biofilm formation, and antimicrobial peptide susceptibility in Klebsiella pneumoniae. MicrobiologyOpen 2019, 8, e00635. [Google Scholar] [CrossRef] [PubMed]
- Benamara, H.; Rihouey, C.; Jouenne, T.; Alexandre, S. Impact of the biofilm mode of growth on the inner membrane phospholipid composition and lipid domains in Pseudomonas aeruginosa. Biochim. Biophys. Acta 2011, 1808, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Benamara, H.; Rihouey, C.; Abbes, I.; Ben Mlouka, M.A.; Hardouin, J.; Jouenne, T.; Alexandre, S. Characterization of membrane lipidome changes in Pseudomonas aeruginosa during biofilm growth on glass wool. PLoS ONE 2014, 9, e108478. [Google Scholar] [CrossRef] [PubMed]
- Subedi, D.; Vijay, A.K.; Kohli, G.S.; Rice, S.A.; Willcox, M. Comparative genomics of clinical strains of Pseudomonas aeruginosa strains isolated from different geographic sites. Sci. Rep. 2018, 8, 15668. [Google Scholar] [CrossRef]
- Gabrielaite, M.; Johansen, H.K.; Molin, S.; Nielsen, F.C.; Marvig, R.L. Gene loss and acquisition in lineages of Pseudomonas aeruginosa evolving in cystic fibrosis patient airways. mBio 2020, 11, e02359-20. [Google Scholar] [CrossRef] [PubMed]
- Bogdanov, M.; Pyrshev, K.; Yesylevskyy, S.; Ryabichko, S.; Boiko, V.; Ivanchenko, P.; Kiyamova, R.; Guan, Z.; Ramseyer, C.; Dowhan, W. Phospholipid distribution in the cytoplasmic membrane of Gram-negative bacteria is highly asymmetric, dynamic, and cell shape-dependent. Sci. Adv. 2020, 6, eaaz6333. [Google Scholar] [CrossRef]
- Nishizaki, T. Dioleoylphosphoethanolamine retains cell surface GLUT4 by inhibiting PKCα-driven internalization. Cell. Physiol. Biochem. 2018, 46, 1985–1998. [Google Scholar] [CrossRef]
- Mochizuki, S.; Kanegae, N.; Nishina, K.; Kamikawa, Y.; Koiwai, K.; Masunaga, H.; Sakurai, K. The role of the helper lipid dioleoylphosphatidylethanolamine (DOPE) for DNA transfection cooperating with a cationic lipid bearing ethylenediamine. Biochim. Biophys. Acta 2013, 1828, 412–418. [Google Scholar] [CrossRef]
- Wang, D.Y.; van der Mei, H.C.; Ren, Y.; Busscher, H.J.; Shi, L. Lipid-based Aantimicrobial delivery-systems for the treatment of bacterial infections. Front. Chem. 2020, 7, 872. [Google Scholar] [CrossRef]
- Huster, D.; Jin, A.J.; Arnold, K.; Gawrisch, K. Water permeability of polyunsaturated lipid membranes measured by 17O NMR. Biophys. J. 1997, 73, 855–864. [Google Scholar] [CrossRef]
| Phospholipidomes Experimentally Determined [Mean %mol. ± SD (Rank)] | In Silico Phospholipidomes [Mean %mol ± SD (Rank)] | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| «Low Attachment Capacity» (LAC) CF Strains | «High Attachment Capacity» (HAC) CF Strains | Collection | ||||||||||||
| 01 [1%] (1) | 02 [2%] | 03 [2%] | 04 [2%] | 05 [3%] | 05 [85%] | 04 [87%] | 03 [89%] | 02 [90%] | 01 [91%] | PAO1 [41%] | LAC Group | HAC Group | CF Group | |
| PC 32:0 | 11.2 ± 1.0 (3) | 10.3 ± 0.4 (3) | 10.6 ± 0.6 (3) | 8.5 ± 1.4 (4) | 7.9 ± 0.6 (3) | 12.2 ± 1.2 (4) | 12.6 ± 0.8 (2) | 13.0 ± 2.0 (2) | 11.9 ± 1.7 (2) | 12.6 ± 1.6 (3) | 4.5 ± 0.4 (6) | 9.7 ± 1.6 (3) | 12.5 ± 1.6 (2) | 11.1 ± 2.1 (3) |
| PC 32:1 | 7.2 ± 0.3 (4) | 8.5 ± 0.2 (4) | 8.7 ± 0.2 (4) | 6.6 ± 0.4 (5) | 6.8 ± 0.6 (4) | 12.3 ± 0.8 (3) | 11.8 ± 0.5 (4) | 11.3 ± 1.5 (4) | 9.6 ± 1.2 (4) | 12.3 ± 0.6 (4) | 5.0 ± 0.2 (5) | 7.6 ± 1.0 (4) | 11.5 ± 1.4 (4) | 9.5 ± 2.3 (4) |
| PC 34:0 | 7.1 ± 0.2 (5) | 6.6 ± 0.1 (5) | 6.4 ± 0.2 (5) | 6.4 ± 0.3 (6) | 6.6 ± 0.2 (5) | 5.4 ± 0.1 (6) | 5.3 ± 0.1 (6) | 6.3 ± 0.4 (5) | 6.6 ± 0.3 (5) | 5.5 ± 0.2 (6) | 6.7 ± 0.2 (4) | 6.6 ± 0.3 (5) | 5.8 ± 0.6 (5) | 6.2 ± 0.6 (5) |
| PC 34:1 | 57.2 ± 1.1 (1) | 57.3 ± 0.5 (1) | 56.5 ± 1.4 (1) | 53.3 ± 1.5 (1) | 57.9 ± 1.2 (1) | 47.7 ± 1.4 (1) | 46.4 ± 1.0 (1) | 55.2 ± 2.6 (1) | 57.1 ± 2.2 (1) | 47.3 ± 1.2 (1) | 55.3 ± 1.4 (1) | 56.5 ± 2.0 (1) | 50.7 ± 4.8 (1) | 53.6 ± 4.7 (1) |
| PC 34:2 | 12.8 ± 0.3 (2) | 15.2 ± 0.6 (2) | 14.4 ± 1.2 (2) | 13.2 ± 1.8 (2) | 15.7 ± 1.4 (2) | 13.4 ± 1.0 (2) | 12.5 ± 0.9 (3) | 11.9 ± 0.8 (3) | 10.9 ± 0.7 (3) | 13.0 ± 0.8 (2) | 11.3 ± 0.8 (3) | 14.2 ± 1.6 (2) | 12.3 ± 1.2 (3) | 13.3 ± 1.7 (2) |
| PC 35:1 | 3.1 ± 1.1 (6) | 1.4 ± 0.3 (6) | 2.3 ± 1.3 (6) | 8.7 ± 2.0 (3) | 3.5 ± 1.0 (6) | 6.0 ± 1.4 (5) | 7.5 ± 2.0 (5) | 01.7 ± 1.0 (6) | 2.9 ± 1.2 (6) | 6.1 ± 1.0 (5) | 12.9 ± 1.6 (2) | 3.8 ± 2.9 (6) | 4.9 ± 2.6 (6) | 4.3 ± 2.8 (6) |
| PC 35:2 | 1.4 ± 0.5 (7) | 0.7 ± 0.1 (7) | 1.0 ± 0.6 (7) | 3.2 ± 0.9 (7) | 1.6 ± 0.4 (7) | 3.1 ± 0.7 (7) | 3.9 ± 0.9 (7) | 0.7 ± 0.3 (7) | 1.0 ± 0.4 (7) | 3.1 ± 0.4 (7) | 4.4 ± 0.6 (7) | 1.6 ± 1.0 (7) | 2.4 ± 1.4 (7) | 2.0 ± 1.3 (7) |
| PE 14:0-18:1 | 1.4 ± 0.1 (9) | 1.3 ± 0.0 (7) | 1.4 ± 0.0 (9) | 1.5 ± 0.1 (11) | 1.5 ± 0.1 (10) | 1.5 ± 0.1 (10) | 1.4 ± 0.1 (10) | 1.2 ± 0.1 (7) | 1.2 ± 0.1 (8) | 1.5 ± 0.1 (10) | 1.5 ± 0.2 (11) | 1.4 ± 0.1 (10) | 1.4 ± 0.2 (10) | 1.4 ± 0.1 (10) |
| PE 15:0-18:1 | 0.1 ± 0.0 (12) | 0.6 ± 0.0 (11) | 0.6 ± 0.0 (11) | 1.0 ± 0.2 (12) | 0.6 ± 0.0 (12) | 1.2 ± 0.0 (11) | 1.2 ± 0.0 (11) | 0.4 ± 0.1 (11) | 0.5 ± 0.1 (11) | 1.2 ± 0.0 (11) | 0.7 ± 0.1 (12) | 0.6 ± 0.3 (12) | 0.9 ± 0.4 (11) | 0.7 ± 0.4 (12) |
| PE 16:0-16:0 | 8.2 ± 1.0 (3) | 7.4 ± 0.4 (3) | 7.4 ± 0.4 (3) | 7.2 ± 0.9 (4) | 6.0 ± 0.6 (4) | 8.4 ± 1.2 (4) | 9.8 ± 1.1 (4) | 10.0 ± 1.8 (3) | 8.5 ± 1.6 (3) | 8.6 ± 1.6 (4) | 3.8 ± 0.3 (6) | 7.2 ± 1.0 (3) | 9.1 ± 1.7 (3) | 8.1 ± 1.7 (3) |
| PE 16:0-16:1 | 4.9 ± 0.4 (5) | 5.8 ± 0.4 (4) | 5.3 ± 0.9 (4) | 3.2 ± 0.6 (6) | 3.2 ± 0.6 (5) | 6.9 ± 1.3 (5) | 5.4 ± 1.6 (6) | 8.8 ± 1.8 (4) | 7.2 ± 1.7 (4) | 6.4 ± 0.8 (5) | 2.5 ± 0.4 (8) | 4.5 ± 1.2 (5) | 6.9 ± 1.9 (5) | 5.7 ± 2.0 (5) |
| PE 16:0-17:1 | 1.3 ± 0.4 (10) | 0.9 ± 0.2 (9) | 1.6 ± 0.9 (8) | 2.3 ± 0.3 (8) | 1.6 ± 0.2 (9) | 6.0 ± 0.6 (6) | 6.2 ± 0.7 (5) | 1.7 ± 0.6 (6) | 2.1 ± 0.5 (6) | 6.1 ± 0.3 (6) | 2.3 ± 0.4 (10) | 1.5 ± 0.7 (9) | 4.4 ± 2.1 (6) | 3.0 ± 2.1 (6) |
| PE 16:0-18:1 | 56.7 ± 2.3 (1) | 58.9 ± 0.7 (1) | 56.8 ± 2.7 (1) | 48.5 ± 2.5 (1) | 54.1 ± 1.2 (1) | 39.2 ± 1.7 (1) | 38.0 ± 1.3 (1) | 55.6 ± 2.2 (1) | 56.5 ± 1.3 (1) | 38.8 ± 1.0 (1) | 43.2 ± 2.4 (1) | 55.0 ± 4.1 (1) | 45.6 ± 8.7 (1) | 50.3 ± 8.2 (1) |
| PE 16:0-19:1 | 5.5 ± 2.0 (4) | 2.3 ± 0.8 (5) | 4.7 ± 3.5 (5) | 13.9 ± 3.3 (2) | 8.4 ± 2.0 (3) | 11 ± 2.6 (3) | 13.2 ± 3.4 (2) | 2.8 ± 1.8 (5) | 4.8 ± 2.2 (5) | 11.7 ± 1.6 (3) | 16.0 ± 1.8 (2) | 7.0 ± 4.7 (4) | 8.7 ± 4.8 (4) | 7.8 ± 4.8 (4) |
| PE 16:1-18:1 | 16.1 ± 0.8 (2) | 18.5 ± 0.7 (2) | 16.9 ± 2.6 (2) | 12.2 ± 2.0 (3) | 15.8 ± 1.7 (2) | 14.3 ± 2.3 (2) | 12 ± 2.5 (3) | 16.3 ± 1.4 (2) | 14.7 ± 1.9 (2) | 13.8 ± 1.3 (2) | 13.4 ± 1.9 (3) | 15.9 ± 2.7 (2) | 14.2 ± 2.4 (2) | 15.1 ± 2.7 (2) |
| PE 16:1-19:1 | 1.6 ± 0.5 (7) | 0.8 ± 0.2 (10) | 1.4 ± 0.7 (9) | 2.3 ± 0.3 (8) | 2.4 ± 0.4 (7) | 3.2 ± 0.4 (8) | 3.3 ± 0.5 (9) | 0.7 ± 0.4 (10) | 1.2 ± 0.4 (8) | 3.3 ± 0.2 (8) | 3.8 ± 0.4 (6) | 1.7 ± 0.8 (8) | 2.4 ± 1.2 (8) | 2.0 ± 1.1 (8) |
| PE 17:1-18:1 | 2.0 ± 0.7 (6) | 1.2 ± 0.3 (8) | 1.9 ± 1.0 (6) | 3.3 ± 0.5 (5) | 3.2 ± 0.6 (5) | 4.8 ± 0.6 (7) | 5.1 ± 0.8 (7) | 1.0 ± 0.5 (9) | 1.6 ± 0.5 (7) | 4.9 ± 0.2 (7) | 5.4 ± 0.6 (4) | 2.3 ± 1.1 (6) | 3.5 ± 1.9 (7) | 2.9 ± 1.6 (7) |
| PE 17:1-19:1 | 0.6 ± 0.3 (11) | 0.2 ± 0.0 (12) | 0.4 ± 0.3 (12) | 2.5 ± 0.7 (7) | 0.8 ± 0.3 (11) | 2.8 ± 0.7 (9) | 3.6 ± 1.0 (8) | 0.3 ± 0.2 (12) | 0.5 ± 0.2 (11) | 2.9 ± 0.4 (9) | 2.5 ± 0.4 (8) | 0.9 ± 0.9 (11) | 2.0 ± 1.5 (9) | 1.5 ± 1.4 (9) |
| PE 18:1-18:1 | 1.6 ± 0.1 (7) | 2.1 ± 0.1 (6) | 1.7 ± 0.3 (7) | 2.1 ± 0.3 (10) | 2.4 ± 0.3 (7) | 0.7 ± 0.0 (12) | 0.7 ± 0.0 (12) | 1.1 ± 0.1 (8) | 1.1 ± 0.1 (10) | 0.8 ± 0.1 (12) | 4.8 ± 0.3 (5) | 2.0 ± 0.4 (7) | 0.9 ± 0.2 (11) | 1.4 ± 0.6 (10) |
| PG 16:0-16:0 | 7.5 ± 1.0 (4) | 6.3 ± 0.4 (3) | 6.5 ± 0.4 (4) | 7.1 ± 0.8 (4) | 5.4 ± 0.4 (5) | 8.2 ± 0.8 (4) | 7.7 ± 1.0 (4) | 6.9 ± 1.8 (3) | 5.5 ± 1.3 (4) | 7.9 ± 1.0 (5) | 3.4 ± 0.4 (8) | 6.6 ± 1.0 (4) | 7.2 ± 1.6 (5) | 6.9 ± 1.3 (4) |
| PG 16:0-17:1 | 3.6 ± 0.9 (6) | 2.2 ± 0.4 (6) | 3.2 ± 1.2 (6) | 4.7 ± 0.4 (7) | 3.4 ± 0.3 (6) | 8.2 ± 0.5 (4) | 7.3 ± 0.5 (5) | 3.8 ± 1.1 (5) | 4.7 ± 0.5 (5) | 7.7 ± 0.8 (6) | 3.7 ± 0.4 (7) | 3.4 ± 1.1 (6) | 6.3 ± 1.9 (6) | 4.9 ± 2.1 (6) |
| PG 16:0-18:1 | 53.1 ± 3.9 (1) | 63.1 ± 1.5 (1) | 59.4 ± 6.3 (1) | 35.9 ± 5.5 (1) | 49.5 ± 3.4 (1) | 33.6 ± 3.8 (1) | 30.8 ± 4.2 (1) | 58.8 ± 2.4 (1) | 56.0 ± 2.7 (1) | 34.1 ± 2.2 (1) | 41.6 ± 4.5 (1) | 52.2 ± 10.4 (1) | 42.7 ± 12.5 (1) | 47.4 ± 12.5 (1) |
| PG 16:0-19:1 | 10.9 ± 3.9 (3) | 4.1 ± 1.3 (4) | 8.9 ± 6.9 (3) | 28.3 ± 5.9 (2) | 20.4 ± 3.9 (2) | 23.1 ± 4.4 (2) | 27.7 ± 6.1 (2) | 6.6 ± 4.3 (4) | 11.7 ± 4.4 (3) | 23.3 ± 2.1 (2) | 26.4 ± 4.5 (2) | 14.5 ± 9.9 (2) | 18.5 ± 9.1 (2) | 16.5 ± 9.7 (2) |
| PG 16:1-18:1 | 15.3 ± 2.5 (2) | 19.7 ± 1.7 (2) | 15.2 ± 4.9 (2) | 8.7 ± 2.7 (3) | 9.5 ± 1.6 (3) | 8 ± 2.5 (6) | 7.3 ± 2.4 (5) | 18.5 ± 4.1 (2) | 13.7 ± 3.4 (2) | 8.1 ± 0.6 (4) | 7.1 ± 1.8 (4) | 13.7 ± 5.0 (3) | 11.1 ± 5.2 (3) | 12.4 ± 5.3 (3) |
| PG 16:1-19:1 | 3.0 ± 0.6 (7) | 1.5 ± 0.3 (7) | 2.1 ± 0.6 (7) | 2.8 ± 0.4 (8) | 3.1 ± 0.2 (7) | 4.1 ± 0.2 (8) | 3.5 ± 0.3 (8) | 1.6 ± 0.7 (7) | 2.5 ± 0.6 (7) | 4.0 ± 0.3 (8) | 3.8 ± 0.2 (6) | 2.5 ± 0.8 (7) | 3.2 ± 1.1 (8) | 2.8 ± 1.0 (7) |
| PG 17:1-18:1 | 5.1 ± 1.2 (5) | 2.5 ± 0.6 (5) | 3.7 ± 1.5 (5) | 6.4 ± 0.7 (5) | 6.5 ± 0.5 (4) | 9.8 ± 0.7 (3) | 9.2 ± 0.5 (3) | 3.0 ± 1.4 (6) | 4.7 ± 1.1 (5) | 9.8 ± 0.6 (3) | 9.3 ± 0.9 (3) | 4.8 ± 1.8 (5) | 7.3 ± 3.0 (4) | 6.1 ± 2.8 (5) |
| PG 17:1-19:1 | 1.2 ± 0.5 (8) | 0.3 ± 0.1 (8) | 0.7 ± 0.7 (8) | 5.9 ± 1.6 (6) | 1.9 ± 0.6 (8) | 4.9 ± 1.3 (7) | 6.3 ± 1.7 (7) | 0.6 ± 0.4 (8) | 1.0 ± 0.4 (8) | 4.9 ± 0.6 (7) | 4.2 ± 0.8 (5) | 2.0 ± 2.2 (8) | 3.5 ± 2.5 (7) | 2.8 ± 2.5 (7) |
| PG 18:0-18:1 | 0.2 ± 0.0 (9) | 0.2 ± 0.0 (9) | 0.2 ± 0.0 (9) | 0.3 ± 0.0 (9) | 0.3 ± 0.1 (9) | 0.1 ± 0 (9) | 0.1 ± 0.0 (9) | 0.2 ± 0.0 (9) | 0.3 ± 0 (9) | 0.1 ± 0.0 (9) | 0.5 ± 0.1 (9) | 0.2 ± 0.1 (9) | 0.2 ± 0.1 (9) | 0.2 ± 0.1 (9) |
| CF Group vs. PAO1 (CF/PAO1) | LAC Group vs. PAO1 (LAC/PAO1) | HAC Group vs. PAO1 (HAC/PAO1) | LAC vs. HAC Groups (LAC/HAC) | |||||
|---|---|---|---|---|---|---|---|---|
| Ratio | p-Value | Ratio | p-Value | Ratio | p-Value | Ratio | p-Value | |
| PC 32:0 | 2.5 | 3.5 × 10−6 | 2.2 | 1.0 × 10−5 | 2.8 | 1.0 × 10−5 | 0.8 | 5.4 × 10−10 |
| PC 32:1 | 1.9 | 3.5 × 10−6 | 1.5 | 1.0 × 10−5 | 2.3 | 1.0 × 10−5 | 0.7 | 2.5 × 10−13 |
| PC 34:0 | 0.9 | 3.1 × 10−2 | 1.0 | 3.9 × 10−1 | 0.9 | 1.2 × 10−3 | 1.1 | 1.7 × 10−7 |
| PC 34:1 | 1.0 | 8.6 × 10−1 | 1.0 | 6.8 × 10−2 | 0.9 | 3.0 × 10−2 | 1.1 | 4.3 × 10−6 |
| PC 34:2 | 1.2 | 5.2 × 10−4 | 1.3 | 2.8 × 10−5 | 1.1 | 1.6 × 10−2 | 1.2 | 6.9 × 10−7 |
| PC 35:1 | 0.3 | 3.8 × 10−6 | 0.3 | 1.2 × 10−5 | 0.4 | 1.0 × 10−5 | 0.8 | 4.5 × 10−2 |
| PC 35:2 | 0.5 | 3.4 × 10−5 | 0.4 | 2.5 × 10−5 | >0.5 | 2.5 × 10−4 | 0.7 | 3.9 × 10−2 |
| PE 14:0-18:1 | 0.9 | 1.2 × 10−1 | 0.9 | 2.3 × 10−1 | 0.9 | 7.9 × 10−2 | 1.0 | 9.7 × 10−1 |
| PE 15:0-18:1 | 1.0 | 3.5 × 10−1 | 0.8 | 7.8 × 10−3 | 1.2 | 3.8 × 10−1 | 0.7 | 6.6 × 10−3 |
| PE 16:0-16:0 | 2.2 | 3.5 × 10−6 | 1.9 | 1.0 × 10−5 | 2.4 | 1.0 × 10−5 | 0.8 | 3.4 × 10−7 |
| PE 16:0-16:1 | 2.3 | 1.2 × 10−5 | 1.8 | 9.6 × 10−5 | 2.8 | 1.0 × 10−5 | 0.6 | 1.1 × 10−8 |
| PE 16:0-17:1 | 1.3 | 6.1 × 10−1 | 0.7 | 2.3 × 10−3 | 1.9 | 3.9 × 10−2 | 0.4 | 1.4 × 10−8 |
| PE 16:0-18:1 | 1.2 | 5.1 × 10−2 | 1.3 | 1.5 × 10−5 | 1.1 | 5.5 × 10−1 | 1.2 | 4.1 × 10−6 |
| PE 16:0-19:1 | 0.5 | 5.6 × 10−5 | 0.4 | 1.4 × 10−4 | >0.5 | 1.1 × 10−4 | 0.8 | 9.6 × 10−2 |
| PE 16:1-18:1 | 1.1 | 8.0 × 10−2 | 1.2 | 1.7 × 10−2 | 1.1 | 3.5 × 10−1 | 1.1 | 4.7 × 10−3 |
| PE 16:1-19:1 | >0.5 | 1.8 × 10−5 | 0.4 | 1.3 × 10−5 | 0.6 | 1.4 × 10−4 | 0.7 | 8.1 × 10−3 |
| PE 17:1-18:1 | >0.5 | 7.6 × 10−5 | 0.4 | 1.2× 10−5 | 0.6 | 1.7 × 10−3 | 0.7 | 6.0 × 10−3 |
| PE 17:1-19:1 | 0.6 | 4.7 × 10−2 | 0.4 | 3.8 × 10−4 | 0.8 | 8.2 × 10−1 | 0.4 | 8.3 × 10−4 |
| PE 18:1-18:1 | 0.3 | 3.5 × 10−6 | 0.4 | 1.0 × 10−5 | 0.2 | 1.0 × 10−5 | 2.3 | 3.5 × 10−14 |
| PG 16:0-16:0 | 2.0 | 3.5 × 10−6 | 1.9 | 1.0 × 10−5 | 2.1 | 1.0 × 10−5 | 0.9 | 1.3 × 10−2 |
| PG 16:0-17:1 | 1.3 | 2.1 × 10−1 | 0.9 | 3.1 × 10−1 | 1.7 | 6.3 × 10−4 | >0.5 | 6.5 × 10−9 |
| PG 16:0-18:1 | 1.1 | 2.6 × 10−1 | 1.3 | 6.4 × 10−3 | 1.0 | 5.7 × 10−1 | 1.2 | 1.0 × 10−3 |
| PG 16:0-19:1 | 0.6 | 5.4 × 10−3 | >0.5 | 3.2 × 10−3 | 0.7 | 1.9 × 10−2 | 0.8 | 6.0 × 10−2 |
| PG 16:1-18:1 | 1.8 | 3.9 × 10−3 | 1.9 | 1.2 × 10−3 | 1.6 | 2.5 × 10−2 | 1.2 | 1.8 × 10−2 |
| PG 16:1-19:1 | 0.7 | 5.0 × 10−3 | 0.7 | 5.1 × 10−5 | 0.8 | 2.0 × 10−1 | 0.8 | 6.8 × 10−4 |
| PG 17:1-18:1 | 0.7 | 2.9 × 10−3 | >0.5 | 1.0 × 10−5 | 0.8 | 2.1 × 10−1 | 0.7 | 4.6 × 10−4 |
| PG 17:1-19:1 | 0.7 | 9.1 × 10−2 | 0.5 | 3.5 × 10−3 | 0.8 | 7.7 × 10−1 | 0.6 | 7.2 × 10−3 |
| PG 18:0-18:1 | 0.5 | 3.4 × 10−6 | >0.5 | 9.7 × 10−6 | 0.4 | 9.5 × 10−6 | 1.2 | 4.9 × 10−4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Le Sénéchal, C.; Puges, M.; Barthe, C.; Costaglioli, P.; Tokarski, C.; Buré, C.; Vilain, S. Analysis of the Phospholipid Profile of the Collection Strain PAO1 and Clinical Isolates of Pseudomonas aeruginosa in Relation to Their Attachment Capacity. Int. J. Mol. Sci. 2021, 22, 4003. https://doi.org/10.3390/ijms22084003
Le Sénéchal C, Puges M, Barthe C, Costaglioli P, Tokarski C, Buré C, Vilain S. Analysis of the Phospholipid Profile of the Collection Strain PAO1 and Clinical Isolates of Pseudomonas aeruginosa in Relation to Their Attachment Capacity. International Journal of Molecular Sciences. 2021; 22(8):4003. https://doi.org/10.3390/ijms22084003
Chicago/Turabian StyleLe Sénéchal, Caroline, Mathilde Puges, Christophe Barthe, Patricia Costaglioli, Caroline Tokarski, Corinne Buré, and Sébastien Vilain. 2021. "Analysis of the Phospholipid Profile of the Collection Strain PAO1 and Clinical Isolates of Pseudomonas aeruginosa in Relation to Their Attachment Capacity" International Journal of Molecular Sciences 22, no. 8: 4003. https://doi.org/10.3390/ijms22084003
APA StyleLe Sénéchal, C., Puges, M., Barthe, C., Costaglioli, P., Tokarski, C., Buré, C., & Vilain, S. (2021). Analysis of the Phospholipid Profile of the Collection Strain PAO1 and Clinical Isolates of Pseudomonas aeruginosa in Relation to Their Attachment Capacity. International Journal of Molecular Sciences, 22(8), 4003. https://doi.org/10.3390/ijms22084003







