Prebiotic Pathway from Ribose to RNA Formation
Abstract
1. Introduction
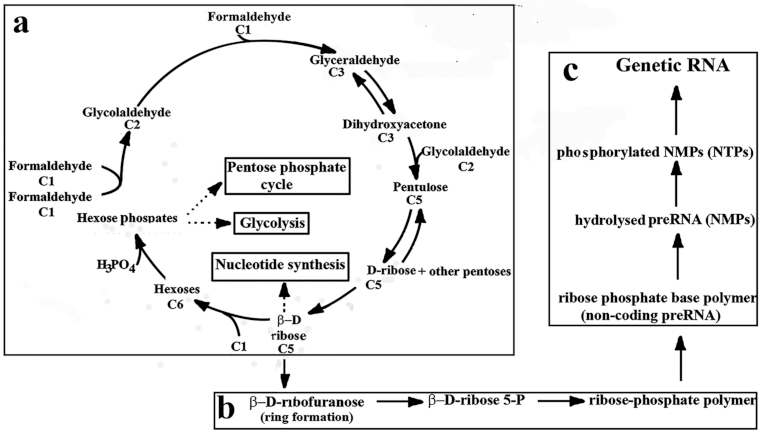
2. Prebiotic Synthesis of Ribose
3. Prebiotic Synthesis of Purine and Pyrimidine Nucleobases
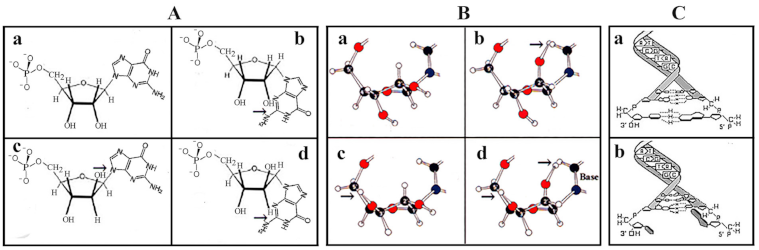
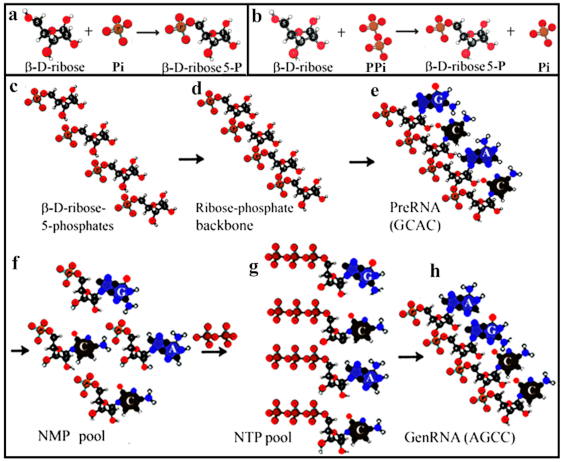
4. Phosphorylation, Polymerization, and Nucleotide Formation
5. From preRNA to genRNA Formation
6. Protection of Ribose
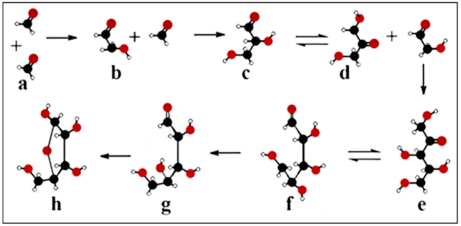
6.1. Catalysis in the Formose Reaction
6.2. Formose–Ribose–RNA Pathway
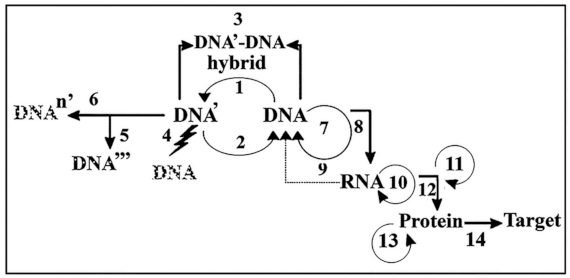
7. Hierarchical Processes in the Transfer of Cellular Information
8. Conclusions
9. Brief Summary
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dodd, M.S.; Papineau, M.S.D.D.; Grenne, T.; Slack, J.F.; Rittner, M.S.D.D.P.M.; Pirajno, F.; O’Neil, J.; Little, C.T.S. Evidence for early life in Earth’s oldest hydrothermal vent precipitates. Nat. Cell Biol. 2017, 543, 60–64. [Google Scholar] [CrossRef] [PubMed]
- Sojo, V.; Herschy, B.; Whicher, A.; Camprubí, E.; Lane, N. The Origin of Life in Alkaline Hydrothermal Vents. Astrobioogy 2016, 16, 181–197. [Google Scholar] [CrossRef] [PubMed]
- Calvin, M. Chemical evolution and the origin of life. Am. Sci. 1956, 44, 248–263. [Google Scholar]
- Fränzle, S.; Markert, B.; Wünschmann, S. Dynamics of trace metals in organisms and ecosystems: Prediction of metal bioconcentration in different organisms and estimation of exposure risks. Environ. Pollut. 2007, 150, 23–33. [Google Scholar] [CrossRef]
- Markert, B.; Fränzle, S.; Wünschmann, S. Chemical Evolution: The Biological System of Elements; Springer International: Cham, Switzerland, 2015; ISBN 9783319143552. [Google Scholar]
- Matsumoto, T.; Inoue, S. Formose reactions. Part 3. Selective formose reaction catalyzed by organic bases. J. Chem. Soc. Perkin Trans. 1982, 1, 1975–1979. [Google Scholar] [CrossRef]
- Kim, H.-J.; Ricardo, A.; Illangkoon, H.I.; Kim, M.J.; Carrigan, M.A.; Frye, F.; Benner, S.A. Synthesis of carbohydrates in mineral-guided prebiotic cycles. J. Am. Chem. Soc. 2011, 133, 9457–9468. [Google Scholar] [CrossRef]
- Jeilani, Y.A.; Nguyen, M.T. Autocatalysis in Formose Reaction and Formation of RNA Nucleosides. J. Phys. Chem. B 2020, 124, 11324–11336. [Google Scholar] [CrossRef]
- Omran, A.; Menor-Salvan, C.; Springsteen, G.; Pasek, M. The Messy Alkaline Formose Reaction and Its Link to Metabolism. Life 2020, 10, 125. [Google Scholar] [CrossRef]
- Zellner, N.E.; McCaffrey, V.P.; Butler, J.H. Cometary Glycolaldehyde as a Source of pre-RNA Molecules. Astrobiology 2020, 20, 1377–1388. [Google Scholar] [CrossRef]
- Omran, A. Plausibility of the Formose Reaction in Alkaline Hydrothermal Vent Environments. Orig. Life Evol. Biosph. 2020, 1–13. [Google Scholar] [CrossRef]
- Furukawa, Y.; Chikaraishi, Y.; Ohkouchi, N.; Ogawa, N.O.; Glavin, D.P.; Dworkin, J.P.; Abe, C.; Nakamura, T. Extraterrestrial ribose and other sugars in primitive meteorites. Proc. Natl. Acad. Sci. USA 2019, 116, 24440–24445. [Google Scholar] [CrossRef]
- Furukawa, Y.; Horiuchi, M.; Kakegawa, T. Selective Stabilization of Ribose by Borate. Orig. Life Evol. Biosph. 2013, 43, 353–361. [Google Scholar] [CrossRef]
- Nitta, S.; Furukawa, Y.; Kakegawa, T. Effects of Silicate, Phosphate, and Calcium on the Stability of Aldopentoses. Orig. Life Evol. Biosphere 2015, 46, 189–202. [Google Scholar] [CrossRef]
- Gollihar, J.; Levy, M.; Ellington, A.D. Many Paths to the Origin of Life. Science 2014, 343, 259–260. [Google Scholar] [CrossRef]
- Vázquez-Mayagoitia, Á.; Horton, S.R.; Sumpter, B.G.; Šponer, J.; Šponer, J.E.; Fuentes-Cabrera, M. On the Stabilization of Ribose by Silicate Minerals. Astrobiology 2011, 11, 115–121. [Google Scholar] [CrossRef]
- Theobald, D.L. A formal test of the theory of universal common ancestry. Nat. Cell Biol. 2010, 465, 219–222. [Google Scholar] [CrossRef]
- Banfalvi, G. Ribose Selected as Precursor to Life. DNA Cell Biol. 2020, 39, 177–186. [Google Scholar] [CrossRef]
- Ferris, J.P.; Hagan, W.J. HCN and chemical evolution: The possible role of cyano compounds in prebiotic synthesis. Tetrahedron 1984, 40, 1093–1120. [Google Scholar] [CrossRef]
- Sutherland, J.D. The Origin of Life-Out of the Blue. Angew. Chem. Int. Ed. 2016, 55, 104–121. [Google Scholar] [CrossRef] [PubMed]
- Ferus, M.; Kubelík, P.; Knížek, A.; Pastorek, A.; Sutherland, J.; Civiš, S. High Energy Radical Chemistry Formation of HCN-rich Atmospheres on early Earth. Sci. Rep. 2017, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Ritson, D.J.; Ranjan, S.; Todd, Z.R.; Sasselov, D.D.; Sutherland, J.D. Photochemical reductive homologation of hydrogen cyanide using sulfite and ferrocyanide. Chem. Commun. 2018, 54, 5566–5569. [Google Scholar] [CrossRef]
- Ranjan, S.; Todd, Z.R.; Sutherland, J.D.; Sasselov, D.D. Sulfidic Anion Concentrations on Early Earth for Surficial Origins-of-Life Chemistry. Astrobiology 2018, 18, 1023–1040. [Google Scholar] [CrossRef]
- Breslow, R.; Appayee, C. Transketolase reaction under credible prebiotic conditions. Proc. Natl. Acad. Sci. USA 2013, 110, 4184–4187. [Google Scholar] [CrossRef]
- Miller, S.L.; Orgel, L.E. The Origins of Life on the Earth; Prentice-Hall: Englewood Cliffs, NJ, USA, 1973; p. 229. [Google Scholar]
- Bar-Nun, A.; Chang, S.J. Photochemical reactions of carbon monoxide and water in Earth’s primitive atmosphere. Geophys. Res. 1983, 88, 6662–6672. [Google Scholar] [CrossRef]
- Bekker, A.; Holland, H.D.; Wang, P.L.; Rumbl, D., III; Stein, H.J.; Hannah, J.L.; Coetzee, L.L.; Beukes, N.J. Dating the rise of the atmospheric oxygen. Nature 2004, 427, 117–120. [Google Scholar] [CrossRef]
- Schirrmeister, B.E.; Gugger, M.; Donoghue, P.C.J. Cyanobacteria and the Great Oxidation Event: Evidence from genes and fossils. Palaeontology 2015, 58, 769–785. [Google Scholar] [CrossRef]
- Benner, S.A.; Bell, E.A.; Biondi, E.; Brasser, R.; Carell, T.; Kim, H.; Mojzsis, S.J.; Omran, A.; Pasek, M.A.; Trail, D. When Did Life Likely Emerge on Earth in an RNA-First Process? ChemSystemsChem 2020, 2, 1900035. [Google Scholar] [CrossRef]
- Bánfalvi, G. Heavy Metals, Trace Elements and Their Cellular Effects; Springe: Dordrecht, The Netherlands, 2011; pp. 3–28. [Google Scholar]
- Boutlerow, A. Formation Synthétique d’une Substance Sucrée. CR Acad. Sci. 1861, 53, 145–147. [Google Scholar]
- Zafar, I.; Senad, N. The formose reaction: A tool to produce synthetic carbohydrates within a regenerative life support system. Curr. Org. Chem. 2012, 16, 769–788. [Google Scholar] [CrossRef]
- Delidovich, I.V.; Simonov, A.N.; Taran, O.P.; Parmon, V.N. Catalytic Formation of Monosaccharides: From the Formose Reaction towards Selective Synthesis. ChemSusChem 2014, 7, 1833–1846. [Google Scholar] [CrossRef]
- Breslow, R. On the mechanism of the formose reaction. Tetrahedron Lett. 1959, 1, 22–26. [Google Scholar] [CrossRef]
- Huskey, W.P.; Epstein, I.R.J. Auto-catalysis and apparent bistability in the formose reaction. Am. Chem. Soc. 1989, 111, 3157–3163. [Google Scholar] [CrossRef]
- Müller, D.; Pitsch, S.; Kittaka, A.; Wagner, E.; Wintner, C.E.; Eschenmoser, A.; Ohlofjgewidmet, G. Chemie von a-Aminonitrilen. Aldomerisierung von Glycolaldehyd-phosphat zu racemischen Hexose-2,4,6-triphosphaten und (in Gegenwart von Formaldehyd) racemischen Pentose-2,4-diphosphaten: Rac-Allose-2,4,6-triphosphat und rac-Ribose-2,4-diphosphat sind die R. Helv. Chim. Acta 1990, 73, 1410–1468. [Google Scholar] [CrossRef]
- Eschenmoser, A. The search for the chemistry of life’s origin. Tetrahedron 2007, 63, 12821–12844. [Google Scholar] [CrossRef]
- Kopetzky, D.; Antonietti, M. Hydrothermal formose reaction. New J. Chem. 2011, 35, 1787–1794. [Google Scholar] [CrossRef]
- Lamour, S.; Pallmann, S.; Haas, M.; Trapp, O. Prebiotic Sugar Formation Under Nonaqueous Conditions and Mechanochemical Acceleration. Life 2019, 9, 52. [Google Scholar] [CrossRef]
- Gardner, P.M.; Winzer, K.; Davis, B.G. Sugar synthesis in a protocellular model leads to a cell signalling response in bacteria. Nat. Chem. 2009, 1, 377–383. [Google Scholar] [CrossRef]
- Banfalvi, G. Why ribose was selected as the sugar component of nucleic acids. DNA Cell Biol. 2006, 25, 189–196. [Google Scholar] [CrossRef]
- Okano, K. Synthesis and pharmaceutical application of l-ribose. Tetrahedron 2009, 65, 1937–1949. [Google Scholar] [CrossRef]
- Oró, J.; Kimball, A. Synthesis of purines under possible primitive earth conditions. I. Adenine from hydrogen cyanide. Arch. Biochem. Biophys. 1961, 94, 217–227. [Google Scholar] [CrossRef]
- Ferris, J.P.; Sanchez, R.A.; Orgel, L.E. Studies in prebiotic synthesis. 3. synthesis of pyrimidines from cyanoacetylene and cyanate. J. Mol. Biol. 1968, 33, 693–704. [Google Scholar] [CrossRef]
- Fischer, G.; Geith, J.; Klapötke, T.M.; Krumm, B. Synthesis, Properties and Dimerization Study of Isocyanic Acid. Z. Nat. B 2002, 57, 19–24. [Google Scholar] [CrossRef]
- Tozzi, M.G.; Camici, M.; Mascia, L.; Sgarrella, F.; Ipata, P.L. Pentose hosphates in nucleoside interconversion and catabolism. FEBS. J. 2006, 273, 1089–1101. [Google Scholar] [CrossRef] [PubMed]
- Gross, M. How life can arise from chemistry. Curr. Biol. 2016, 26, 1247–1249. [Google Scholar] [CrossRef]
- Drew, K.N.; Zajicek, J.; Bondo, G.; Bose, B.; Serianni, A.S. 13C-labeled aldopentoses: Detection and quantitation of cyclic and acyclic forms by heteronuclear 1D and 2D NMR spectroscopy. Carbohydr. Res. 1998, 307, 199–209. [Google Scholar] [CrossRef]
- Calvin, M. The Bakerian Lecture—Chemical evolution. Proc. R. Soc. 1965, 288, 41. [Google Scholar]
- Halmann, M.; Sanchez, R.A.; Orgel, L.E. Phosphorylation of d-ribose in aqueous solution. J. Org. Chem. 1969, 34, 3702–3703. [Google Scholar] [CrossRef]
- Hu, J.; Lei, W.; Wang, J.; Chen, H.-Y.; Xu, J.-J. Regioselective 5′-position phosphorylation of ribose and ribonucleosides: Phosphate transfer in the activated pyrophosphate complex in the gas phase. Chem. Commun. 2018, 55, 310–313. [Google Scholar] [CrossRef]
- Bánfalvi, G.; Sarkar, N. Origin and degradation of the RNA primers at the 5′ termini of nascent DNA chains in Bacillus subtilis. J. Mol. Biol. 1985, 186, 275–282. [Google Scholar] [CrossRef]
- Crick, F. The origin of the genetic code. J. Mol. Biol. 1968, 38, 367–379. [Google Scholar] [CrossRef]
- Yakhnin, A.V. A Model for the Origin of Life through Rearrangements among Prebiotic Phosphodiester Polymers. Orig. Life Evol. Biosph. 2013, 43, 39–47. [Google Scholar] [CrossRef]
- Bernhardt, H.S.; Sandwick, R.K. Purine Biosynthetic Intermediate-Containing Ribose-Phosphate Polymers as Evolutionary Precursors to RNA. J. Mol. Evol. 2014, 79, 91–104. [Google Scholar] [CrossRef]
- Larralde, R.; Robertson, M.P.; Miller, S.L. Rates of decomposition of ribose and other sugars: Implications for chemical evolution. Proc. Natl. Acad. Sci. USA 1995, 92, 8158–8160. [Google Scholar] [CrossRef]
- Akouche, M.; Jaber, M.; Zins, E.-L.; Maurel, M.-C.; Lambert, J.-F.; Georgelin, T. Thermal Behavior of d-Ribose Adsorbed on Silica: Effect of Inorganic Salt Coadsorption and Significance for Prebiotic Chemistry. Chem. Eur. J. 2016, 22, 15834–15846. [Google Scholar] [CrossRef]
- Westheimer, F.H. Why nature chose phosphates. Science 1987, 235, 1173–1178. [Google Scholar] [CrossRef]
- Menor-Salván, C.; Ruiz-Bermejo, D.M.; Guzmán, M.I.; Osuna-Esteban, S.; Veintemillas-Verdaguer, S. Synthesis of Pyrimidines and Triazines in Ice: Implications for the Prebiotic Chemistry of Nucleobases. Chem. Eur. J. 2009, 15, 4411–4418. [Google Scholar] [CrossRef]
- Liu, Z.; Wu, L.-F.; Xu, J.; Bonfio, C.; Russell, D.A.; Sutherland, J.D. Harnessing chemical energy for the activation and joining of prebiotic building blocks. Nat. Chem. 2020, 12, 1023–1028. [Google Scholar] [CrossRef]
- Gilbert, W. The RNA World. Nature 1986, 319, 618. [Google Scholar] [CrossRef]
- Robertson, M.P.; Joyce, G.F. The Origins of the RNA World. Cold Spring Harb. Perspect. Biol. 2010, 4, a003608. [Google Scholar] [CrossRef]
- Reuveni, S.; Ehrenberg, M.; Paulsson, J. Ribosomes are optimized for autocatalytic production. Nat. Cell Biol. 2017, 547, 293–297. [Google Scholar] [CrossRef]
- Yokobayashi, Y. Aptamer-based and aptazyme-based riboswitches in mammalian cells. Curr. Opin. Chem. Biol. 2019, 52, 72–78. [Google Scholar] [CrossRef]
- Vasas, V.; Fernando, C.; Santos, M.; Kauffman, S.; Szathmáry, E. Evolution before genes. Biol. Direct 2012, 7, 1. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Banfalvi, G. Prebiotic Pathway from Ribose to RNA Formation. Int. J. Mol. Sci. 2021, 22, 3857. https://doi.org/10.3390/ijms22083857
Banfalvi G. Prebiotic Pathway from Ribose to RNA Formation. International Journal of Molecular Sciences. 2021; 22(8):3857. https://doi.org/10.3390/ijms22083857
Chicago/Turabian StyleBanfalvi, Gaspar. 2021. "Prebiotic Pathway from Ribose to RNA Formation" International Journal of Molecular Sciences 22, no. 8: 3857. https://doi.org/10.3390/ijms22083857
APA StyleBanfalvi, G. (2021). Prebiotic Pathway from Ribose to RNA Formation. International Journal of Molecular Sciences, 22(8), 3857. https://doi.org/10.3390/ijms22083857





