Optimisation of Recombinant Myrosinase Production in Pichia pastoris
Abstract
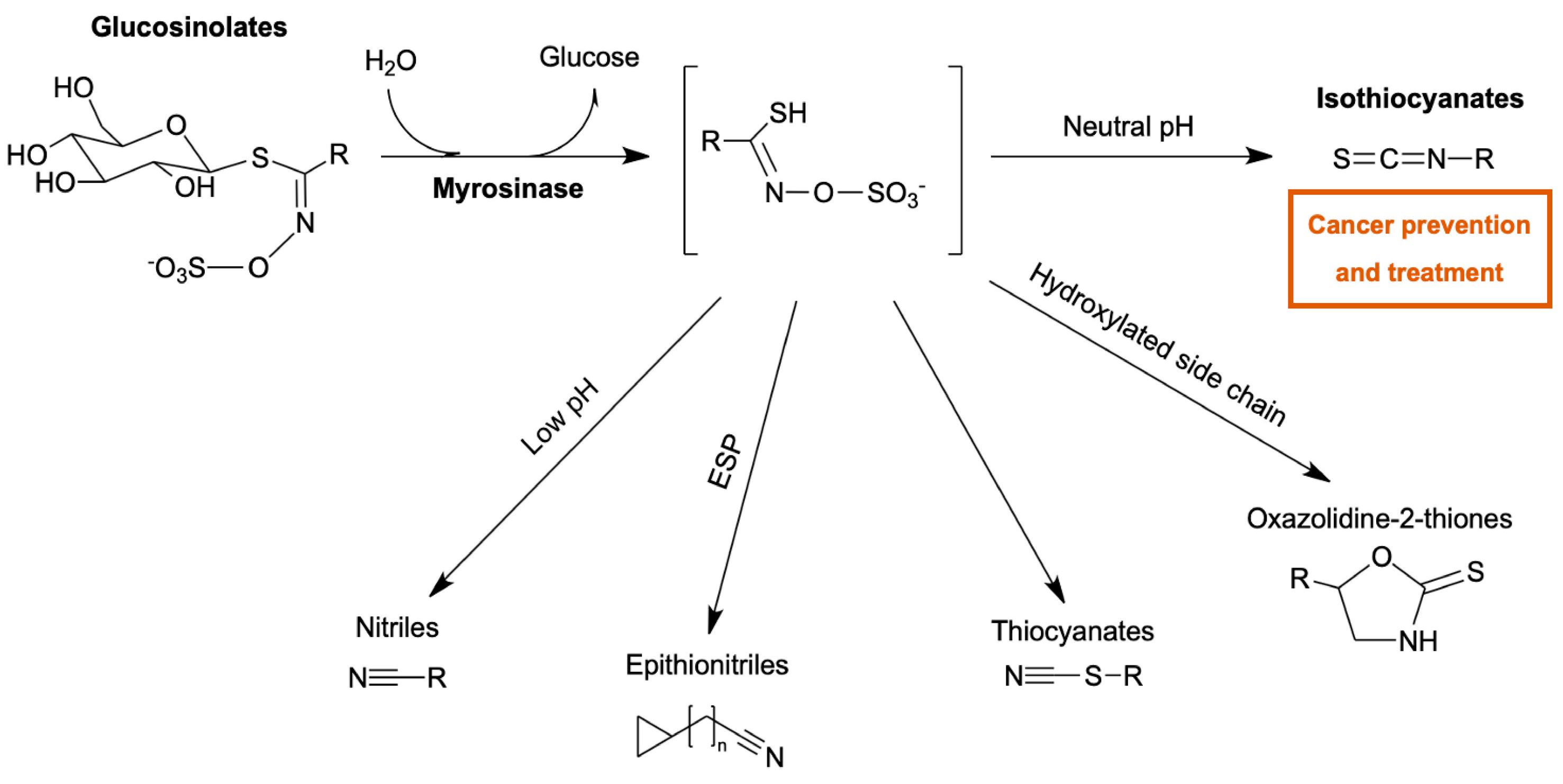
1. Introduction
2. Results and Discussion
2.1. Recombinant Strain Selection and Myrosinase Activity Assay
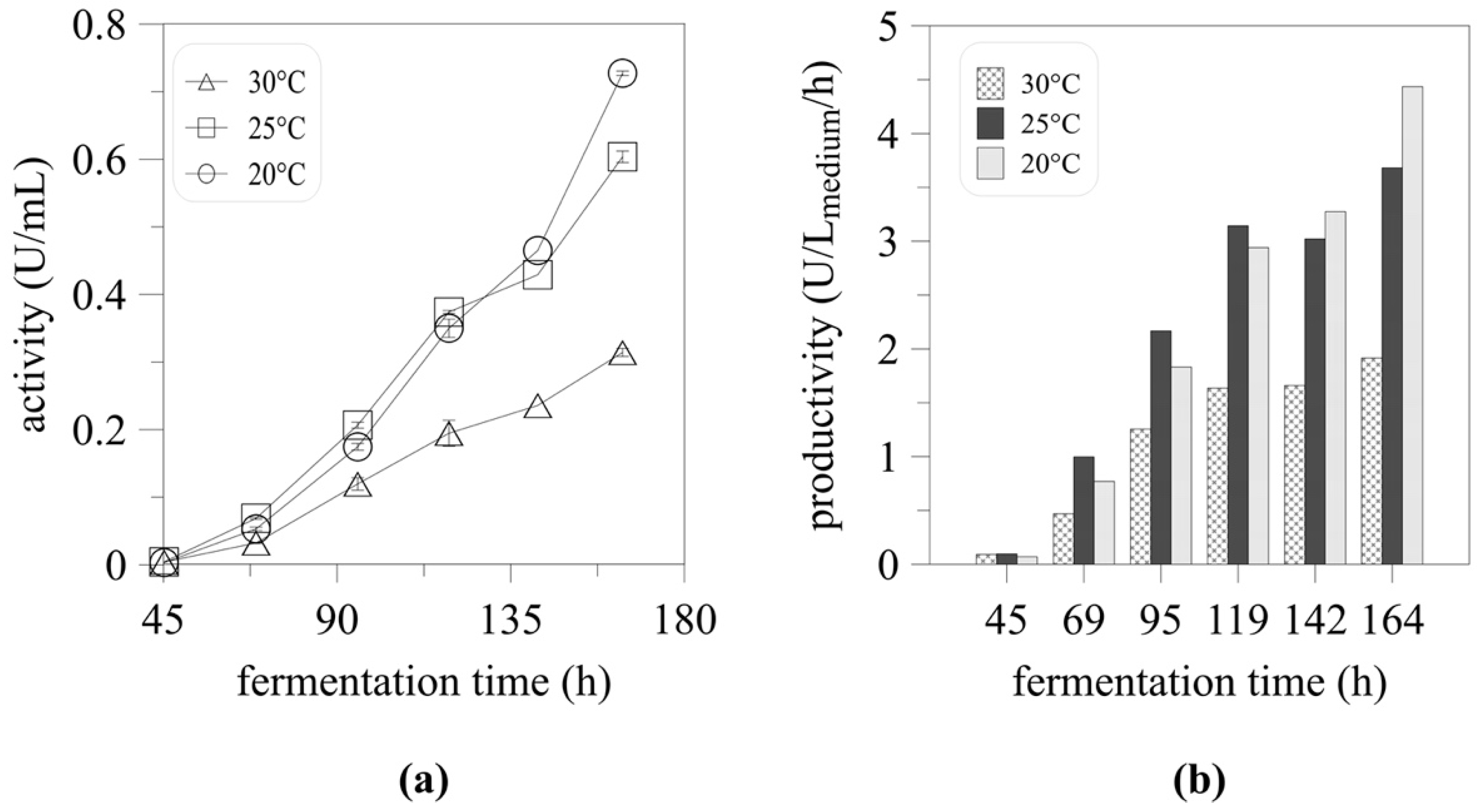
2.2. High-Cell Density Cultivation and the Effect of Temperature on Myrosinase Expression
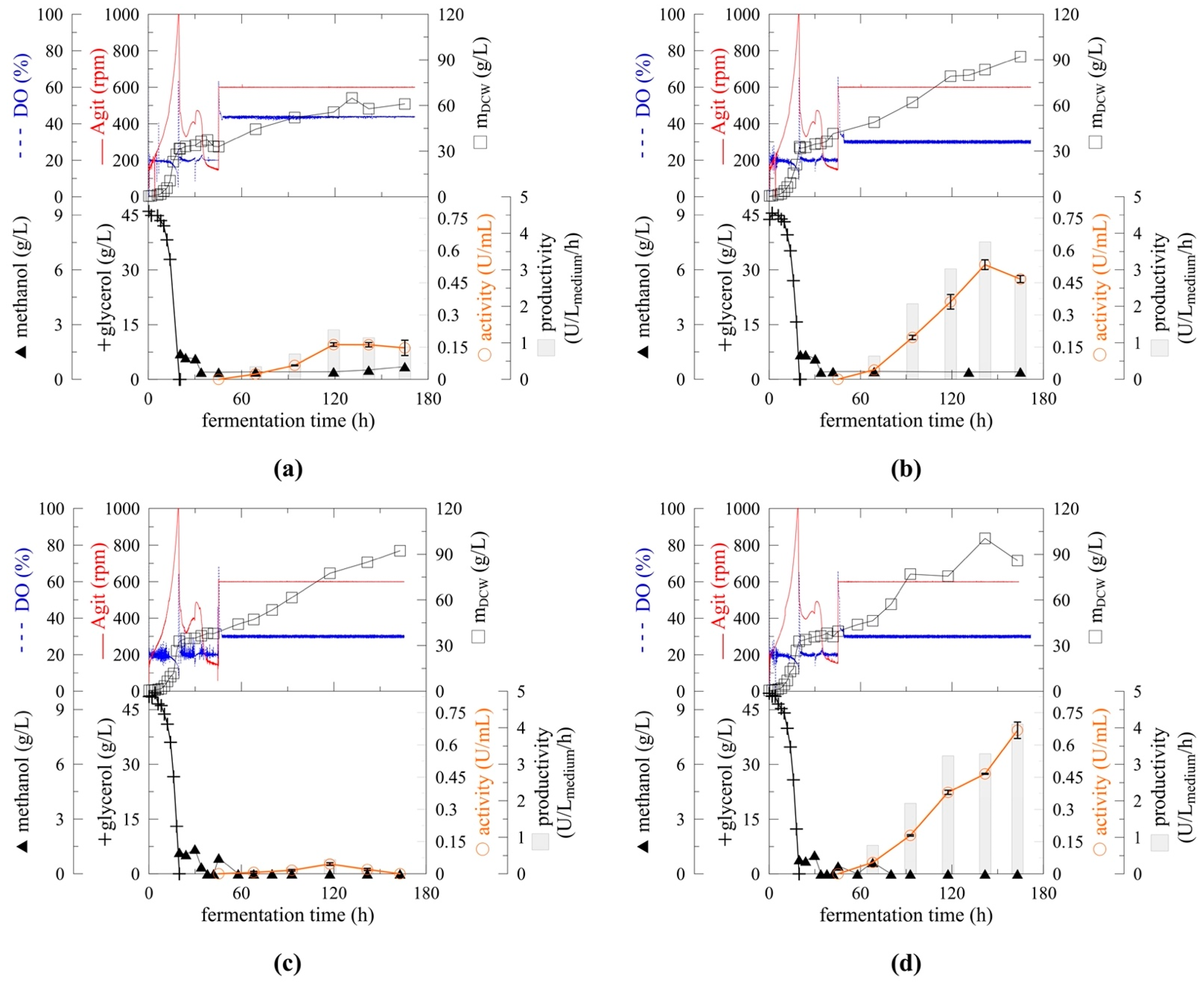
2.3. Cultivation Upscale
2.4. Effect of pH on Myrosinase Expression
2.5. Enzyme Parameters
2.5.1. Temperature and pH Optimum
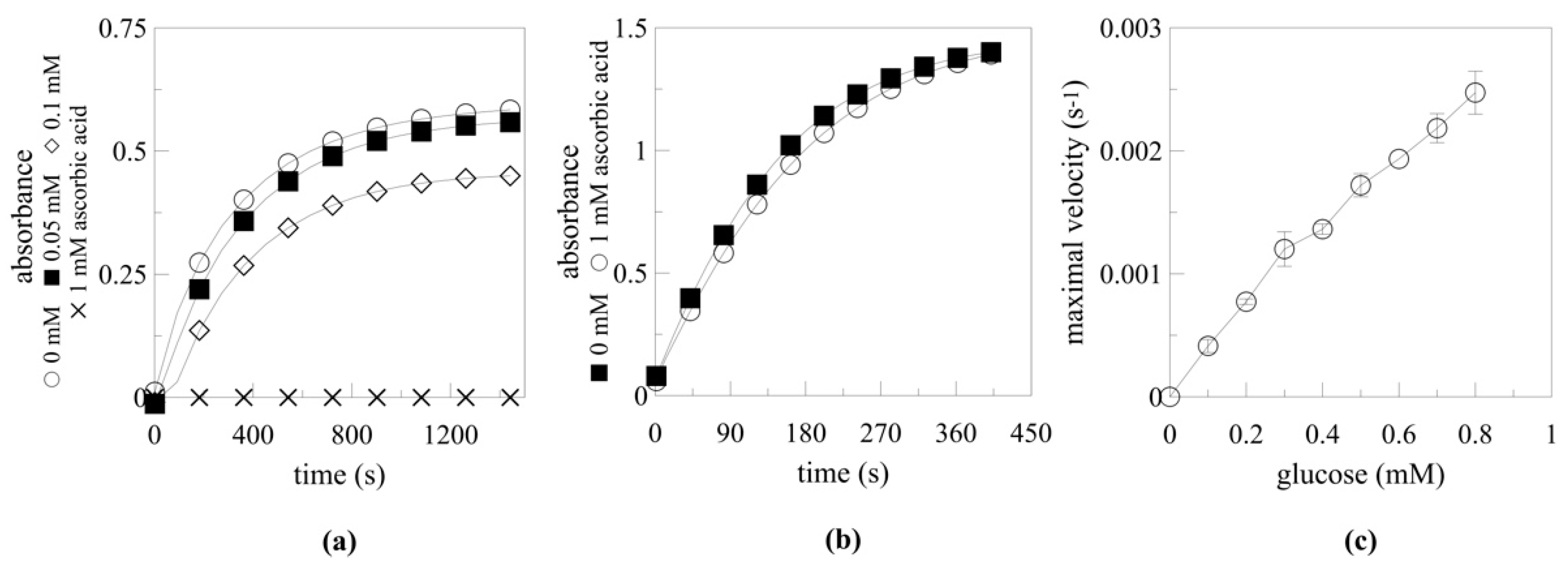
2.5.2. Myrosinase Activation by Ascorbic Acid
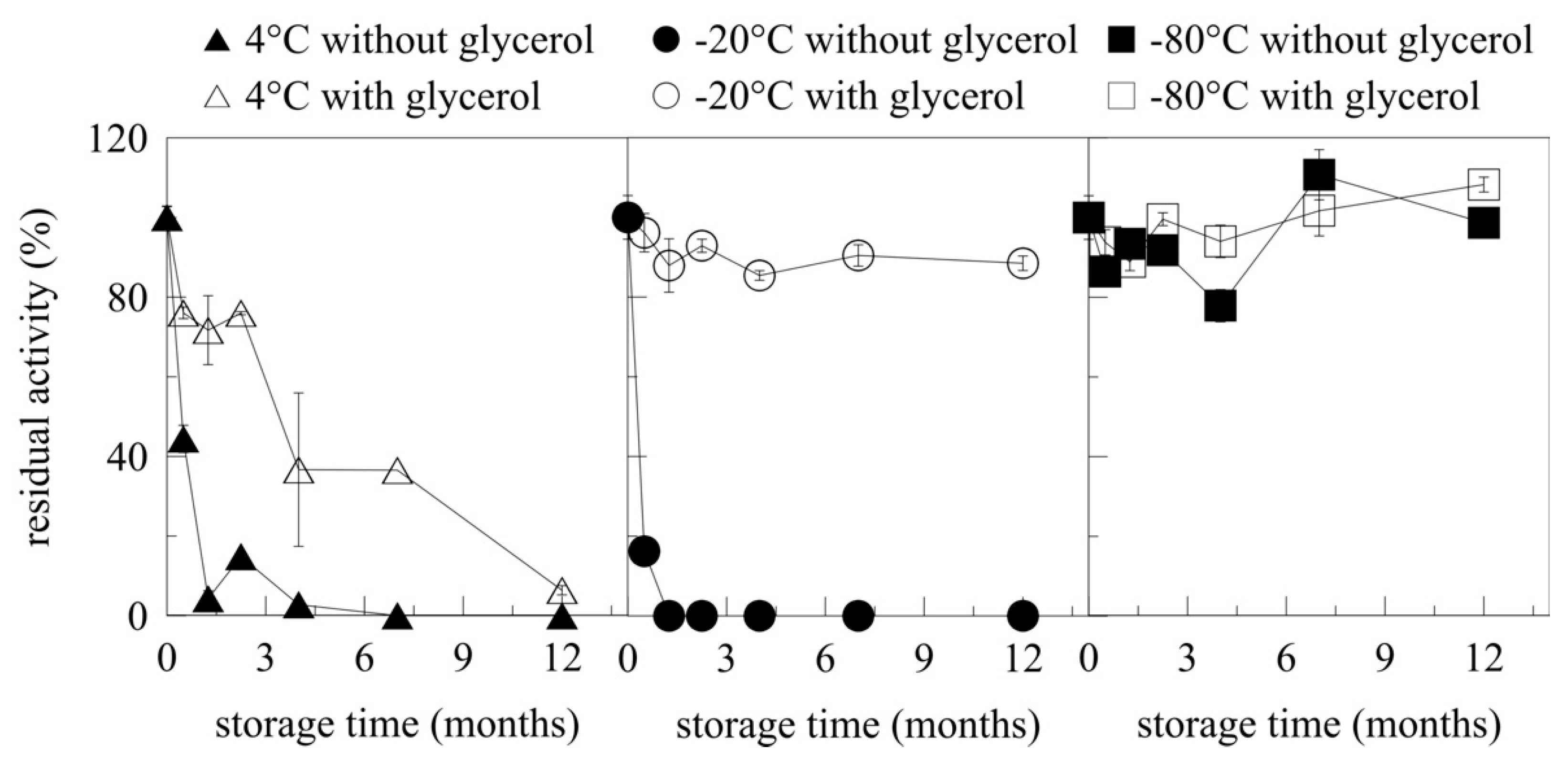
2.6. Storage of Myrosinase
3. Materials and Methods
3.1. Microorganism and Media
3.2. Recombinant Strain Preparation
3.3. Cultivation in Deep Well Plates
3.4. Microplate Assay
3.5. Flask Cultivation
3.6. High-Cell Density Cultivation and Scale-Up
3.7. Myrosinase Activity Assay
3.8. Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kissen, R.; Rossiter, J.T.; Bones, A.M. The ‘Mustard Oil Bomb’: Not so Easy to Assemble?! Localization, Expression and Distribution of the Components of the Myrosinase Enzyme System. Phytochem. Rev. 2009, 8, 69–86. [Google Scholar] [CrossRef]
- Navarro, S.L.; Li, F.; Lampe, J.W. Mechanisms of Action of Isothiocyanates in Cancer Chemoprevention: An Update. Food Funct. 2011, 2, 579–587. [Google Scholar] [CrossRef] [PubMed]
- Mitsiogianni, M.; Koutsidis, G.; Mavroudis, N.; Trafalis, D.T.; Botaitis, S.; Franco, R.; Zoumpourlis, V.; Amery, T.; Galanis, A.; Pappa, A.; et al. The Role of Isothiocyanates as Cancer Chemo-Preventive, Chemo-Therapeutic and Anti-Melanoma Agents. Antioxidants 2019, 8, 106. [Google Scholar] [CrossRef]
- Turrini, E.; Ferruzzi, L.; Fimognari, C. Natural Compounds to Overcome Cancer Chemoresistance: Toxicological and Clinical Issues. Expert Opin. Drug Metab. Toxicol. 2014, 10, 1677–1690. [Google Scholar] [CrossRef] [PubMed]
- Bhat, R.; Vyas, D. Myrosinase: Insights on Structural, Catalytic, Regulatory, and Environmental Interactions. Crit. Rev. Biotechnol. 2019, 39, 508–523. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Sun, X.; Tan, D.; Gong, S.; Meijer, J.; Zhang, J. The Two Non-Functional Myrosinase Genes TGG3 and TGG6 in Arabidopsis Are Expressed Predominantly in Pollen. Plant. Sci. 2009, 177, 371–375. [Google Scholar] [CrossRef]
- Chen, S.; Halkier, B.A. Functional Expression and Characterization of the Myrosinase MYR1 from Brassica napus in Saccharomyces cerevisiae. Protein Expr. Purif. 1999, 17, 414–420. [Google Scholar] [CrossRef] [PubMed]
- Härtel, F.V.; Brandt, A. Characterization of a Brassica napus Myrosinase Expressed and Secreted by Pichia pastoris. Protein Expr. Purif. 2002, 24, 221–226. [Google Scholar] [CrossRef]
- Cereghino, J.L.; Cregg, J.M. Heterologous Protein Expression in the Methylotrophic Yeast. FEMS Microbiol. Rev. 2000, 24, 45–66. [Google Scholar] [CrossRef]
- Daly, R.; Hearn, M.T.W. Expression of Heterologous Proteins in Pichia pastoris: A Useful Experimental Tool in Protein Engineering and Production. J. Mol. Recognit. 2005, 18, 119–138. [Google Scholar] [CrossRef]
- Cereghino, G.P.L.; Cereghino, J.L.; Ilgen, C.; Cregg, J.M. Production of Recombinant Proteins in Fermenter Cultures of the Yeast Pichia pastoris. Curr. Opin. Biotechnol. 2002, 13, 329–332. [Google Scholar] [CrossRef]
- Chung, W.C.; Huang, H.C.; Chiang, B.T.; Huang, H.C.; Huang, J.W. Inhibition of Soil-Borne Plant Pathogens by the Treatment of Sinigrin and Myrosinases Released from Reconstructed Escherichia coli and Pichia pastoris. Biocontrol. Sci. Technol. 2005, 15, 455–465. [Google Scholar] [CrossRef]
- Andersson, D.; Chakrabarty, R.; Bejai, S.; Zhang, J.; Rask, L.; Meijer, J. Myrosinases from Root and Leaves of Arabidopsis thaliana Have Different Catalytic Properties. Phytochemistry 2009, 70, 1345–1354. [Google Scholar] [CrossRef]
- Loebers, A.; Müller-Uri, F.; Kreis, W. A Young Root-Specific Gene (ArMY2) from Horseradish Encoding a MYR II Myrosinase with Kinetic Preference for the Root-Specific Glucosinolate Gluconasturtiin. Phytochemistry 2014, 99, 26–35. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Li, D.; Sun, X.; Zhu, Y.J.; Nong, H.; Zhang, J. Characterization of a Root-Specific β-Thioglucoside Glucohydrolase Gene in Carica papaya and Its Recombinant Protein Expressed in Pichia pastoris. Plant Sci. 2009, 177, 716–723. [Google Scholar] [CrossRef]
- Nong, H.; Zhang, J.-M.; Li, D.-Q.; Wang, M.; Sun, X.-P.; Zhu, Y.J.; Meijer, J.; Wang, Q.-H. Characterization of a Novel β-Thioglucosidase CpTGG1 in Carica papaya and Its Substrate-Dependent and Ascorbic Acid-Independent O-β-Glucosidase Activity. J. Integr. Plant. Biol. 2010, 52, 879–890. [Google Scholar] [CrossRef]
- Kleinwächter, M.; Selmar, D. A Novel Approach for Reliable Activity Determination of Ascorbic Acid Depending Myrosinases. J. Biochem. Biophys. Methods 2004, 59, 253–265. [Google Scholar] [CrossRef] [PubMed]
- Burmeister, W.P.; Cottaz, S.; Rollin, P.; Vasella, A.; Henrissat, B. High Resolution X-Ray Crystallography Shows That Ascorbate Is a Cofactor for Myrosinase and Substitutes for the Function of the Catalytic Base. J. Biol. Chem. 2000, 275, 39385–39393. [Google Scholar] [CrossRef] [PubMed]
- Petrovičová, T.; Markošová, K.; Hegyi, Z.; Smonou, I.; Rosenberg, M.; Rebroš, M. Co-Immobilization of Ketoreductase and Glucose Dehydrogenase. Catalysts 2018, 8, 168. [Google Scholar] [CrossRef]
- Cregg, J.M.; Cereghino, J.L.; Shi, J.; Higgins, D.R. Recombinant Protein Expression in Pichia pastoris. Mol. Biotechnol. 2000, 16, 23–52. [Google Scholar] [CrossRef]
- Markošová, K.; Weignerová, L.; Rosenberg, M.; Křen, V.; Rebroš, M. Upscale of Recombinant α-L-Rhamnosidase Production by Pichia pastoris MutS Strain. Front. Microbiol. 2015, 6, 1140. [Google Scholar] [CrossRef] [PubMed]
- Dragosits, M.; Stadlmann, J.; Albiol, J.; Baumann, K.; Maurer, M.; Gasser, B.; Sauer, M.; Altmann, F.; Ferrer, P.; Mattanovich, D. The Effect of Temperature on the Proteome of Recombinant Pichia pastoris. J. Proteome Res. 2009, 8, 1380–1392. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Yang, L.; Guo, Y.; Fang, F.; Wang, D.; Li, R.; Jiang, M.; Kang, W.; Ma, J.; Sun, J.; et al. High-Temperature Cultivation of Recombinant Pichia pastoris increases Endoplasmic Reticulum Stress and Decreases Production of Human Interleukin-10. Microb. Cell Fact. 2014, 13, 163. [Google Scholar] [CrossRef]
- Hong, F.; Meinander, N.Q.; Jönsson, L.J. Fermentation Strategies for Improved Heterologous Expression of Laccase in Pichia pastoris: Heterologous Expression of Laccase in Pichia pastoris. Biotechnol. Bioeng. 2002, 79, 438–449. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Z.; Xu, Q.; Du, G.; Hua, Z.; Liu, L.; Li, J.; Chen, J. Lowering Induction Temperature for Enhanced Production of Polygalacturonate Lyase in Recombinant Pichia pastoris. Process. Biochem. 2009, 44, 949–954. [Google Scholar] [CrossRef]
- Shi, X.; Karkut, T.; Chamankhah, M.; Alting-Mees, M.; Hemmingsen, S.M.; Hegedus, D. Optimal Conditions for the Expression of a Single-Chain Antibody (ScFv) Gene in Pichia pastoris. Protein Expr. Purif. 2003, 28, 321–330. [Google Scholar] [CrossRef]
- Mattanovich, D.; Gasser, B.; Hohenblum, H.; Sauer, M. Stress in Recombinant Protein Producing Yeasts. J. Biotechnol. 2004, 113, 121–135. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, R.; Wu, X. The Proteolytic Systems and Heterologous Proteins Degradation in the Methylotrophic Yeast Pichia pastoris. Ann. Microbiol. 2007, 57, 553–560. [Google Scholar] [CrossRef]
- Clare, J.J.; Romanes, M.A.; Rayment, F.B.; Rowedder, J.E.; Smith, M.A.; Payne, M.M.; Sreekrishna, K.; Henwood, C.A. Production of Mouse Epidermal Growth Factor in Yeast: High-Level Secretion Using Pichia pastoris Strains Containing Multiple Gene Copies. Gene 1991, 105, 205–212. [Google Scholar] [CrossRef]
- Lin, H.; Kim, T.; Xiong, F.; Yang, X. Enhancing the Production of Fc Fusion Protein in Fed-Batch Fermentation of Pichia pastoris by Design of Experiments. Biotechnol. Prog. 2007, 23, 621–625. [Google Scholar] [CrossRef]
- Zhou, C.; Tokuhisa, J.G.; Bevan, D.R.; Esen, A. Properties of β-Thioglucoside Hydrolases (TGG1 and TGG2) from Leaves of Arabidopsis thaliana. Plant. Sci. 2012, 191–192, 82–92. [Google Scholar] [CrossRef] [PubMed]
- Grossmann, G.A.; Terra, W.R. α-Galactosidases from the Larval Midgut of Tenebrio molitor (Coleoptera) and Spodoptera frugiperda (Lepidoptera). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2001, 128, 109–122. [Google Scholar] [CrossRef]
- Chen, C.C.; Guo, W.J.; Isselbacher, K.J. Rat Intestinal Trehalase. Studies of the Active Site. Biochem. J. 1987, 247, 715–724. [Google Scholar] [CrossRef] [PubMed]
- Terra, W.R.; Ferreira, C.; de Bianchi, A.G. Physical Properties and Tris Inhibition of an Insect Trehalase and a Thermodynamic Approach to the Nature of Its Active Site. Biochim. Biophys. Acta Enzymol. 1978, 524, 131–141. [Google Scholar] [CrossRef]
- Semenza, G.; Balthazar, A.-K. Steady-State Kinetics of Rabbit-Intestinal Sucrase. Kinetic Mechanism, Na+ Activation, Inhibition by Tris(Hydroxymethyl)Aminomethane at the Glucose Subsite. Eur. J. Biochem. 1974, 41, 149–162. [Google Scholar] [CrossRef] [PubMed]
- Fernández, D.; Boix, E.; Pallarès, I.; Avilés, F.X.; Vendrell, J. Structural and Functional Analysis of the Complex between Citrate and the Zinc Peptidase Carboxypeptidase A. Enzyme Res. 2011, 2011, 1–8. [Google Scholar] [CrossRef]
- Arsiccio, A.; Giorsello, P.; Marenco, L.; Pisano, R. Considerations on Protein Stability During Freezing and Its Impact on the Freeze-Drying Cycle: A Design Space Approach. J. Pharm. Sci. 2020, 109, 464–475. [Google Scholar] [CrossRef]
- Kolhe, P.; Amend, E.; Singh, S.K. Impact of Freezing on PH of Buffered Solutions and Consequences for Monoclonal Antibody Aggregation. Biotechnol. Prog. 2010, 26, 727–733. [Google Scholar] [CrossRef]
- Weis, R.; Luiten, R.; Skranc, W.; Schwab, H.; Wubbolts, M.; Glieder, A. Reliable High-Throughput Screening with by Limiting Yeast Cell Death Phenomena. FEMS Yeast Res. 2004, 5, 179–189. [Google Scholar] [CrossRef]
- Lin-Cereghino, J.; Wong, W.W.; Xiong, S.; Giang, W.; Luong, L.T.; Vu, J.; Johnson, S.D.; Lin-Cereghino, G.P. Condensed Protocol for Competent Cell Preparation and Transformation of the Methylotrophic Yeast Pichia pastoris. Biotechniques 2005, 38, 44–48. [Google Scholar] [CrossRef]
- Markošová, K.; Camattari, A.; Rosenberg, M.; Glieder, A.; Turner, N.J.; Rebroš, M. Cloning and Upscale Production of Monoamine Oxidase N (MAO-N D5) by Pichia pastoris. Biotechnol. Lett. 2018, 40, 127–133. [Google Scholar] [CrossRef]
- Tsao, R.; Yu, Q.; Potter, J.; Chiba, M. Direct and Simultaneous Analysis of Sinigrin and Allyl Isothiocyanate in Mustard Samples by High-Performance Liquid Chromatography. J. Agric. Food Chem. 2002, 50, 4749–4753. [Google Scholar] [CrossRef]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]

| Gene Donor | Gene | Expression Vector | Expression Host | Reference |
|---|---|---|---|---|
| Arabidopsis thaliana | TGG1, TGG2 | pHIL-S1 (extracellular) | P. pastoris GS115 (Mut+) | [12] |
| Arabidopsis thaliana | TGG1, TGG2 TGG4, TGG5 | pPIC3.5K (intracellular) | P. pastoris GS115 (Mut+) | [13] |
| Armoracia rusticana | ArMY2 | pPIC3.5K (intracellular) | P. pastoris GS115 (Mut+) | [14] |
| Brassica napus | MYR1 | pHIL-S1 (extracellular) | P. pastoris GS115 (Mut+) | [8] |
| Carica papaya | CpTGG2 | pPIC3.5 (intracellular) | P. pastoris GS115 (Mut+) | [15] |
| Carica papaya | CpTGG1 | pPIC3.5 (intracellular) | P. pastoris GS115 (Mut+) | [16] |
| Conditions | Total Protein (mg/L) | Maximal DCW (g/L) | Maximal Activity (U/mL) | Activity after 163 h (U/mL) | Maximal Productivity (U/Lmedium/h) | Productivity after 163 h (U/Lmedium/h) | |
|---|---|---|---|---|---|---|---|
| 1 | 30 °C, pH 5 | 267.3 | 64.9 | 0.16 | 0.15 | 1.35 | 0.89 |
| 2 | 20 °C, pH 5 | 256.1 | 92.05 | 0.53 | 0.47 | 3.02 | 2.83 |
| 3 | 20 °C, pH 4 | 383.0 | 92.35 | 0.045 | 0 | 0.39 | 0 |
| 4 | 20 °C, pH 6 | 286.5 | 100.5 | 0.67 | 0.67 | 4.10 | 4.10 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rosenbergová, Z.; Kántorová, K.; Šimkovič, M.; Breier, A.; Rebroš, M. Optimisation of Recombinant Myrosinase Production in Pichia pastoris. Int. J. Mol. Sci. 2021, 22, 3677. https://doi.org/10.3390/ijms22073677
Rosenbergová Z, Kántorová K, Šimkovič M, Breier A, Rebroš M. Optimisation of Recombinant Myrosinase Production in Pichia pastoris. International Journal of Molecular Sciences. 2021; 22(7):3677. https://doi.org/10.3390/ijms22073677
Chicago/Turabian StyleRosenbergová, Zuzana, Kristína Kántorová, Martin Šimkovič, Albert Breier, and Martin Rebroš. 2021. "Optimisation of Recombinant Myrosinase Production in Pichia pastoris" International Journal of Molecular Sciences 22, no. 7: 3677. https://doi.org/10.3390/ijms22073677
APA StyleRosenbergová, Z., Kántorová, K., Šimkovič, M., Breier, A., & Rebroš, M. (2021). Optimisation of Recombinant Myrosinase Production in Pichia pastoris. International Journal of Molecular Sciences, 22(7), 3677. https://doi.org/10.3390/ijms22073677






