Marliolide Derivative Induces Melanosome Degradation via Nrf2/p62-Mediated Autophagy
Abstract
1. Introduction
2. Results
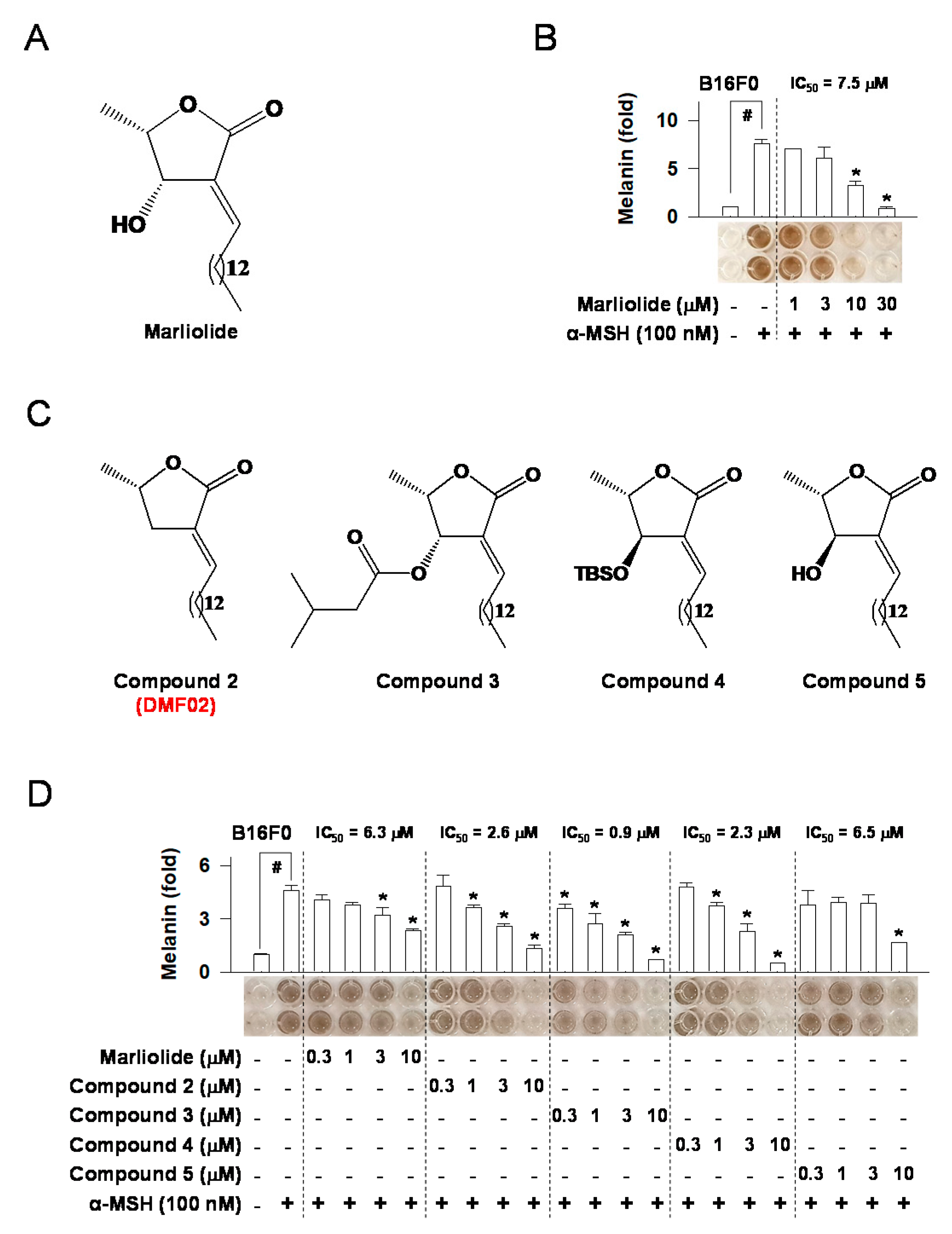
2.1. Marliolide or Marliolide Derivatives Reduced Melanin Products in B16F0 Melanoma Cells
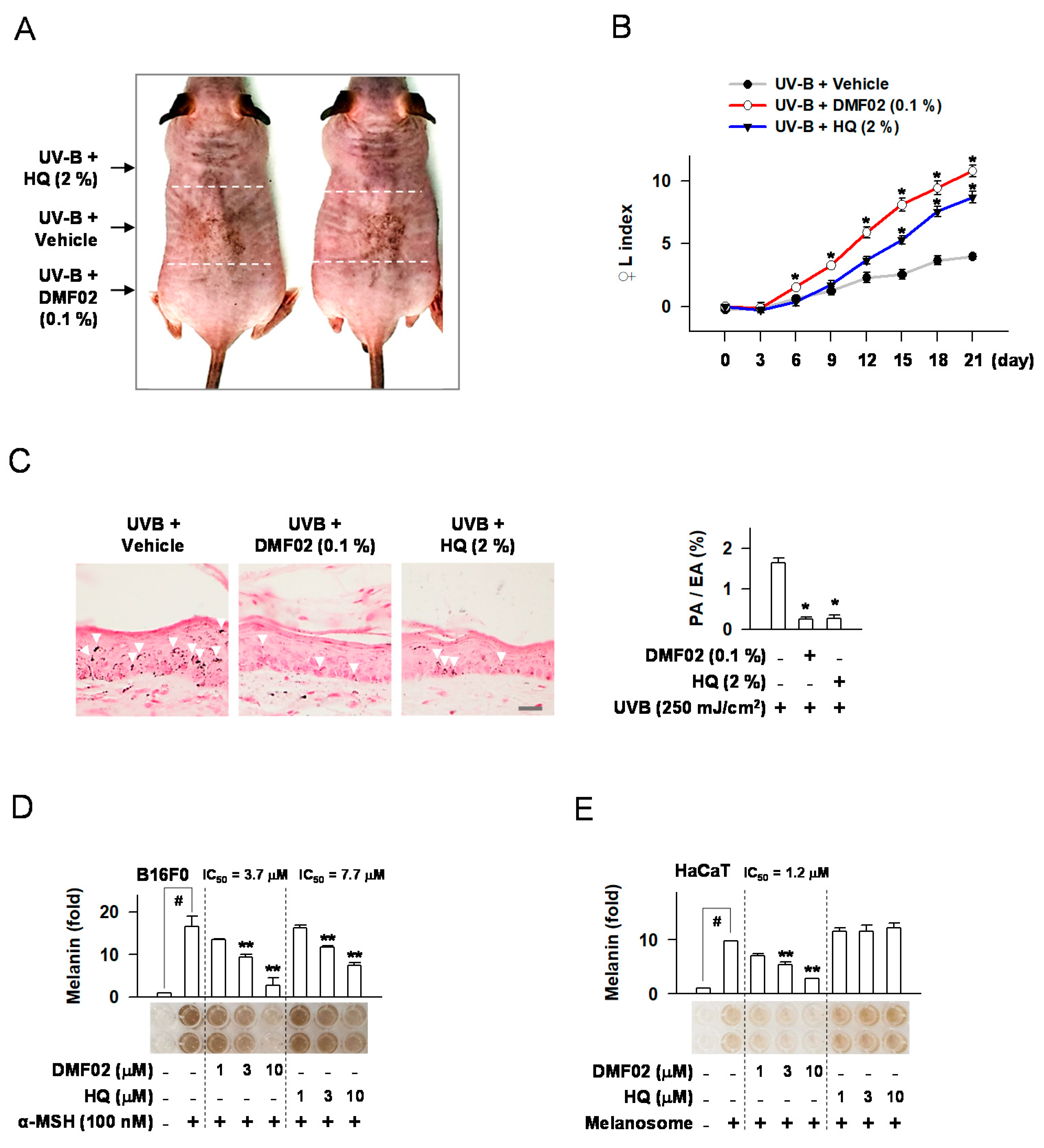
2.2. DMF02 Reduced UVB-Induced Pigmentation in HRM-2 Mice Skin
2.3. DMF02 Decreased Melanin Production in Melanocytes and Keratinocytes Containing Transferred Melanosomes
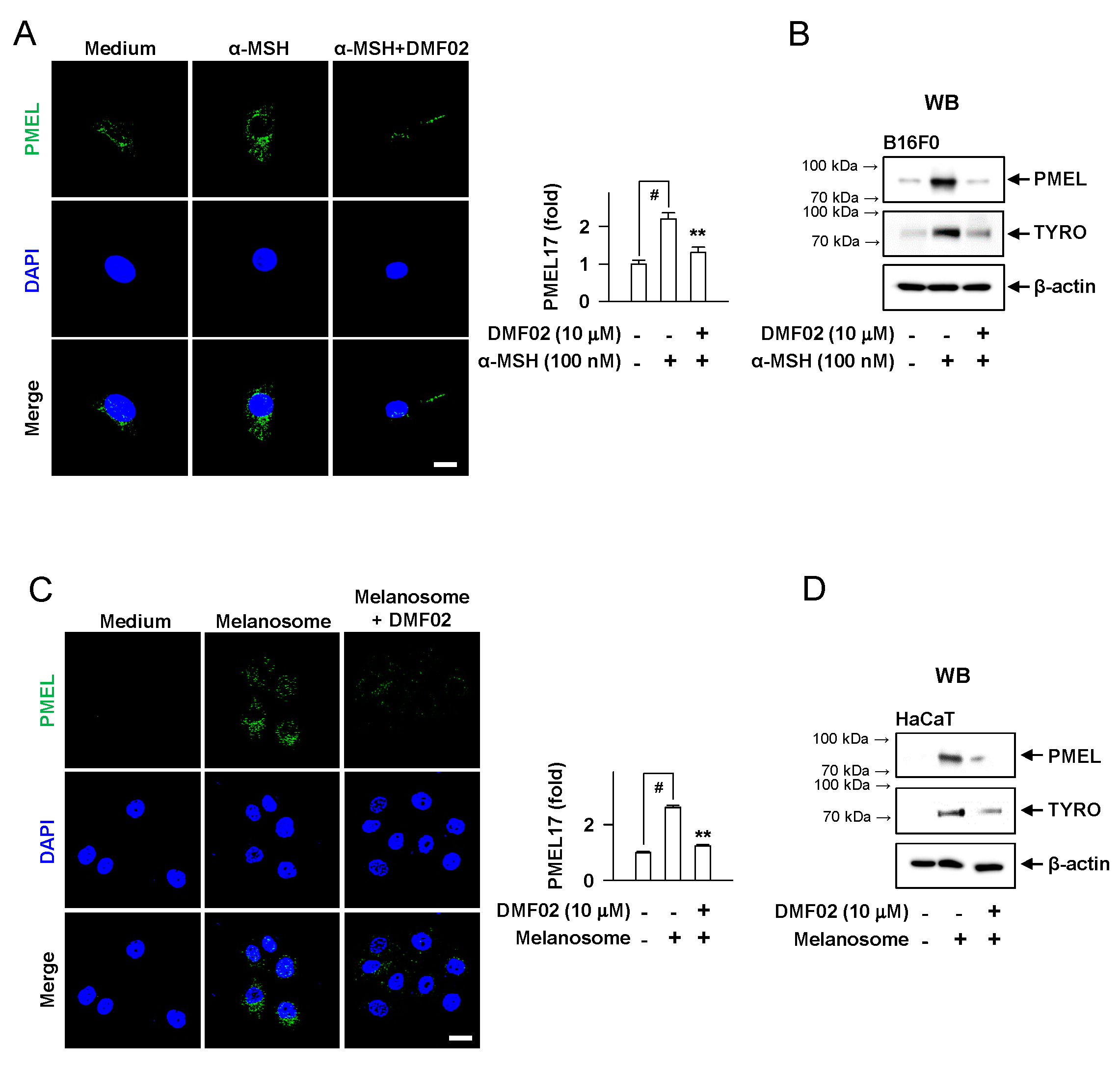
2.4. DMF02 Decreased Melanosome in Melanocytes and Keratinocytes
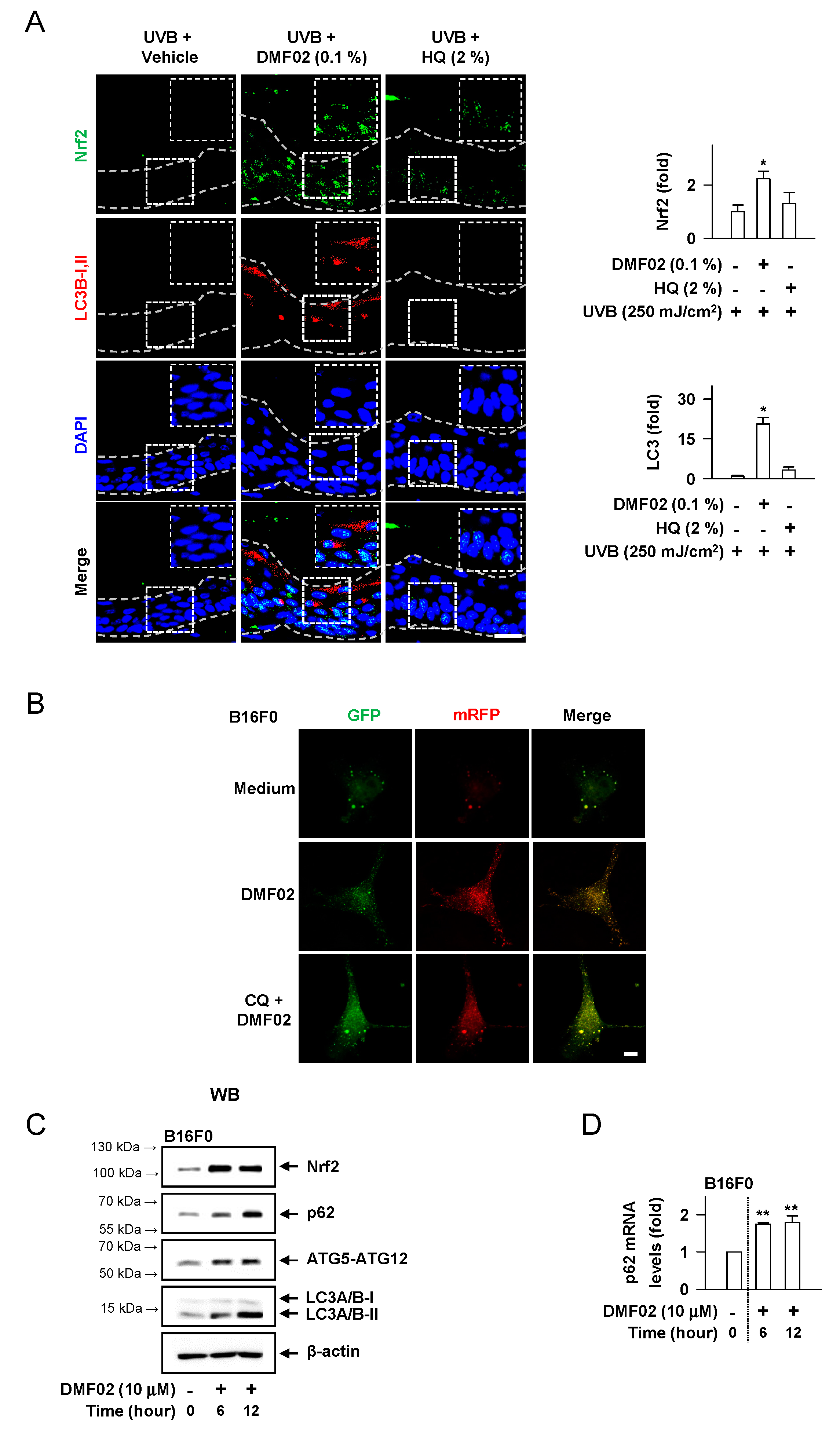
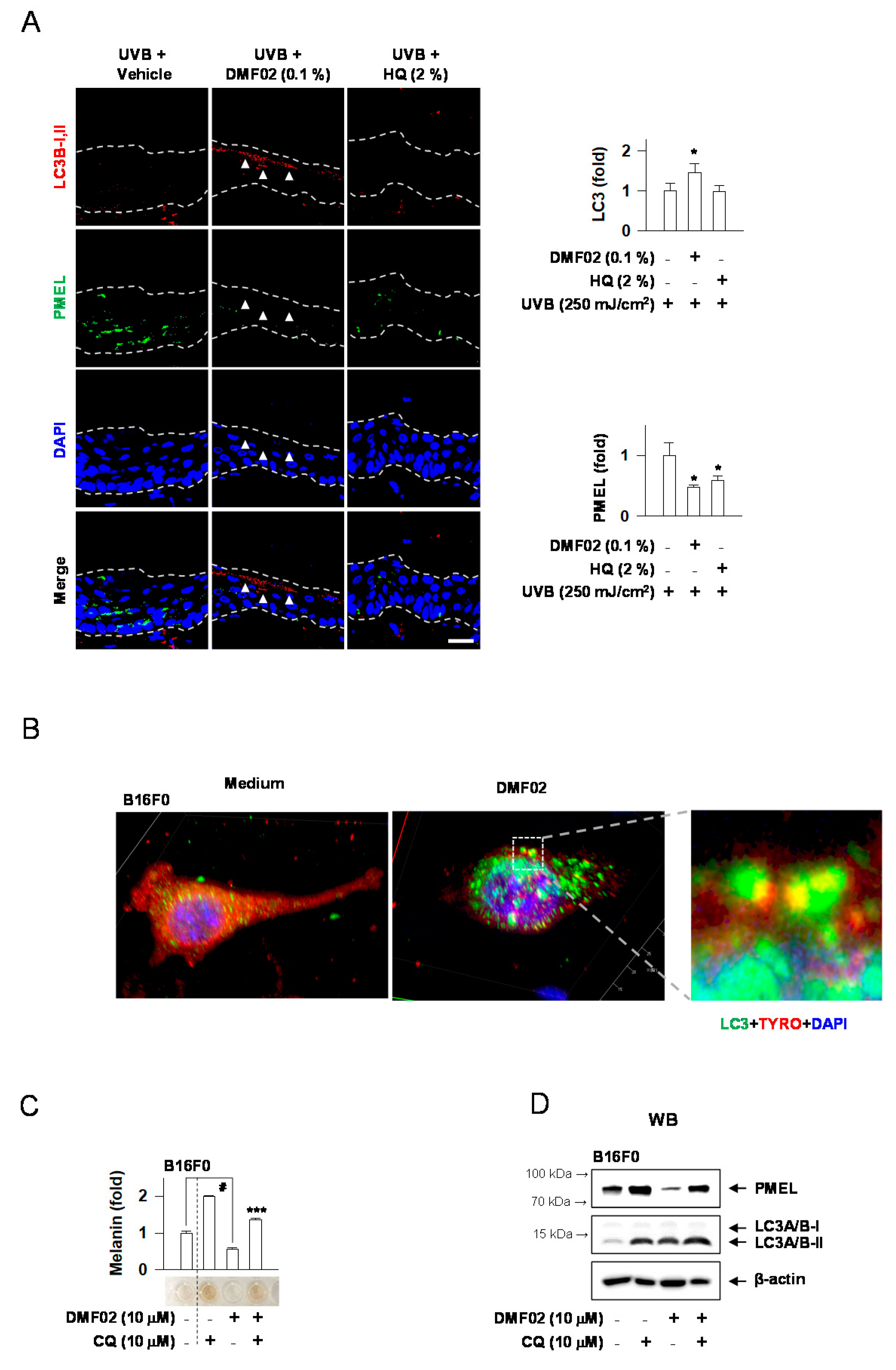
2.5. DMF02 Increased Autophagy in HRM-2 Mice Skin
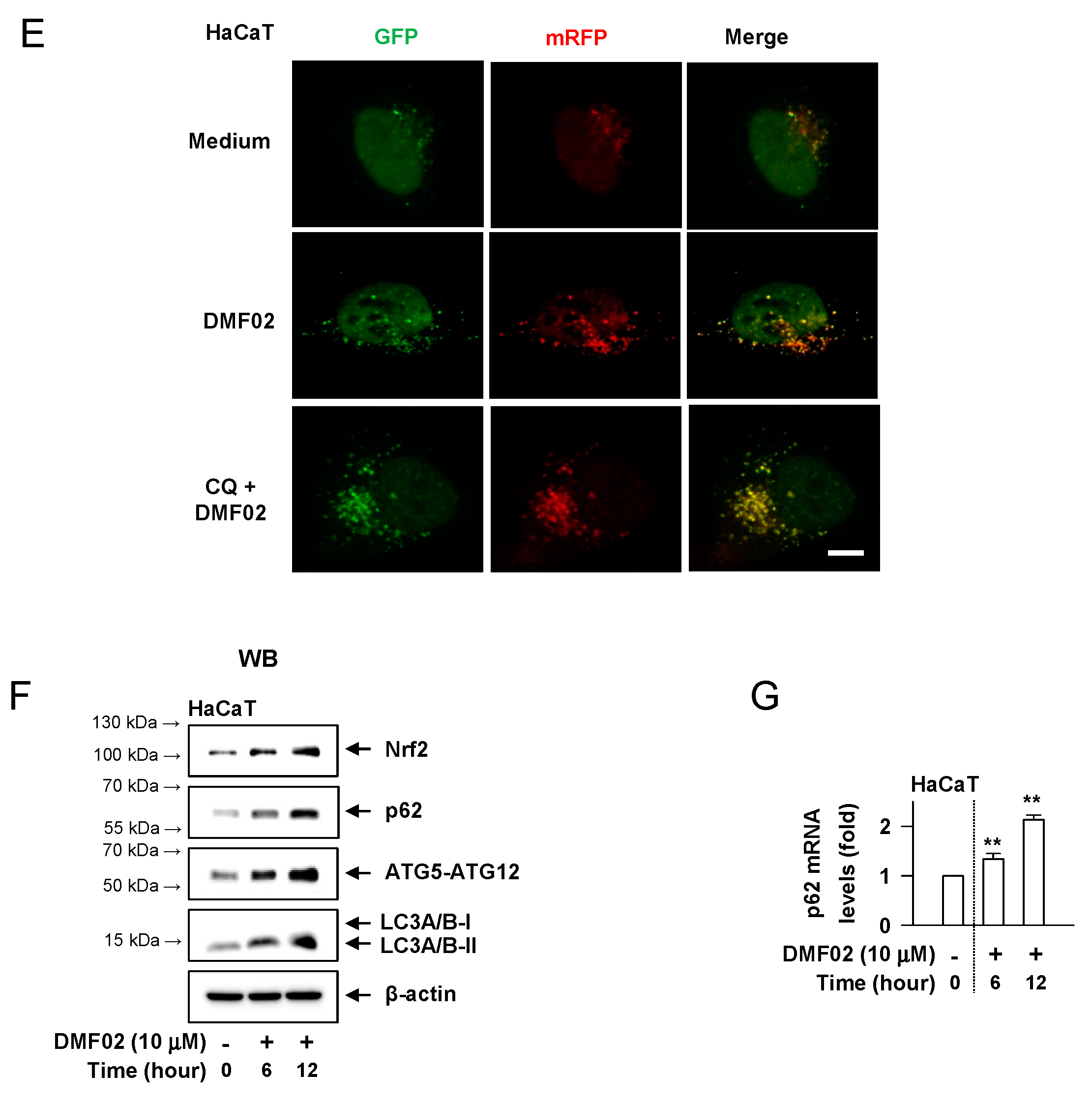
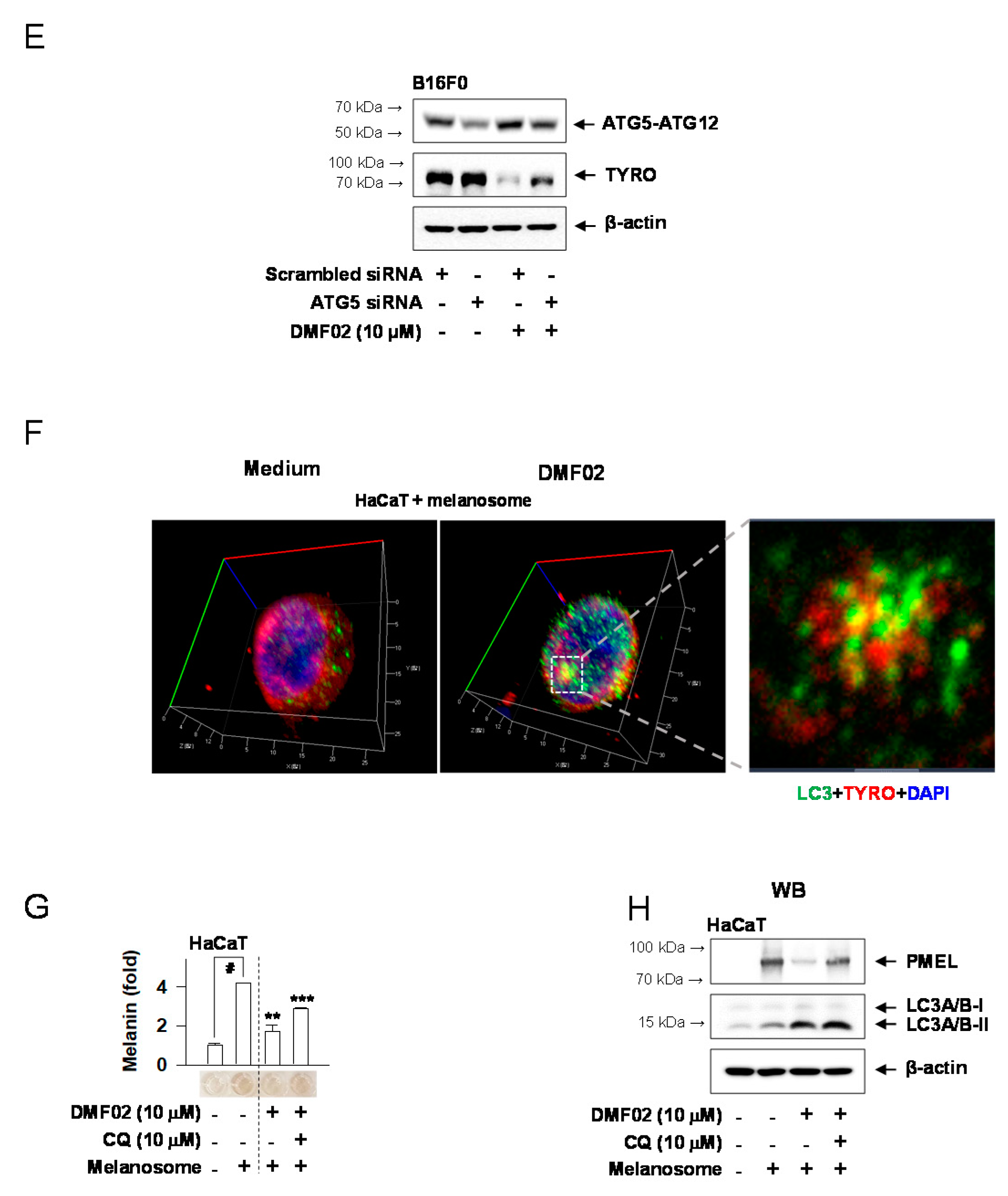
2.6. DMF02 Reduced Melanin-Containing Melanosome via Autophagy in Melanocytes and Keratinocytes
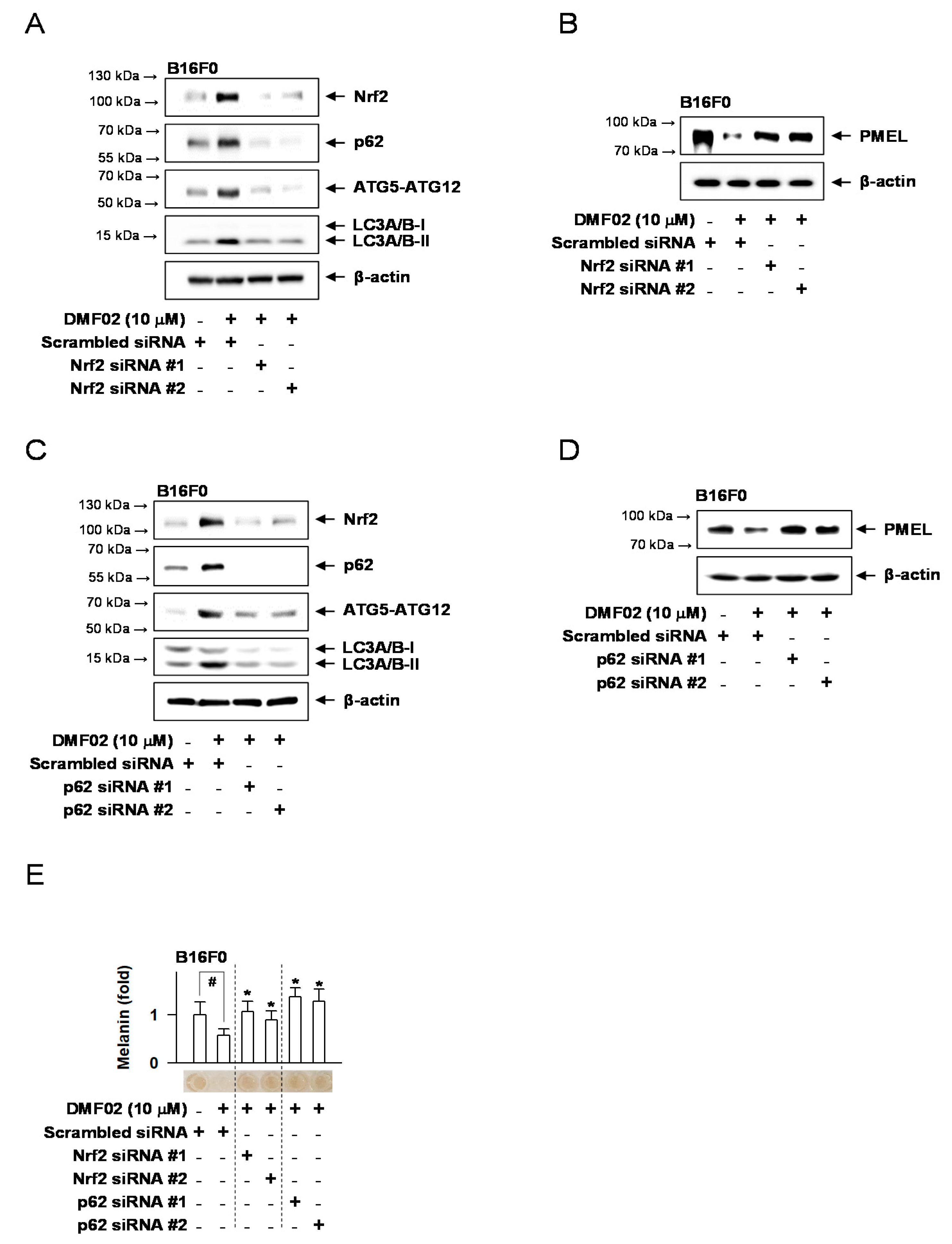
2.7. Nrf2 Knockdown Decreased DMF02-Induced Autophagy, Blocking Melanosome Degradation in Melanocytes
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Cell Culture
4.3. Melanin Quantification
4.4. UVB-Induced Skin Hyperpigmentation
4.5. Isolation of Melanosomes and Incubation of Isolated Melanosomes in HaCaT Cells
4.6. Transfection and Plasmids
4.7. Western Blot Analysis
4.8. Real-Time PCR Analysis
4.9. Small Interfering RNA Knockdown
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hearing, V.J. Determination of melanin synthetic pathways. J. Investig. Dermatol. 2011, 131, E8–E11. [Google Scholar] [CrossRef]
- Kondo, T.; Hearing, V.J. Update on the regulation of mammalian melanocyte function and skin pigmentation. Expert Rev. Dermatol. 2011, 6, 97–108. [Google Scholar] [CrossRef]
- Fu, C.; Chen, J.; Lu, J.; Yi, L.; Tong, X.; Kang, L.; Pei, S.; Ouyang, Y.; Jiang, L.; Ding, Y.; et al. Roles of inflammation factors in melanogenesis. Mol. Med. Rep. 2020, 21, 1421–1430. [Google Scholar] [CrossRef] [PubMed]
- D’Mello, S.A.; Finlay, G.J.; Baguley, B.C.; Askarian-Amiri, M.E. Signaling Pathways in Melanogenesis. Int. J. Mol. Sci. 2016, 17, 1144. [Google Scholar] [CrossRef]
- Brenner, M.; Hearing, V.J. The protective role of melanin against UV damage in human skin. Photochem. Photobiol. 2008, 84, 539–549. [Google Scholar] [CrossRef]
- Kurita, M.; Kato, H.; Yoshimura, K. A therapeutic strategy based on histological assessment of hyperpigmented skin lesions in Asians. J. Plast. Reconstr. Aesthet. Surg. 2009, 62, 955–963. [Google Scholar] [CrossRef]
- Galluzzi, L.; Pietrocola, F.; Bravo-San Pedro, J.M.; Amaravadi, R.K.; Baehrecke, E.H.; Cecconi, F.; Codogno, P.; Debnath, J.; Gewirtz, D.A.; Karantza, V.; et al. Autophagy in malignant transformation and cancer progression. EMBO J. 2015, 34, 856–880. [Google Scholar] [CrossRef]
- Levine, B.; Kroemer, G. Autophagy in the pathogenesis of disease. Cell 2008, 132, 27–42. [Google Scholar] [CrossRef]
- Yun, W.J.; Kim, E.Y.; Park, J.E.; Jo, S.Y.; Bang, S.H.; Chang, E.J.; Chang, S.E. Microtubule-associated protein light chain 3 is involved in melanogenesis via regulation of MITF expression in melanocytes. Sci. Rep. 2016, 6, 19914. [Google Scholar] [CrossRef] [PubMed]
- Murase, D.; Hachiya, A.; Takano, K.; Hicks, R.; Visscher, M.O.; Kitahara, T.; Hase, T.; Takema, Y.; Yoshimori, T. Autophagy has a significant role in determining skin color by regulating melanosome degradation in keratinocytes. J. Investig. Dermatol. 2013, 133, 2416–2424. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.F.; Gruber, F.; Ni, C.; Mildner, M.; Koenig, U.; Karner, S.; Barresi, C.; Rossiter, H.; Narzt, M.S.; Nagelreiter, I.M.; et al. Suppression of autophagy dysregulates the antioxidant response and causes premature senescence of melanocytes. J. Investig. Dermatol. 2015, 135, 1348–1357. [Google Scholar] [CrossRef]
- Ni, C.; Narzt, M.S.; Nagelreiter, I.M.; Zhang, C.F.; Larue, L.; Rossiter, H.; Grillari, J.; Tschachler, E.; Gruber, F. Autophagy deficient melanocytes display a senescence associated secretory phenotype that includes oxidized lipid mediators. Int. J. Biochem. Cell Biol. 2016, 81, 375–382. [Google Scholar] [CrossRef]
- Villeneuve, N.F.; Lau, A.; Zhang, D.D. Regulation of the Nrf2-Keap1 antioxidant response by the ubiquitin proteasome system: An insight into cullin-ring ubiquitin ligases. Antioxid. Redox Signal. 2010, 13, 1699–1712. [Google Scholar] [CrossRef]
- Ma, Q. Role of nrf2 in oxidative stress and toxicity. Annu. Rev. Pharmacol. Toxicol. 2013, 53, 401–426. [Google Scholar] [CrossRef] [PubMed]
- Stewart, D.; Killeen, E.; Naquin, R.; Alam, S.; Alam, J. Degradation of transcription factor Nrf2 via the ubiquitin-proteasome pathway and stabilization by cadmium. J. Biol. Chem. 2003, 278, 2396–2402. [Google Scholar] [CrossRef]
- Lee, J.; Kim, K.S.; Lee, H.G.; Park, C.H.; Ku, M.S.; Keum, Y.S. Skin Protective Effect of Methylated Marliolide through Induction of NRF2/ARE. J. Soc. Cosmet. Sci. Korea 2018, 44, 375–379. [Google Scholar]
- Reddy, N.M.; Potteti, H.R.; Vegiraju, S.; Chen, H.J.; Tamatam, C.M.; Reddy, S.P. PI3K-AKT Signaling via Nrf2 Protects against Hyperoxia-Induced Acute Lung Injury, but Promotes Inflammation Post-Injury Independent of Nrf2 in Mice. PLoS ONE 2015, 10, e0129676. [Google Scholar] [CrossRef]
- Zou, W.; Chen, C.; Zhong, Y.; An, J.; Zhang, X.; Yu, Y.; Yu, Z.; Fu, J. PI3K/Akt pathway mediates Nrf2/ARE activation in human L02 hepatocytes exposed to low-concentration HBCDs. Environ. Sci. Technol. 2013, 47, 12434–12440. [Google Scholar] [CrossRef]
- Jian, Z.; Li, K.; Liu, L.; Zhang, Y.; Zhou, Z.; Li, C.; Gao, T. Heme oxygenase-1 protects human melanocytes from H2O2-induced oxidative stress via the Nrf2-ARE pathway. J. Investig. Dermatol. 2011, 131, 1420–1427. [Google Scholar] [CrossRef] [PubMed]
- Jian, Z.; Li, K.; Song, P.; Zhu, G.; Zhu, L.; Cui, T.; Liu, B.; Tang, L.; Wang, X.; Wang, G.; et al. Impaired activation of the Nrf2-ARE signaling pathway undermines H2O2-induced oxidative stress response: A possible mechanism for melanocyte degeneration in vitiligo. J. Investig. Dermatol. 2014, 134, 2221–2230. [Google Scholar] [CrossRef]
- Chaiprasongsuk, A.; Onkoksoong, T.; Pluemsamran, T.; Limsaengurai, S.; Panich, U. Photoprotection by dietary phenolics against melanogenesis induced by UVA through Nrf2-dependent antioxidant responses. Redox Biol. 2016, 8, 79–90. [Google Scholar] [CrossRef] [PubMed]
- Park, S.H.; Choi, E.; Kim, S.; Kim, D.S.; Kim, J.H.; Chang, S.; Choi, J.S.; Park, K.J.; Roh, K.B.; Lee, J.; et al. Oxidative Stress-Protective and Anti-Melanogenic Effects of Loliolide and Ethanol Extract from Fresh Water Green Algae, Prasiola japonica. Int. J. Mol. Sci. 2018, 19, 2825. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.M.; Kim, M.Y.; Sohn, K.C.; Jung, S.Y.; Lee, H.E.; Lim, J.W.; Kim, S.; Lee, Y.H.; Im, M.; Seo, Y.J.; et al. Nrf2 negatively regulates melanogenesis by modulating PI3K/Akt signaling. PLoS ONE 2014, 9, e96035. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Mailar, K.; Yoo, O.K.; Choi, W.J.; Keum, Y.S. Marliolide inhibits skin carcinogenesis by activating NRF2/ARE to induce heme oxygenase-1. Eur. J. Med. Chem. 2018, 150, 113–126. [Google Scholar] [CrossRef] [PubMed]
- Bartolini, D.; Dallaglio, K.; Torquato, P.; Piroddi, M.; Galli, F. Nrf2-p62 autophagy pathway and its response to oxidative stress in hepatocellular carcinoma. Transl. Res. 2018, 193, 54–71. [Google Scholar] [CrossRef] [PubMed]
- Negi, C.K.; Jena, G. Nrf2, a novel molecular target to reduce type 1 diabetes associated secondary complications: The basic considerations. Eur. J. Pharmacol. 2019, 843, 12–26. [Google Scholar] [CrossRef]
- Bahn, G.; Park, J.S.; Yun, U.J.; Lee, Y.J.; Choi, Y.; Baek, S.H.; Choi, B.Y.; Cho, Y.S.; Kim, H.K.; Han, J.; et al. NRF2/ARE pathway negatively regulates BACE1 expression and ameliorates cognitive deficits in mouse Alzheimer’s models. Proc. Natl. Acad. Sci. USA 2019, 116, 12516–12523. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.M.; Chen, C.Y.; Wen, Z.H. Identifying melanogenesis inhibitors from Cinnamomum subavenium with in vitro and in vivo screening systems by targeting the human tyrosinase. Exp. Dermatol. 2011, 20, 242–248. [Google Scholar] [CrossRef]
- Kim, J.Y.; Kim, J.; Ahn, Y.; Lee, E.J.; Hwang, S.; Almurayshid, A.; Park, K.; Chung, H.J.; Kim, H.J.; Lee, S.H.; et al. Autophagy induction can regulate skin pigmentation by causing melanosome degradation in keratinocytes and melanocytes. Pigment Cell Melanoma Res. 2020, 33, 403–415. [Google Scholar] [CrossRef]
- Jain, A.; Lamark, T.; Sjottem, E.; Larsen, K.B.; Awuh, J.A.; Overvatn, A.; McMahon, M.; Hayes, J.D.; Johansen, T. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J. Biol. Chem. 2010, 285, 22576–22591. [Google Scholar] [CrossRef]
- Liu, W.J.; Ye, L.; Huang, W.F.; Guo, L.J.; Xu, Z.G.; Wu, H.L.; Yang, C.; Liu, H.F. p62 links the autophagy pathway and the ubiqutin-proteasome system upon ubiquitinated protein degradation. Cell. Mol. Biol. Lett. 2016, 21, 29. [Google Scholar] [CrossRef]
- Lippai, M.; Low, P. The role of the selective adaptor p62 and ubiquitin-like proteins in autophagy. Biomed. Res. Int. 2014, 2014, 832704. [Google Scholar] [CrossRef]
- Cabe, M.; Rademacher, D.J.; Karlsson, A.B.; Cherukuri, S.; Bakowska, J.C. PB1 and UBA domains of p62 are essential for aggresome-like induced structure formation. Biochem. Biophys. Res. Commun. 2018, 503, 2306–2311. [Google Scholar] [CrossRef]
- Ichimura, Y.; Waguri, S.; Sou, Y.S.; Kageyama, S.; Hasegawa, J.; Ishimura, R.; Saito, T.; Yang, Y.; Kouno, T.; Fukutomi, T.; et al. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol. Cell 2013, 51, 618–631. [Google Scholar] [CrossRef] [PubMed]
- Funderburk, F.; Wang, Q.J.; Yue, Z. The Beclin1-VPS34 complex—At the crossroads of autophagy and beyond. Trends Cell Biol. 2010, 20, 355–362. [Google Scholar] [CrossRef]
- Ho, H.; Kapadia, R.; Al-Tahan, S.; Ahmad, S.; Ganesan, A.K. WIPI1 coordinates melanogenic gene transcription and melanosome formation via TORC1 inhibition. J. Biol. Chem. 2011, 286, 12509–12523. [Google Scholar] [CrossRef]
- Wataya-Kaneda, M.; Tanaka, M.; Yang, L.; Yang, F.; Tsuruta, D.; Nakamura, A.; Matsumoto, S.; Hamasaki, T.; Tanemura, A.; Katayama, I. Clinical and Histologic Analysis of the Efficacy of Topical Rapamycin Therapy Against Hypomelanotic Macules in Tuberous Sclerosis Complex. JAMA Dermatol. 2015, 151, 722–730. [Google Scholar] [CrossRef]
- Kalie, E.; Razi, M.; Tooze, S.A. ULK1 regulates melanin levels in MNT-1 cells independently of mTORC1. PLoS ONE 2013, 8, e75313. [Google Scholar] [CrossRef] [PubMed]
- Ho, H.; Ganesan, A.K. The pleiotropic roles of autophagy regulators in melanogenesis. Pigment Cell Melanoma Res. 2011, 24, 594–604. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yun, C.-Y.; Choi, N.; Lee, J.U.; Lee, E.J.; Kim, J.Y.; Choi, W.J.; Oh, S.H.; Sung, J.-H. Marliolide Derivative Induces Melanosome Degradation via Nrf2/p62-Mediated Autophagy. Int. J. Mol. Sci. 2021, 22, 3995. https://doi.org/10.3390/ijms22083995
Yun C-Y, Choi N, Lee JU, Lee EJ, Kim JY, Choi WJ, Oh SH, Sung J-H. Marliolide Derivative Induces Melanosome Degradation via Nrf2/p62-Mediated Autophagy. International Journal of Molecular Sciences. 2021; 22(8):3995. https://doi.org/10.3390/ijms22083995
Chicago/Turabian StyleYun, Cheong-Yong, Nahyun Choi, Jae Un Lee, Eun Jung Lee, Ji Young Kim, Won Jun Choi, Sang Ho Oh, and Jong-Hyuk Sung. 2021. "Marliolide Derivative Induces Melanosome Degradation via Nrf2/p62-Mediated Autophagy" International Journal of Molecular Sciences 22, no. 8: 3995. https://doi.org/10.3390/ijms22083995
APA StyleYun, C.-Y., Choi, N., Lee, J. U., Lee, E. J., Kim, J. Y., Choi, W. J., Oh, S. H., & Sung, J.-H. (2021). Marliolide Derivative Induces Melanosome Degradation via Nrf2/p62-Mediated Autophagy. International Journal of Molecular Sciences, 22(8), 3995. https://doi.org/10.3390/ijms22083995





