Hemodynamic Forces Regulate Cardiac Regeneration-Responsive Enhancer Activity during Ventricle Regeneration
Abstract
1. Introduction
2. Results
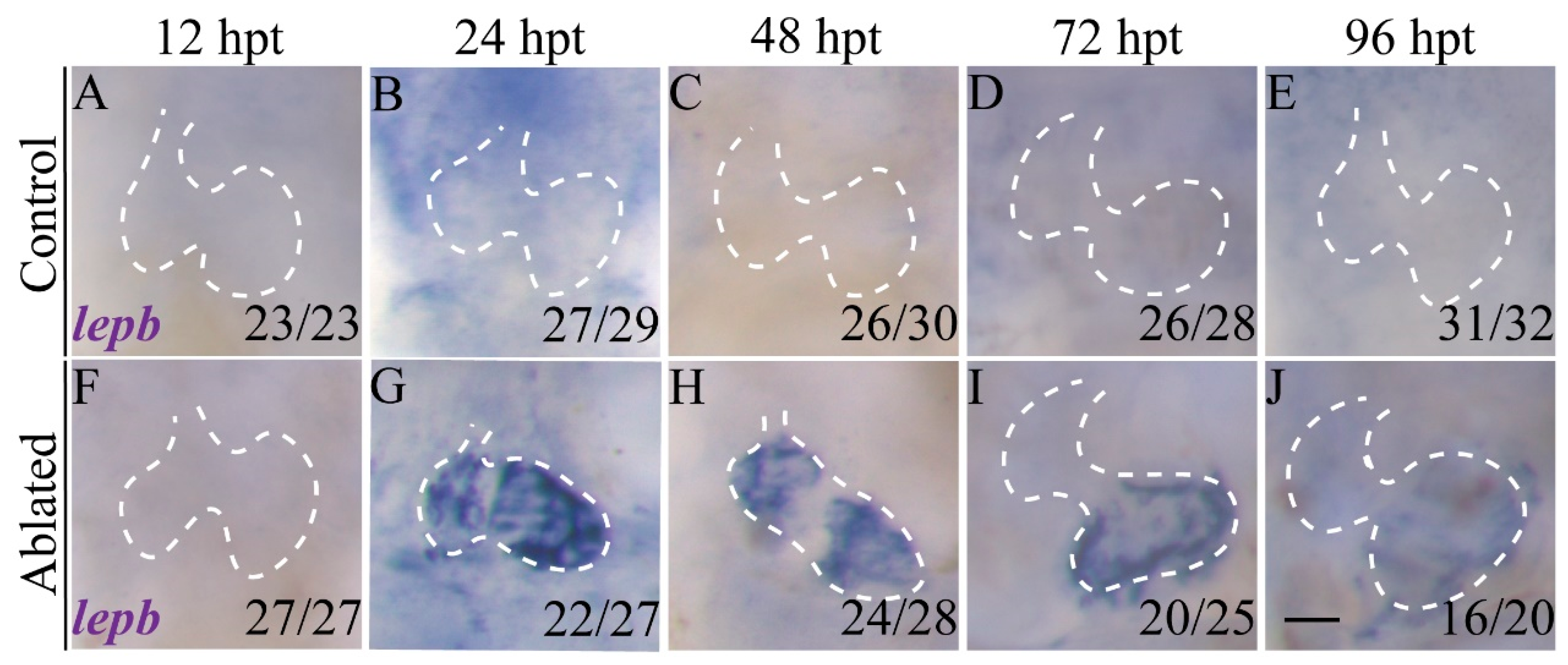
2.1. lepb Is Induced during Zebrafish Larval Ventricle Regeneration
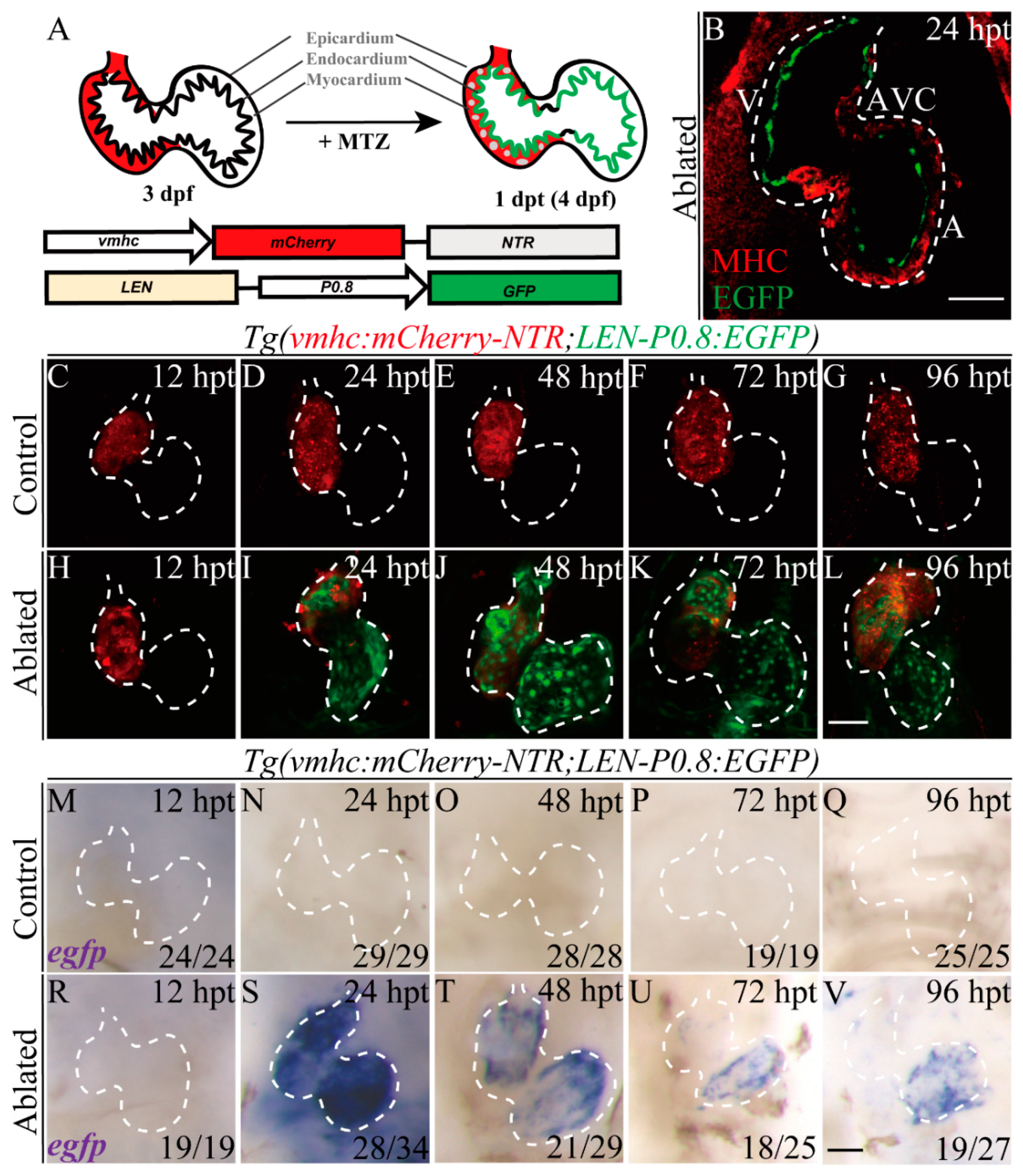
2.2. LEN Is Activated in the Endocardium during Ventricle Regeneration
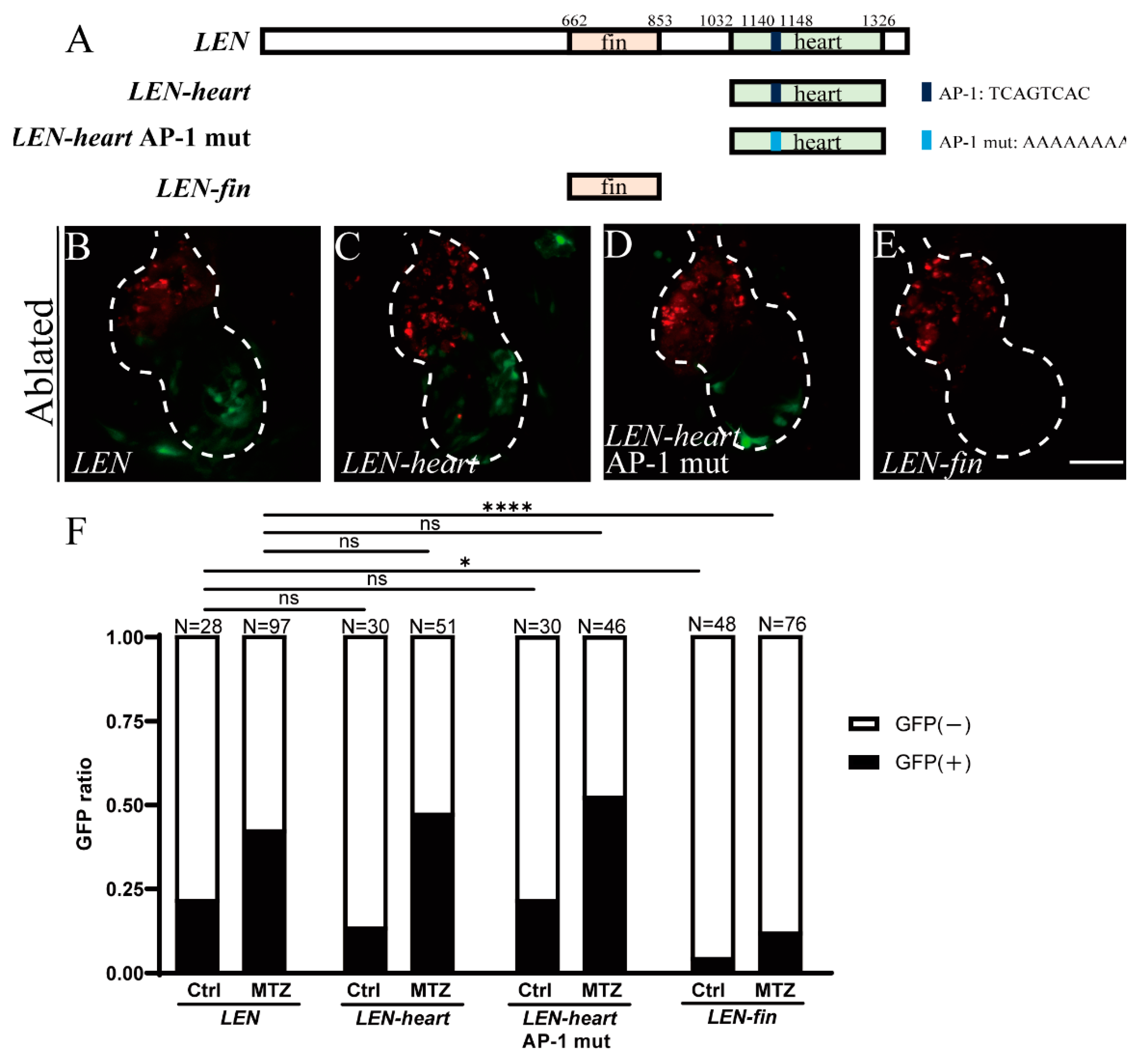
2.3. Mutation of Predicted AP-1 Binding Site Has No Effect on LEN Activity
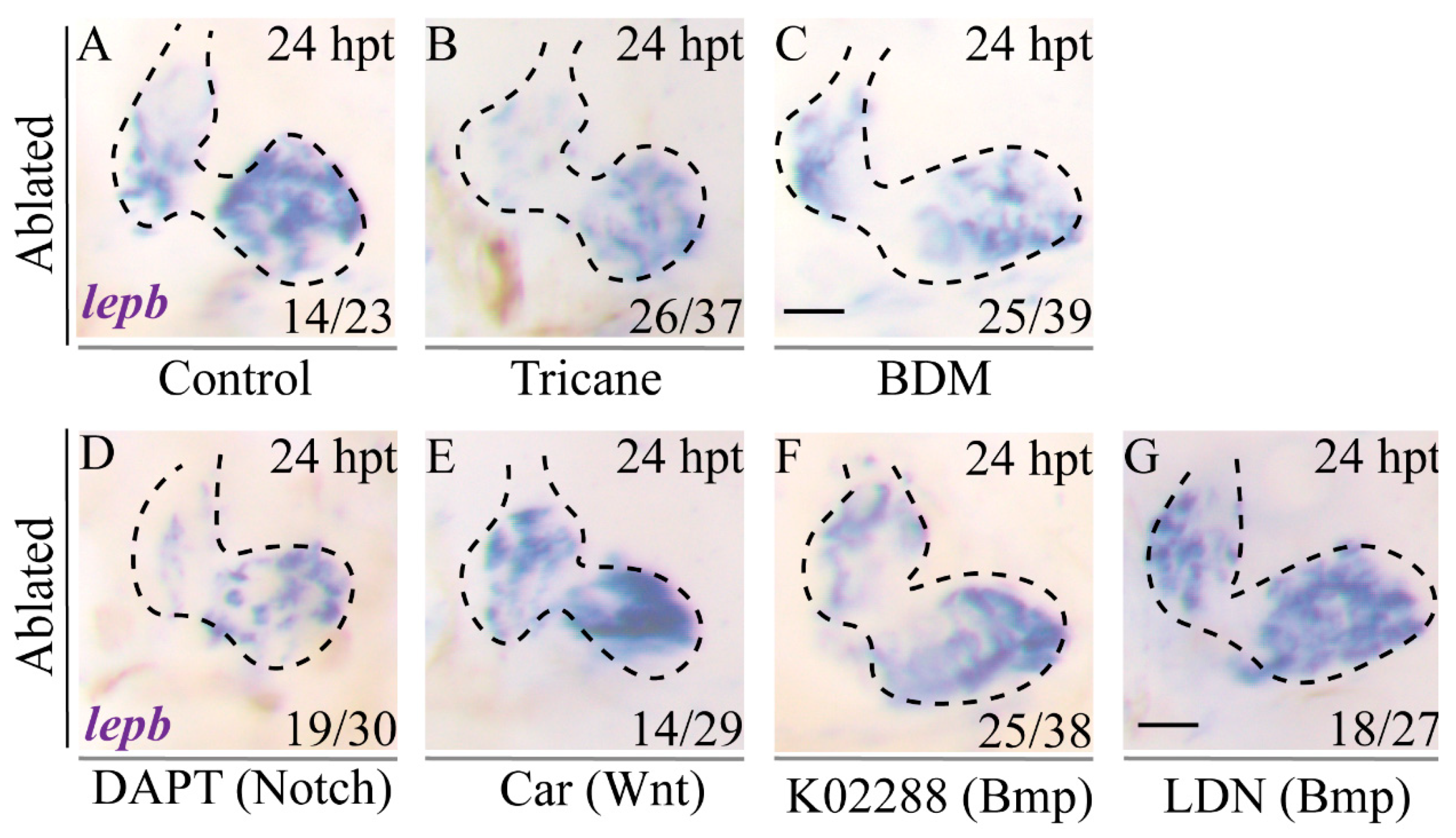
2.4. Activation of LEN Is Attenuated by Hemodynamic Force Alteration
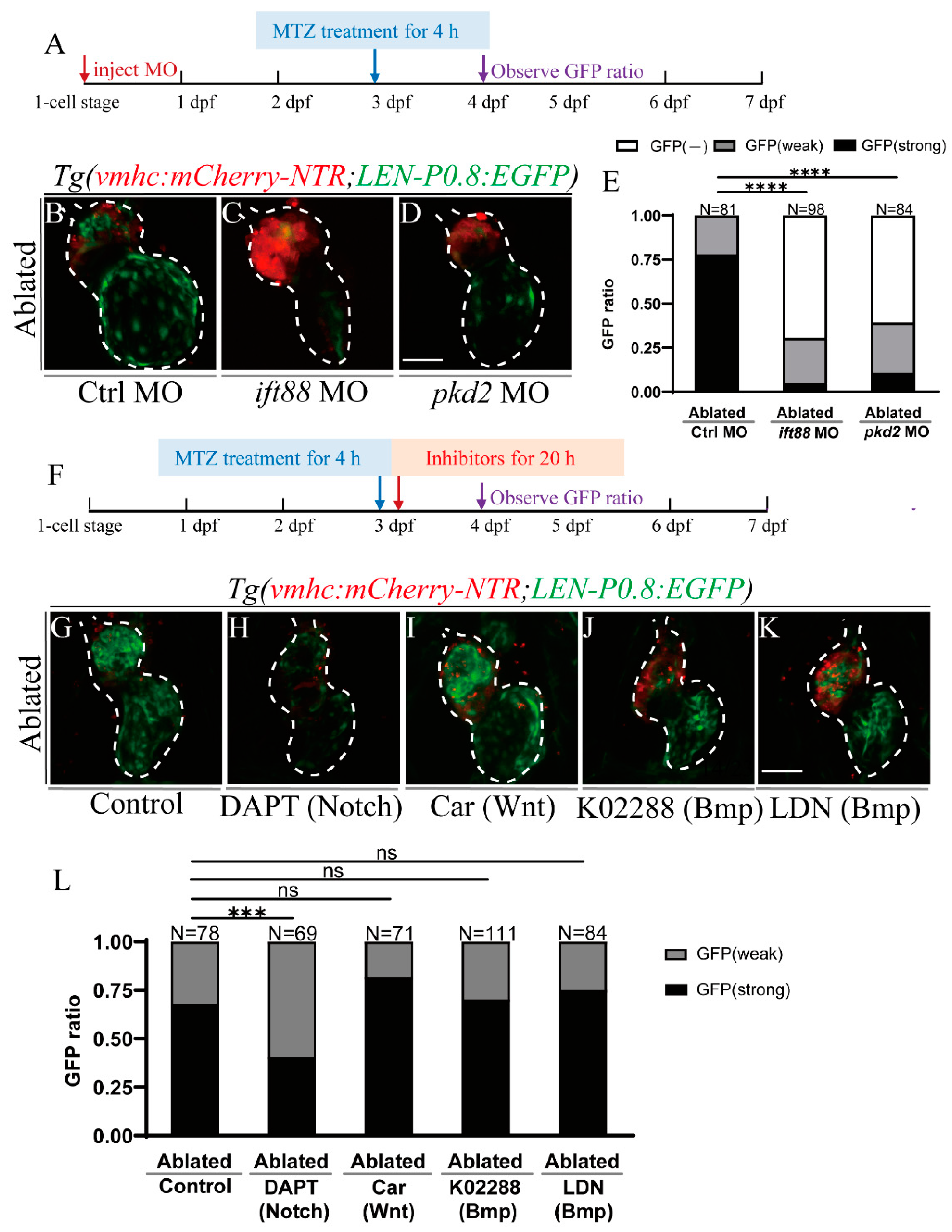
2.5. Primary Cilia and Mechanosensitive Ion Channel Affect LEN Activity
2.6. Notch Signaling Influences LEN Activity
2.7. Inhibition of Blood Flow and Notch Signaling Attenuates lepb Induction during Ventricle Regeneration
3. Discussion
4. Materials and Methods
4.1. Zebrafish Husbandry
4.2. Generation of Transgenic Zebrafish Lines
4.3. Ventricle Ablation
4.4. Morpholino Injection
4.5. Drug Treatment
4.6. In Situ Hybridization
4.7. Immunofluorescence
4.8. Quantification and Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Virani, S.S.; Alonso, A.; Benjamin, E.J.; Bittencourt, M.S.; Callaway, C.W.; Carson, A.P.; Chamberlain, A.M.; Chang, A.R.; Cheng, S.; Delling, F.N.; et al. Heart Disease and Stroke Statistics-2020 Update: A Report from the American Heart Association. Circulation 2020, 141, e139–e596. [Google Scholar] [CrossRef] [PubMed]
- Derks, W.; Bergmann, O. Polyploidy in Cardiomyocytes: Roadblock to Heart Regeneration? Circ. Res. 2020, 126, 552–565. [Google Scholar] [CrossRef]
- Thygesen, K.; Alpert, J.S.; Jaffe, A.S.; Simoons, M.L.; Chaitman, B.R.; White, H.D.; Katus, H.A.; Apple, F.S.; Lindahl, B.; Morrow, D.A.; et al. Third universal definition of myocardial infarction. J. Am. Coll. Cardiol. 2012, 60, 1581–1598. [Google Scholar] [CrossRef]
- Poss, K.D.; Wilson, L.C.; Keating, M.T. Heart Regeneration in Zebrafish. Science 2002, 298, 2188–2190. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Han, P.; Yang, H.; Ouyang, K.; Lee, D.; Lin, Y.F.; Ocorr, K.; Kang, G.; Chen, J.; Stainier, D.Y.; et al. In vivo cardiac reprogramming contributes to zebrafish heart regeneration. Nature 2013, 498, 497–501. [Google Scholar] [CrossRef]
- Gonzalez-Rosa, J.M.; Martin, V.; Peralta, M.; Torres, M.; Mercader, N. Extensive scar formation and regression during heart regeneration after cryoinjury in zebrafish. Development 2011, 138, 1663–1674. [Google Scholar] [CrossRef]
- González-Rosa, J.M.; Burns, C.E.; Burns, C.G. Zebrafish heart regeneration: 15 years of discoveries. Regeneration 2017, 4, 105–123. [Google Scholar] [CrossRef]
- Chablais, F.; Jazwinska, A. The regenerative capacity of the zebrafish heart is dependent on TGF signaling. Development 2012, 139, 1921–1930. [Google Scholar] [CrossRef]
- Lien, C.L.; Schebesta, M.; Makino, S.; Weber, G.J.; Keating, M.T. Gene expression analysis of zebrafish heart regeneration. PLoS Biol. 2006, 4, e260. [Google Scholar] [CrossRef]
- Choi, W.Y.; Gemberling, M.; Wang, J.; Holdway, J.E.; Shen, M.C.; Karlstrom, R.O.; Poss, K.D. In vivo monitoring of cardiomyocyte proliferation to identify chemical modifiers of heart regeneration. Development 2013, 140, 660–666. [Google Scholar] [CrossRef] [PubMed]
- Itou, J.; Oishi, I.; Kawakami, H.; Glass, T.J.; Richter, J.; Johnson, A.; Lund, T.C.; Kawakami, Y. Migration of cardiomyocytes is essential for heart regeneration in zebrafish. Development 2012, 139, 4133–4142. [Google Scholar] [CrossRef]
- Fernandez, C.E.; Bakovic, M.; Karra, R. Endothelial Contributions to Zebrafish Heart Regeneration. J. Cardiovasc. Dev. Dis. 2018, 5, 56. [Google Scholar] [CrossRef]
- Shawber, C.J.; Kitajewski, J. Notch function in the vasculature: Insights from zebrafish, mouse and man. Bioessays 2004, 26, 225–234. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Sinha, T.; Wynshaw-Boris, A. Wnt signaling in mammalian development: Lessons from mouse genetics. Cold Spring Harb. Perspect. Biol. 2012, 4, a007963. [Google Scholar] [CrossRef] [PubMed]
- Zinski, J.; Tajer, B.; Mullins, M.C. TGF-β Family Signaling in Early Vertebrate Development. Cold Spring Harb. Perspect. Biol. 2018, 10, a033274. [Google Scholar] [CrossRef]
- Lowery, J.W.; De Caestecker, M.P. BMP signaling in vascular development and disease. Cytokine Growth Factor Rev. 2010, 21, 287–298. [Google Scholar] [CrossRef] [PubMed]
- Ong, C.T.; Corces, V.G. Enhancer function: New insights into the regulation of tissue-specific gene expression. Nat. Rev. Genet. 2011, 12, 283–293. [Google Scholar] [CrossRef]
- Bulger, M.; Groudine, M. Functional and mechanistic diversity of distal transcription enhancers. Cell 2011, 144, 327–339. [Google Scholar] [CrossRef]
- Long, H.K.; Prescott, S.L.; Wysocka, J. Ever-Changing Landscapes: Transcriptional Enhancers in Development and Evolution. Cell 2016, 167, 1170–1187. [Google Scholar] [CrossRef]
- Benveniste, D.; Sonntag, H.J.; Sanguinetti, G.; Sproul, D. Transcription factor binding predicts histone modifications in human cell lines. Proc. Natl. Acad. Sci. USA 2014, 111, 13367–13372. [Google Scholar] [CrossRef] [PubMed]
- Lettice, L.A.; Heaney, S.J.; Purdie, L.A.; Li, L.; De Beer, P.; Oostra, B.A.; Goode, D.; Elgar, G.; Hill, R.E.; De Graaff, E. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum. Mol. Genet. 2003, 12, 1725–1735. [Google Scholar] [CrossRef]
- Visel, A.; Rubin, E.M.; Pennacchio, L.A.J.N. Genomic views of distant-acting enhancers. Nature 2009, 461, 199–205. [Google Scholar] [CrossRef] [PubMed]
- Tippens, N.D.; Vihervaara, A.; Lis, J.T. Enhancer transcription: What, where, when and why? Genes Dev. 2018, 32, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Shlyueva, D.; Stampfel, G.; Stark, A. Transcriptional enhancers: From properties to genome-wide predictions. Nat. Rev. Genet. 2014, 15, 272–286. [Google Scholar] [CrossRef]
- Andersson, R.; Sandelin, A. Determinants of enhancer and promoter activities of regulatory elements. Nat. Rev. Genet. 2020, 21, 71–87. [Google Scholar] [CrossRef]
- Buenrostro, J.D.; Giresi, P.G.; Zaba, L.C.; Chang, H.Y.; Greenleaf, W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 2013, 10, 1213–1218. [Google Scholar] [CrossRef]
- Goldberg, A.D.; Banaszynski, L.A.; Noh, K.M.; Lewis, P.W.; Elsaesser, S.J.; Stadler, S.; Dewell, S.; Law, M.; Guo, X.; Li, X.; et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 2010, 140, 678–691. [Google Scholar] [CrossRef]
- Visel, A.; Prabhakar, S.; Akiyama, J.A.; Shoukry, M.; Lewis, K.D.; Holt, A.; Plajzer-Frick, I.; Afzal, V.; Rubin, E.M.; Pennacchio, L.A. Ultraconservation identifies a small subset of extremely constrained developmental enhancers. Nat. Genet. 2008, 40, 158–160. [Google Scholar] [CrossRef]
- Kvon, E.Z. Using transgenic reporter assays to functionally characterize enhancers in animals. Genomics 2015, 106, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Navratilova, P.; Fredman, D.; Hawkins, T.A.; Turner, K.; Lenhard, B.; Becker, T.S. Systematic human/zebrafish comparative identification of cis-regulatory activity around vertebrate developmental transcription factor genes. Dev. Biol. 2009, 327, 526–540. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Hu, J.; Karra, R.; Dickson, A.L.; Tornini, V.A.; Nachtrab, G.; Gemberling, M.; Goldman, J.A.; Black, B.L.; Poss, K.D. Modulation of tissue repair by regeneration enhancer elements. Nature 2016, 532, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Pfefferli, C.; Jazwinska, A. The careg element reveals a common regulation of regeneration in the zebrafish myocardium and fin. Nat. Commun. 2017, 8, 15151. [Google Scholar] [CrossRef] [PubMed]
- Hung, H.A.; Sun, G.; Keles, S.; Svaren, J. Dynamic regulation of Schwann cell enhancers after peripheral nerve injury. J. Biol. Chem. 2015, 290, 6937–6950. [Google Scholar] [CrossRef]
- Gehrke, A.R.; Neverett, E.; Luo, Y.J.; Brandt, A.; Ricci, L.; Hulett, R.E.; Gompers, A.; Ruby, J.G.; Rokhsar, D.S.; Reddien, P.W.; et al. Acoel genome reveals the regulatory landscape of whole-body regeneration. Science 2019, 363, eaau6173. [Google Scholar] [CrossRef]
- Goldman, J.A.; Kuzu, G.; Lee, N.; Karasik, J.; Gemberling, M.; Foglia, M.J.; Karra, R.; Dickson, A.L.; Sun, F.; Tolstorukov, M.Y.; et al. Resolving Heart Regeneration by Replacement Histone Profiling. Dev. Cell 2017, 40, 392–404. [Google Scholar] [CrossRef]
- Harris, R.E.; Setiawan, L.; Saul, J.; Hariharan, I.K. Localized epigenetic silencing of a damage-activated WNT enhancer limits regeneration in mature Drosophila imaginal discs. Elife 2016, 5, e11588. [Google Scholar] [CrossRef]
- Guenther, C.A.; Wang, Z.; Li, E.; Tran, M.C.; Logan, C.Y.; Nusse, R.; Pantalena-Filho, L.; Yang, G.P.; Kingsley, D.M. A distinct regulatory region of the Bmp5 locus activates gene expression following adult bone fracture or soft tissue injury. Bone 2015, 77, 31–41. [Google Scholar] [CrossRef]
- Suzuki, N.; Hirano, K.; Ogino, H.; Ochi, H. Arid3a regulates nephric tubule regeneration via evolutionarily conserved regeneration signal-response enhancers. Elife 2019, 8, e43186. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Hu, C.K.; Zeng, A.; Alegre, D.; Hu, D.; Gotting, K.; Ortega Granillo, A.; Wang, Y.; Robb, S.; Schnittker, R.; et al. Changes in regeneration-responsive enhancers shape regenerative capacities in vertebrates. Science 2020, 369, eaaz3090. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, A.M.; Kang, J. Regeneration enhancers: Starting a journey to unravel regulatory events in tissue regeneration. Semin. Cell Dev. Biol. 2020, 97, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Goldman, J.A.; Poss, K.D. Gene regulatory programmes of tissue regeneration. Nat. Rev. Genet. 2020, 21, 511–525. [Google Scholar] [CrossRef]
- Beisaw, A.; Kuenne, C.; Stefan, G.; Dallmann, J.; Stainier, D.Y.R. AP-1 Contributes to Chromatin Accessibility to Promote Sarcomere Disassembly and Cardiomyocyte Protrusion During Zebrafish Heart Regeneration. Circ. Res. 2020, 126, 1760–1778. [Google Scholar] [CrossRef]
- Li, X.; Lu, Q.; Peng, Y.; Geng, F.; Shao, X.; Zhou, H.; Cao, Y.; Zhang, R. Primary cilia mediate Klf2-dependant Notch activation in regenerating heart. Protein Cell 2020, 11, 433–445. [Google Scholar] [CrossRef] [PubMed]
- Gálvez-Santisteban, M.; Chen, D.; Zhang, R.; Serrano, R.; Nguyen, C.; Zhao, L.; Nerb, L.; Masutani, E.M.; Vermot, J.; Burns, C.G.; et al. Hemodynamic-mediated endocardial signaling controls in vivo myocardial reprogramming. Elife 2019, 8, e44816. [Google Scholar] [CrossRef] [PubMed]
- Chen, J. Impaired cardiovascular function caused by different stressors elicits a common pathological and transcriptional response in zebrafish embryos. Zebrafish 2013, 10, 389–400. [Google Scholar] [CrossRef]
- Hsu, J.J.; Vedula, V.; Baek, K.I.; Chen, C.; Chen, J.; Chou, M.I.; Lam, J.; Subhedar, S.; Wang, J.; Ding, Y.; et al. Contractile and hemodynamic forces coordinate Notch1b-mediated outflow tract valve formation. JCI Insight 2019, 4, e124460. [Google Scholar] [CrossRef] [PubMed]
- Nauli, S.M.; Kawanabe, Y.; Kaminski, J.J.; Pearce, W.J.; Ingber, D.E.; Zhou, J. Endothelial cilia are fluid shear sensors that regulate calcium signaling and nitric oxide production through polycystin-1. Circulation 2008, 117, 1161–1171. [Google Scholar] [CrossRef] [PubMed]
- Delaval, B.; Bright, A.; Lawson, N.D.; Doxsey, S. The cilia protein IFT88 is required for spindle orientation in mitosis. Nat. Cell Biol. 2011, 13, 461–468. [Google Scholar] [CrossRef]
- Obara, T.; Mangos, S.; Liu, Y.; Zhao, J.; Wiessner, S.; Kramer-Zucker, A.G.; Olale, F.; Schier, A.F.; Drummond, I.A. Polycystin-2 immunolocalization and function in zebrafish. J. Am. Soc. Nephrol. 2006, 17, 2706–2718. [Google Scholar] [CrossRef]
- Zhao, L.; Ben-Yair, R.; Burns, C.E.; Burns, C.G. Endocardial Notch Signaling Promotes Cardiomyocyte Proliferation in the Regenerating Zebrafish Heart through Wnt Pathway Antagonism. Cell Rep. 2019, 26, 546–554. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.; Fu, W.; Yu, G.; Hu, X.; Lai, K.S.; Peng, X.; Zhou, Y.; Zhu, X.; Christov, P.; Sawyer, L.; et al. Discovering small molecules as Wnt inhibitors that promote heart regeneration and injury repair. J. Mol. Cell Biol. 2020, 12, 42–54. [Google Scholar] [CrossRef]
- Wu, C.C.; Kruse, F.; Vasudevarao, M.D.; Junker, J.P.; Zebrowski, D.C.; Fischer, K.; Nol, E.S.; Grün, D.; Berezikov, E.; Engel, F.B. Spatially Resolved Genome-wide Transcriptional Profiling Identifies BMP Signaling as Essential Regulator of Zebrafish Cardiomyocyte Regeneration. Dev. Cell 2016, 36, 36–49. [Google Scholar] [CrossRef]
- Bochenek, M.L.; Leidinger, C.; Rosinus, N.S.; Gogiraju, R.; Schfer, K. Activated Endothelial TGFβ1 Signaling Promotes Venous Thrombus Non-Resolution in Mice Via Endothelin-1: Potential Role for Chronic Thromboembolic Pulmonary Hypertension. Circ. Res. 2019, 126, 162–181. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Zeng, Y.N.; Zhang, K.; Zhao, Y.; Wu, Y.Y.; Li, G.; Cheng, H.Y.; Zhang, M.; Lai, F.; Wang, J.B. Polydatin Increases Radiosensitivity by Inducing Apoptosis of Stem Cells in Colorectal Cancer. Int. J. Biol. Sci. 2019, 15, 430–440. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Duan, K.; Ai, Z.; Niu, B.; Chen, Y.; Kong, R.; Li, T. Generation of cortical neurons through large-scale expanding neuroepithelial stem cell from human pluripotent stem cells. Stem Cell Res. Ther. 2020, 11, 431. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, K.; Holdway, J.E.; Major, R.J.; Blum, N.; Dahn, R.D.; Begemann, G.; Poss, K.D. Retinoic acid production by endocardium and epicardium is an injury response essential for zebrafish heart regeneration. Dev. Cell 2011, 20, 397–404. [Google Scholar] [CrossRef]
- Goishi, K.; Lee, P.; Davidson, A.J.; Nishi, E.; Zon, L.I.; Klagsbrun, M. Inhibition of zebrafish epidermal growth factor receptor activity results in cardiovascular defects. Mech. Dev. 2003, 120, 811–822. [Google Scholar] [CrossRef]
- Yang, K.; Kang, J. Tissue Regeneration Enhancer Elements: A Way to Unlock Endogenous Healing Power. Dev. Dyn. 2019, 248, 34–42. [Google Scholar] [CrossRef]
- Suzuki, N.; Ochi, H. Regeneration enhancers: A clue to reactivation of developmental genes. Dev. Growth Differ. 2020, 62, 343–354. [Google Scholar] [CrossRef]
- Andrés-Delgado, L.; Mercader, N. Interplay between cardiac function and heart development. Biochim. Biophys. Acta 2016, 1863, 1707–1716. [Google Scholar] [CrossRef] [PubMed]
- Goddard, L.M.; Duchemin, A.L.; Ramalingan, H.; Wu, B.; Kahn, M.L. Hemodynamic Forces Sculpt Developing Heart Valves through a KLF2-WNT9B Paracrine Signaling Axis. Dev. Cell 2017, 43, 274. [Google Scholar] [CrossRef] [PubMed]
- Peralta, M.; Ortiz Lopez, L.; Jerabkova, K.; Lucchesi, T.; Vitre, B.; Han, D.; Guillemot, L.; Dingare, C.; Sumara, I.; Mercader, N.; et al. Intraflagellar Transport Complex B Proteins Regulate the Hippo Effector Yap1 during Cardiogenesis. Cell Rep. 2020, 32, 107932. [Google Scholar] [CrossRef] [PubMed]
- Paavola, J.; Schliffke, S.; Rossetti, S.; Kuo, I.Y.; Yuan, S.; Sun, Z.; Harris, P.C.; Torres, V.E.; Ehrlich, B.E. Polycystin-2 mutations lead to impaired calcium cycling in the heart and predispose to dilated cardiomyopathy. J. Mol. Cell Cardiol. 2013, 58, 199–208. [Google Scholar] [CrossRef]
- Westerfield, M. The Zebrafish Book: A Quide for the Laboratory Use of Zebrafish Danio (Brachydanio) Rerio; University of Oregon Press: Eugene, OR, USA, 1994. [Google Scholar]
- Zhang, R.; Xu, X. Transient and transgenic analysis of the zebrafish ventricular myosin heavy chain (vmhc) promoter: An inhibitory mechanism of ventricle-specific gene expression. Dev. Dyn. 2009, 238, 1564–1573. [Google Scholar] [CrossRef] [PubMed]
- Samsa, L.A.; Givens, C.; Tzima, E.; Stainier, D.Y.; Qian, L.; Liu, J. Cardiac contraction activates endocardial Notch signaling to modulate chamber maturation in zebrafish. Development 2015, 142, 4080–4091. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Xu, W.; Jing, Y.; Wu, M.; Song, S.; Cao, Y.; Mei, C. Yes-Associated Protein (Yap) Is Necessary for Ciliogenesis and Morphogenesis during Pronephros Development in Zebrafish (Danio Rerio). Int. J. Biol. Sci. 2015, 11, 935–947. [Google Scholar] [CrossRef]
- Schottenfeld, J.; Sullivan-Brown, J.; Burdine, R.D. Zebrafish curly up encodes a Pkd2 ortholog that restricts left-side-specific expression of southpaw. Development 2007, 134, 1605–1615. [Google Scholar] [CrossRef]
- Campbell, S.A.; McDonald, C.L.; Krentz, N.A.J.; Lynn, F.C.; Hoffman, B.G. TrxG Complex Catalytic and Non-catalytic Activity Play Distinct Roles in Pancreas Progenitor Specification and Differentiation. Cell Rep. 2019, 28, 1830–1844. [Google Scholar] [CrossRef]
- Ho, J.; Yang, X.; Nikou, S.A.; Kichik, N.; Donkin, A.; Ponde, N.O.; Richardson, J.P.; Gratacap, R.L.; Archambault, L.S.; Zwirner, C.P.; et al. Candidalysin activates innate epithelial immune responses via epidermal growth factor receptor. Nat. Commun. 2019, 10, 2297. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Geng, F.; Ma, J.; Li, X.; Hu, Z.; Zhang, R. Hemodynamic Forces Regulate Cardiac Regeneration-Responsive Enhancer Activity during Ventricle Regeneration. Int. J. Mol. Sci. 2021, 22, 3945. https://doi.org/10.3390/ijms22083945
Geng F, Ma J, Li X, Hu Z, Zhang R. Hemodynamic Forces Regulate Cardiac Regeneration-Responsive Enhancer Activity during Ventricle Regeneration. International Journal of Molecular Sciences. 2021; 22(8):3945. https://doi.org/10.3390/ijms22083945
Chicago/Turabian StyleGeng, Fang, Jinmin Ma, Xueyu Li, Zhengyue Hu, and Ruilin Zhang. 2021. "Hemodynamic Forces Regulate Cardiac Regeneration-Responsive Enhancer Activity during Ventricle Regeneration" International Journal of Molecular Sciences 22, no. 8: 3945. https://doi.org/10.3390/ijms22083945
APA StyleGeng, F., Ma, J., Li, X., Hu, Z., & Zhang, R. (2021). Hemodynamic Forces Regulate Cardiac Regeneration-Responsive Enhancer Activity during Ventricle Regeneration. International Journal of Molecular Sciences, 22(8), 3945. https://doi.org/10.3390/ijms22083945





