CRISPR/Cas9-Mediated Knockout of GmFATB1 Significantly Reduced the Amount of Saturated Fatty Acids in Soybean Seeds
Abstract
1. Introduction
2. Results
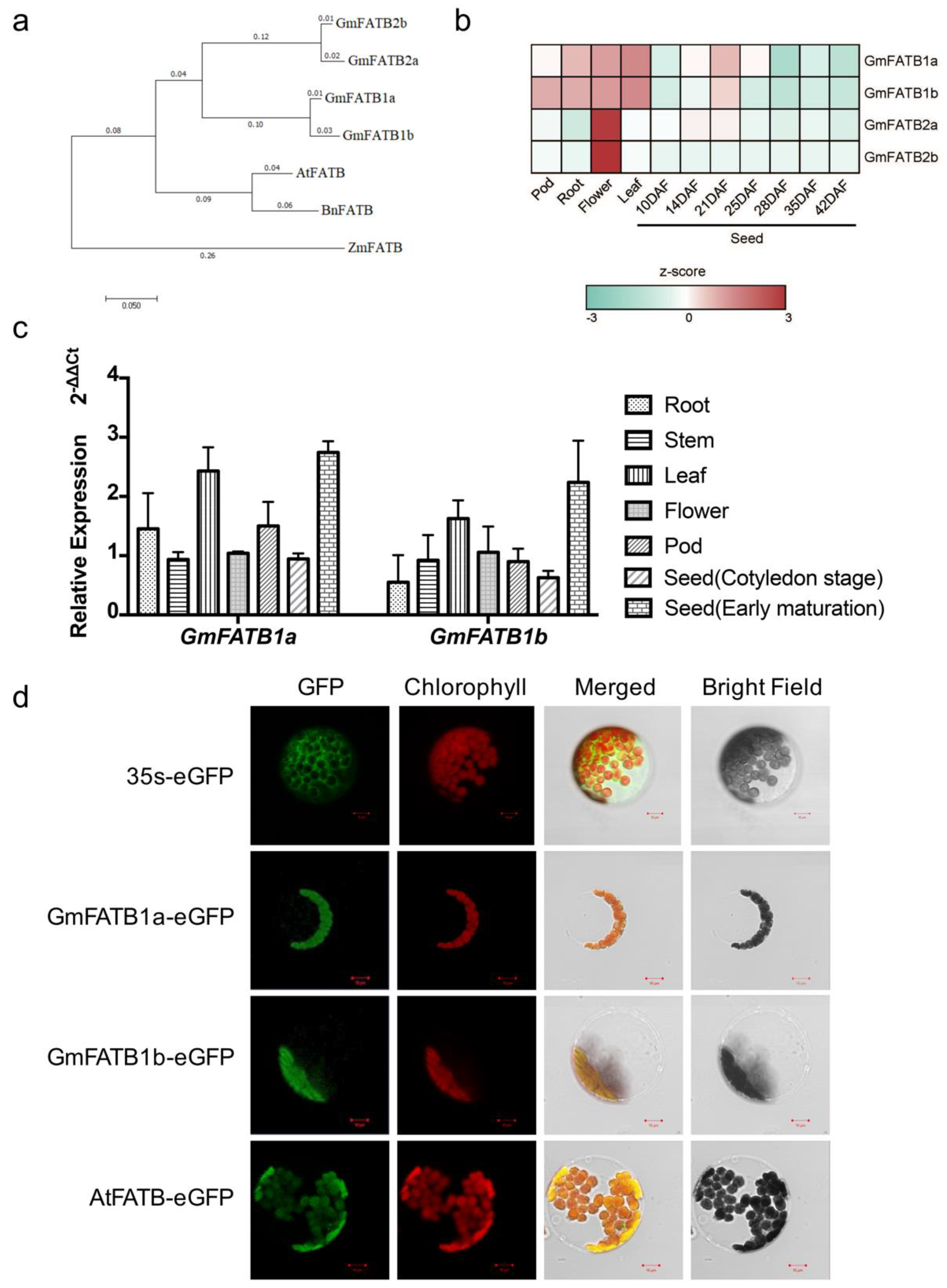
2.1. Expression of GmFATB1a and GmFATB1b
2.2. Knock-Out of GmFATB1a and GmFATB1b by CRISPR/CAS9
2.3. Effect of GmFATB1a and GmFATB1b on Leaf FA Profiles
2.4. Double Mutation of GmFATB1 Causes Male Sterility
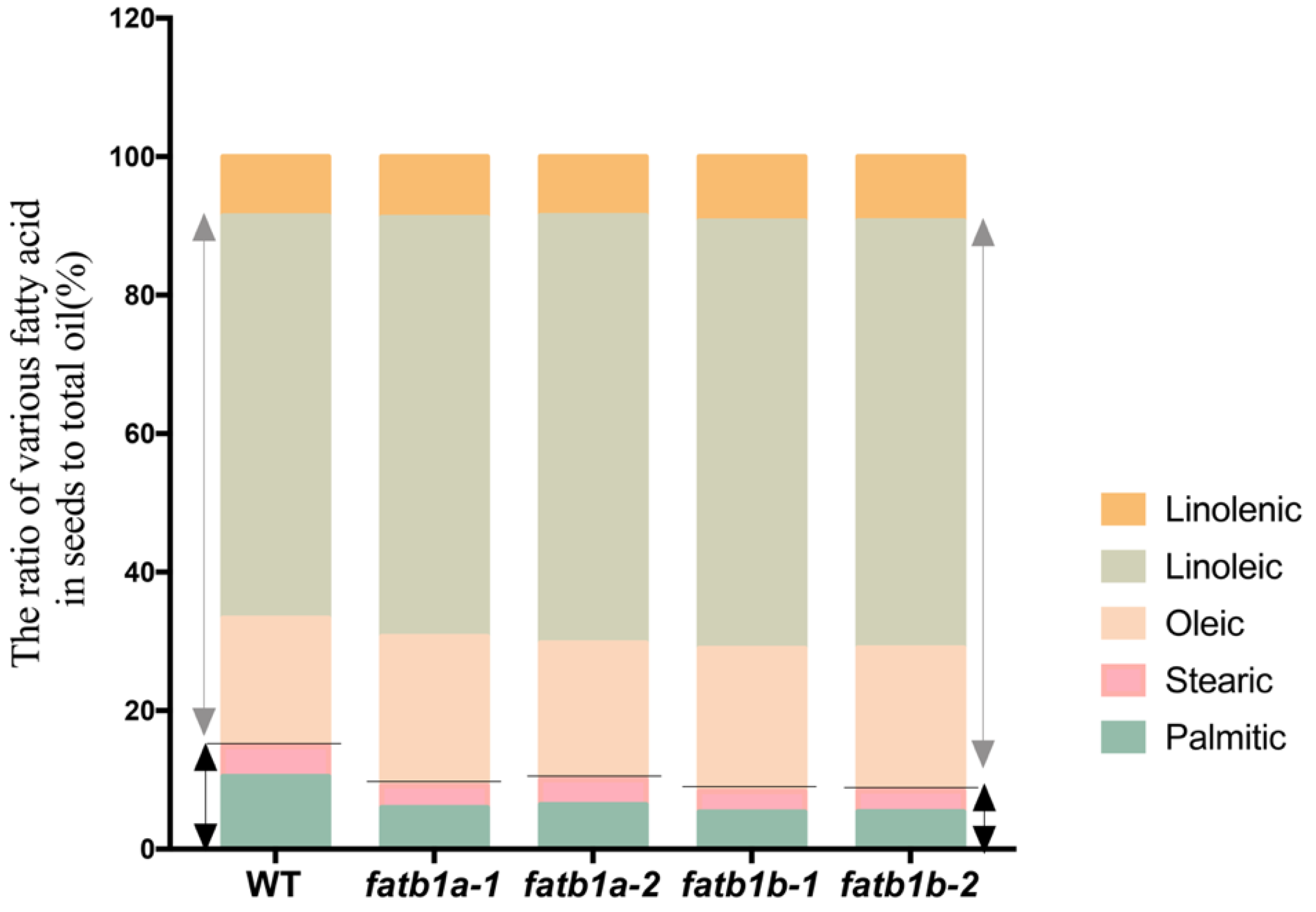
2.5. Mutation of GmFATB1 Significantly Improves the Soybean Nutritional Quality
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Vector Construction and Soybean Transformation
4.3. Phylogenetic Tree Construction
4.4. RNA Extraction and qRT-PCR
4.5. Subcellular Localization in Arabidopsis Mesophyll Protoplasts
4.6. Pollen Grain Staining
4.7. Scanning Electron Microscopy (SEM)
4.8. Transmission Electron Microscopy (TEM)
4.9. FA Analysis
4.10. Seed Protein Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACP | acyl carrier protein |
| FA | fatty acid |
| FATA(B) | acyl-acyl carrier protein thioesterase A(B) |
| I2-KI | iodine potassium iodide |
| LDL-C | low-density lipoprotein cholesterol |
| SEM | scanning electron microscopy |
| TEM | transmission electron microscopy |
References
- SoyStats® 2020. Available online: http://soystats.com/wp-content/uploads/SoyStats2020_Web.pdf (accessed on 8 April 2021).
- Clemente, T.E.; Cahoon, E.B. Soybean Oil: Genetic approaches for modification of functionality and total content. Plant Physiol. 2009, 151, 1030–1040. [Google Scholar] [CrossRef]
- Mancini, A.; Imperlini, E.; Nigro, E.; Montagnese, C.; Daniele, A.; Orru, S.; Buono, P. Biological and nutritional properties of palm oil and palmitic acid: Effects on health. Molecules 2015, 20, 17339–17361. [Google Scholar] [CrossRef]
- Moon, M.L.; Joesting, J.J.; Lawson, M.A.; Chiu, G.S.; Blevins, N.A.; Kwakwa, K.A.; Freund, G.G. The saturated fatty acid, palmitic acid, induces anxiety-like behavior in mice. Metabolism 2014, 63, 1131–1140. [Google Scholar] [CrossRef] [PubMed]
- Ismail, S.R.; Maarof, S.K.; Siedar Ali, S.; Ali, A. Systematic review of palm oil consumption and the risk of cardiovascular disease. PLoS ONE 2018, 13, e0193533. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, H.; Li, Q.; Yang, X.; Zheng, D.; Warburton, M.; Chai, Y.; Zhang, P.; Guo, Y.; Yan, J.; et al. An 11-bp insertion in Zea mays fatb reduces the palmitic acid content of fatty acids in maize grain. PLoS ONE 2011, 6, e24699. [Google Scholar] [CrossRef] [PubMed]
- Farvid, M.S.; Ding, M.; Pan, A.; Sun, Q.; Chiuve, S.E.; Steffen, L.M.; Willett, W.C.; Hu, F.B. Dietary linoleic acid and risk of coronary heart disease: A systematic review and meta-analysis of prospective cohort studies. Circulation 2014, 130, 1568–1578. [Google Scholar] [CrossRef] [PubMed]
- Hartigh, L.D. Conjugated linoleic acid effects on cancer, obesity, and atherosclerosis: A review of pre-clinical and human trials with current perspectives. Nutrients 2019, 11, 370. [Google Scholar] [CrossRef] [PubMed]
- Calder, P.C. Docosahexaenoic Acid. Ann. Nutr. Metab. 2016, 69 (Suppl. 1), 8–21. [Google Scholar] [CrossRef]
- Browse, J.; Somerville, C. Glycerolipid synthesis: Biochemistry and regulation. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1991, 42, 467–506. [Google Scholar] [CrossRef]
- Li-Beisson, Y.; Shorrosh, B.; Beisson, F.; Andersson, M.X.; Arondel, V.; Bates, P.D.; Baud, S.; Bird, D.; Debono, A.; Durrett, T.P.; et al. Acyl-lipid metabolism. Arab. Book 2013, 11, e0161. [Google Scholar] [CrossRef]
- Aubrey, J.; Davies, H.M.; Voelker, T.A. Palmitoyl-Acyl Carrier Protein (ACP) thioesterase and the evolutidnary-origin of plant Acyl-ACP thioesterases. Plant Cell 1995, 7, 359–371. [Google Scholar]
- Voelker, T.A.; Aubrey, J.; Cranmer, A.M.; Davies, H.M.; Knutzon, D.S. Broad-range and binary-range Acyl-Acyl-Carrier-Protein thioesterases suggest an alternative mechanism for medium-chain production in seeds. Plant Physiol. 1997, 114, 669–677. [Google Scholar] [CrossRef] [PubMed]
- Salas, J.J.; Ohlrogge, J.B. Characterization of substrate specificity of plant FatA and FatB acyl-ACP thioesterases. Arch. Biochem. Biophys. 2002, 403, 25–34. [Google Scholar] [CrossRef]
- Shine, W.E.; Mancha, M.; Stumpf, P.K. The function of acyl thioesterases in the metabolism of acyl-coenzymes A and acyl-acyl carrier proteins. Arch. Biochem. Biophys. 1976, 172, 110–116. [Google Scholar] [CrossRef]
- Ohlrogge, J.B.; Shine, W.E.; Stumpf, P.K. Characterization of plant Acyl-ACP and Acyl-CoA Hydrolases. Arch. Biochem. Biophys. 1978, 189, 382–391. [Google Scholar] [CrossRef]
- Mckeon, T.A.; Stumpf, P.K. Purification and characterization of the Stearoyl-Acyl Carrier Protein Desaturase and the Acyl-Acyl Carrier Protein Thioesterase from maturing seeds of Safflower. J. Biol. Chem. 1982, 257, 12141–12147. [Google Scholar] [CrossRef]
- Knutzon, D.S.; Bleibaum, J.L.; Nelsen, J.; Kridl, J.C.; Thompson, G.A. Isolation and characterization of two safflower oleoyl-acyl carrier protein thioesterase cDNA clones. Plant Physiol. 1992, 100, 1751–1758. [Google Scholar] [CrossRef]
- Davies, H.M.; Anderson, L.; Fan, C.; Hawkins, D.J. Developmental induction, purification, and further characterization of 12:0-ACP thioesterase from immature cotyledons of Umbellularia californica. Arch. Biochem. Biophys. 1991, 290, 37–45. [Google Scholar] [CrossRef]
- Voelker, T.A.; Worrell, A.C.; Lana, A.; Janice, B.; Calvin, F.; Hawkins, D.J.; Radke, S.E.; Maelor, D.H. Fatty acid biosynthesis redirected to medium chains in transgenic oilseed plants. Science 1992, 257, 72–73. [Google Scholar] [CrossRef]
- Voelker, T.A.; Davies, H.M. Alteration of the specificity and regulation of fatty-acid synthesis of Escherichia coli by expression of a plant medium-chain Acyl-Acyl Carrier Protein Thioesterase. J. Bacteriol. 1994, 176, 7320–7327. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Voelker, T.A.; Hawkins, D.J. Modification of the substrate-specificity of an Acyl-Acyl Carrier Protein Thioesterase by protein engineering. Proc. Natl. Acad. Sci. USA 1995, 92, 10639–10643. [Google Scholar] [CrossRef]
- Ohlrogge, J.B.; Savage, L.; Jaworski, J.; Voelker, T.A.; Postbeittenmiller, D. Alteration of Acyl-Acyl Carrier Protein pools and Acetyl-CoA Carboxylase expression in Escherichia coli by a plant medium-chain Acyl-Acyl Carrier Protein Thioesterase. Arch. Biochem. Biophys. 1995, 317, 185–190. [Google Scholar] [CrossRef] [PubMed]
- Loader, N.M.; Woolner, E.M.; Hellyer, A.; Slabas, A.R.; Safford, R. Isolation and characterization of two Brassica napus embryo acyl-ACP thioesterase cDNA clones. Plant Mol. Biol. 1993, 23, 769–778. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Force, E.; Cantisan, S.; Serrano-Vega, M.J.; Garces, R. Acyl-acyl carrier protein thioesterase activity from sunflower (Helianthus annuus L.) seeds. Planta 2000, 211, 673–678. [Google Scholar] [CrossRef] [PubMed]
- Serrano-Vega, M.J.; Garces, R.; Martinez-Force, E. Cloning, characterization and structural model of a FatA-type thioesterase from sunflower seeds (Helianthus annuus L.). Planta 2005, 221, 868–880. [Google Scholar] [CrossRef]
- Dormann, P.; Spener, F.; Ohlrogge, J.B. Characterization of two acyl-acyl carrier protein thioesterases from developing Cuphea seeds specific for medium-chain and oleoyl-acyl carrier protein. Planta 1993, 189, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Dormann, P.; Voelker, T.A.; Ohlrogge, J.B. Cloning and expression in Escherichia coli of a novel thioesterase from Arabidopsis thaliana specific for long-chain Acy-Acyl Carrier Proteins. Arch. Biochem. Biophys. 1995, 316, 612–618. [Google Scholar] [CrossRef]
- Bonaventure, G.; Bao, X.; Ohlrogge, J.B.; Pollard, M. Metabolic responses to the reduction in palmitate caused by disruption of the FATB gene in Arabidopsis. Plant Physiol. 2004, 135, 1269–1279. [Google Scholar] [CrossRef][Green Version]
- Bonaventure, G.; Joaquin, J.S.; Michael, R.P.; Ohlrogge, J.B. Disruption of the FATB gene in Arabidopsis demonstrates an essential role of saturated fatty acids in plant growth. Plant Cell 2003, 15, 1020–1033. [Google Scholar] [CrossRef]
- Dormann, P.; Voelker, T.A.; Ohlrogge, J.B. Accumulation of palmitate in arabidopsis mediated by the acyl-acyl carrier protein thioesterase FATB1. Plant Physiol. 2000, 123, 637–643. [Google Scholar] [CrossRef] [PubMed]
- Zheng, P.; Babar, M.D.; Parthasarathy, S.; Gibson, R.; Parliament, K.; Flook, J.; Patterson, T.; Friedemann, P.; Kumpatla, S.; Thompson, S. A truncated FatB resulting from a single nucleotide insertion is responsible for reducing saturated fatty acids in maize seed oil. Theor. Appl. Genet. 2014, 127, 1537–1547. [Google Scholar] [CrossRef]
- Yuan, Y.J.; Gao, L.C.; Sun, R.H.; Yu, T.; Liang, Y.X.; Li, D.D.; Zheng, Y.S. Seed-specific expression of an acyl-acyl carrier protein thioesterase CnFatB3 from coconut (Cocos nucifera L.) increases the accumulation of medium-chain fatty acids in transgenic Arabidopsis seeds. Sci. Hortic. 2017, 223, 5–9. [Google Scholar] [CrossRef]
- Goettel, W.; Ramirez, M.; Upchurch, R.G.; An, Y.Q. Identification and characterization of large DNA deletions affecting oil quality traits in soybean seeds through transcriptome sequencing analysis. Theor. Appl. Genet. 2016, 129, 1577–1593. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef] [PubMed]
- Severin, A.J.; Woody, J.L.; Bolon, Y.T.; Joseph, B.; Diers, B.W.; Farmer, A.D.; Muehlbauer, G.J.; Nelson, R.T.; Grant, D.; Specht, J.E.; et al. RNA-Seq Atlas of Glycine max: A guide to the soybean transcriptome. BMC Plant Biol. 2010, 10, 160. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Sun, J.; Hong, X.; Zhao, H.; Wang, Q. Subcellular Localization of Arabidopsis FATB Protein. Acta Agric. Boreali-occident. Sin. 2013, 22, 163–167. [Google Scholar]
- Li, N.; Gugel, I.L.; Giavalisco, P.; Zeisler, V.; Schreiber, L.; Soll, J.; Philippar, K. FAX1, a novel membrane protein mediating plastid fatty acid export. PLoS Biol. 2015, 13, e1002053. [Google Scholar] [CrossRef]
- Hsiao, A.S.; Haslam, R.P.; Michaelson, L.V.; Liao, P.; Chen, Q.F.; Sooriyaarachchi, S.; Mowbray, S.L.; Napier, J.A.; Tanner, J.A.; Chye, M.L. Arabidopsis cytosolic acyl-CoA-binding proteins ACBP4, ACBP5 and ACBP6 have overlapping but distinct roles in seed development. Biosci. Rep. 2014, 34, 865–877. [Google Scholar] [CrossRef]
- Hsiao, A.S.; Yeung, E.C.; Ye, Z.W.; Chye, M.L. The Arabidopsis cytosolic Acyl-CoA-binding proteins play combinatory roles in pollen development. Plant Cell Physiol. 2015, 56, 322–333. [Google Scholar] [CrossRef]
- Ye, Z.W.; Xu, J.; Shi, J.; Zhang, D.; Chye, M.L. Kelch-motif containing acyl-CoA binding proteins AtACBP4 and AtACBP5 are differentially expressed and function in floral lipid metabolism. Plant Mol. Biol. 2017, 93, 209–225. [Google Scholar] [CrossRef]
- Liu, D.H.; Post-beittenmiller, D. Discovery of an epidermal Stearoyl-Acyl Carrier Protein Thioesterase—Its potential role in wax biosynthesis. J. Biol. Chem. 1995, 270, 16962–16969. [Google Scholar] [CrossRef] [PubMed]
- Post-beittenmiller, D. Biochemistry and molecular biology of wax production in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 405–430. [Google Scholar] [CrossRef] [PubMed]
- Wan, X.; Wu, S.; Li, Z.; An, X.; Tian, Y. Lipid Metabolism: Critical roles in male fertility and other aspects of reproductive development in plants. Mol. Plant 2020, 13, 955–983. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-H.; Zhou, G.; Ma, J.; Jiang, W.; Jin, L.-G.; Zhang, Z.; Guo, Y.; Zhang, J.; Sui, Y.; Zheng, L.; et al. De novo assembly of soybean wild relatives for pan-genome analysis of diversity and agronomic traits. Nat. Biotechnol. 2014, 32, 1045–1052. [Google Scholar] [CrossRef]
- Liu, Y.; Du, H.; Li, P.; Shen, Y.; Peng, H.; Liu, S.; Zhou, G.A.; Zhang, H.; Liu, Z.; Shi, M.; et al. Pan-Genome of Wild and Cultivated Soybeans. Cell 2020, 182, 162–176.e13. [Google Scholar] [CrossRef]
- Zhou, Z.; Jiang, Y.; Wang, Z.; Gou, Z.; Lyu, J.; Li, W.; Yu, Y.; Shu, L.; Zhao, Y.; Ma, Y.; et al. Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat. Biotechnol. 2015, 33, 408–414. [Google Scholar] [CrossRef]
- Buhr, T.; Sato, S.; Ebrahim, F.; Xing, A.; Zhou, Y.; Mathiesen, M.; Schweiger, B.; Kinney, A.; Staswick, P.; Clemente, T. Ribozyme termination of RNA transcripts down-regulate seed fatty acid genes in transgenic soybean. Plant J. 2002, 31, 155–163. [Google Scholar] [CrossRef]
- Sun, J.Y.; Hammerlindl, J.; Forseille, L.; Zhang, H.X.; Smith, M.A. Simultaneous over-expressing of an acyl-ACP thioesterase (FatB) and silencing of acyl-acyl carrier protein desaturase by artificial microRNAs increases saturated fatty acid levels in Brassica napus seeds. Plant Biotechnol. J. 2014, 12, 624–637. [Google Scholar] [CrossRef]
- Song, Z.Y.; Tian, J.L.; Fu, W.Z.; Li, L.; Lu, L.H.; Zhou, L.; Shan, Z.H.; Tang, G.X.; Shou, H.X. Screening Chinese soybean genotypes for Agrobacterium-mediated genetic transformation suitability. J. Zhejiang Univ. Sci. B 2013, 14, 289–298. [Google Scholar] [CrossRef]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.D.; Cho, Y.H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Liu, S.; Wang, J.; Yokosho, K.; Zhou, B.; Yu, Y.-C.; Liu, Z.; Frommer, W.B.; Ma, J.F.; Chen, L.-Q.; et al. Simultaneous changes in seed size, oil content and protein content driven by selection of SWEET homologues during soybean domestication. Natl. Sci. Rev. 2020, 7, 1776–1786. [Google Scholar] [CrossRef]


| Material (mg/g) | Palmitic | Stearic | Oleic | Linoleic | Linolenic | Total |
|---|---|---|---|---|---|---|
| Wild type | 6.89 ± 0.21 a | 2.40 ± 0.06 a | 0.73 ± 0.01 a | 4.22 ± 0.10 a | 33.52 ± 2.36 a | 47.76 ± 2.03 a |
| fatb1a-1 | 6.08 ± 0.42 b | 2.13 ± 0.05 b | 0.76 ± 0.11 a | 4.29 ± 0.19 a | 34.01 ± 0.32 a | 47.27 ± 0.21 a |
| fatb1a-2 | 5.66 ± 0.34 bc | 1.98 ± 0.06 bc | 0.73 ± 0.12 a | 3.91 ± 0.54 a | 31.64 ± 3.04 a | 43.93 ± 2.46 b |
| fatb1b-1 | 5.86 ± 0.14 bc | 2.05 ± 0.09 bc | 0.87 ± 0.19 a | 4.34 ± 0.45 a | 32.53 ± 0.66 a | 45.65 ± 1.42 ab |
| fatb1b-2 | 5.43 ± 0.12 c | 1.89 ± 0.05 c | 0.73 ± 0.04 a | 4.29 ± 0.22 a | 31.26 ± 1.08 a | 43.59 ± 1.44 b |
| fatb1a: 1b | 3.98 ± 0.22 d | 1.55 ± 0.11 d | 0.86 ± 0.16 a | 4.56 ± 1.00 a | 35.21 ± 2.38 a | 43.88 ± 2.09 b |
| Materials | Saturated Fatty Acid (mg/g) | Unsaturated Fatty Acid (mg/g) | Total (mg/g) | |||||
|---|---|---|---|---|---|---|---|---|
| Palmitic | Stearic | Total | Oleic | Linoleic | Linolenic | Total | ||
| Wild type | 22.85 ± 0.98 a | 9.42 ± 0.56a | 32.28 ± 1.53a | 40.45 ± 1.70 bc | 126.73 ± 6.09 ab | 18.72 ± 0.93 ab | 185.89 ± 8.56 a | 218.17 ± 10.09 a |
| fatb1a-1 | 12.23 ± 0.82 b | 6.43 ± 0.44 c | 18.66 ± 1.26 c | 43.97 ± 4.08 a | 123.51 ± 4.53 b | 17.90 ± 1.44 c | 185.38 ± 6.53 a | 204.04 ± 6.13 b |
| fatb1a-2 | 13.93 ± 0.30 c | 7.79 ± 0.18 b | 21.72 ± 0.43 b | 42.72 ± 1.63 ab | 133.66 ± 4.50 a | 18.47 ± 0.52ab | 194.85 ± 6.62 a | 216.57 ± 7.05 a |
| fatb1b-1 | 10.81 ± 1.17 d | 6.06 ± 0.50 c | 16.87 ± 1.61 cd | 41.75 ± 4.76 bc | 125.00 ± 10.80 ab | 18.81 ± 1.07 ab | 185.55 ± 16.41 a | 202.42 ± 17.14 b |
| fatb1b-2 | 10.64 ± 0.84 d | 5.92 ± 0.16 c | 16.56 ± 0.99 d | 40.54 ± 3.62 bc | 121.27 ± 7.38 b | 18.23 ± 0.48 b | 180.04 ± 11.26 a | 196.60 ± 10.73 b |
| Material | Plant Height (cm) | Number of Branches | Number of Pods/Plant | Number of Seeds/Plant | Seed Weight (g)/Plant | 100-Seed Weight (g) |
|---|---|---|---|---|---|---|
| Wild type | 53.3 ± 3.6 a | 2.7 ± 0.5 a | 57.6 ± 8.5 a | 127.1 ± 17.9 a | 12.8 ± 2.9 a | 13.3 ± 1.6 a |
| fatb1a-1 | 53.5 ± 4.7 a | 3.5 ± 1.5 a | 64.9 ± 23.4 a | 130.7 ± 44.7 a | 12.9 ± 4.6 a | 13.7 ± 1.0 a |
| fatb1a-2 | 50.2 ± 4.7a | 2.8 ± 0.8 a | 63.1 ± 17.4 a | 140.5 ± 41.3 a | 14.6 ± 3.0 a | 13.0 ± 1.4 a |
| fatb1b-1 | 47.3 ± 9.8 a | 2.6 ± 0.8 a | 56.3 ± 16.1 a | 114.1 ± 43.4 a | 13.2 ± 1.3 a | 13.2 ± 1.3 a |
| fatb1b-2 | 50.0 ± 6.1 a | 2.6 ± 1.2 a | 69.4 ± 19.7 a | 107.5 ± 31.6 a | 13.9 ± 4.8 a | 12.7 ± 1.0 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, J.; Sun, S.; Whelan, J.; Shou, H. CRISPR/Cas9-Mediated Knockout of GmFATB1 Significantly Reduced the Amount of Saturated Fatty Acids in Soybean Seeds. Int. J. Mol. Sci. 2021, 22, 3877. https://doi.org/10.3390/ijms22083877
Ma J, Sun S, Whelan J, Shou H. CRISPR/Cas9-Mediated Knockout of GmFATB1 Significantly Reduced the Amount of Saturated Fatty Acids in Soybean Seeds. International Journal of Molecular Sciences. 2021; 22(8):3877. https://doi.org/10.3390/ijms22083877
Chicago/Turabian StyleMa, Jing, Shuo Sun, James Whelan, and Huixia Shou. 2021. "CRISPR/Cas9-Mediated Knockout of GmFATB1 Significantly Reduced the Amount of Saturated Fatty Acids in Soybean Seeds" International Journal of Molecular Sciences 22, no. 8: 3877. https://doi.org/10.3390/ijms22083877
APA StyleMa, J., Sun, S., Whelan, J., & Shou, H. (2021). CRISPR/Cas9-Mediated Knockout of GmFATB1 Significantly Reduced the Amount of Saturated Fatty Acids in Soybean Seeds. International Journal of Molecular Sciences, 22(8), 3877. https://doi.org/10.3390/ijms22083877








