Abstract
The first amino acid of a protein has an important influence on its metabolic stability. A number of ubiquitin ligases contain binding domains for different amino-terminal residues of their substrates, also known as N-degrons, thereby mediating turnover. This review summarizes, in an exemplary way, both older and more recent findings that unveil how destabilizing amino termini are generated. In most cases, a step of proteolytic cleavage is involved. Among the over 500 proteases encoded in the genome of higher eukaryotes, only a few are known to contribute to the generation of N-degrons. It can, therefore, be expected that many processing paths remain to be discovered.
1. Introduction
N-degrons are degradation signals that reside in the amino terminus of proteins [1,2,3,4,5]. Typically, the amino-terminal residue of N-degrons is essential for their recognition, but other residues in the amino-terminal region modulate the strength of the N-degron. Most N-degrons do not contain the start methionine. How these N-degrons are generated is still unclear in many cases. Considering that only a minor fraction of the over 500 proteases encoded by the average higher eukaryotic genome has been studied extensively, many proteolytic processing and maturation steps that generate N-degrons remain to be characterized. Fortunately, methods to analyze the amino-terminal structure of proteins have been developed and are being refined [6], so that substrates of endoproteolytic cleavage can be identified with increasing efficiency. Here, we review some of the existing data about how N-degrons are generated with an emphasis on plants. Since most knowledge in this respect has been accumulated in animal and fungal models, we will frequently refer to findings from these latter organisms as examples of what can be expected for the plant kingdom. N-degrons with acetylated or formylated amino termini are only briefly addressed at the end. Table 1 summarizes the amino termini we discuss regarding generation, further processing, and the expected impact on protein turnover.

Table 1.
Fate of proteins with the indicated amino-terminal residues.
N-degron recognition usually occurs at dedicated ubiquitin ligases that, with the help of ubiquitin-activating enzymes and ubiquitin-conjugating enzymes, covalently link a ubiquitin chain to the N-degron-carrying substrate. This step is followed by degradation through a large multimeric protease called the 26S proteasome. The process can be modulated by soluble ubiquitin chain-binding receptors. As described in Section 3, autophagy receptors have recently entered the stage, so an autophagy route for the turnover of proteins with certain N-degrons is also possible.
We deduced the amino acid frequencies of the first thirty residues in Arabidopsis thaliana proteins using representative gene models from Araport11 (Figure 1A). Regarding the second residue of open reading frames (ORFs), Table 2 lists the absolute numbers of proteins and relative frequency, which usually deviates from the general abundance of a particular amino acid. The subset of proteins with predicted cytoplasmic or nuclear localization (i.e., proteins with no amino-terminal targeting sequence; Figure 1B) differs from the complete protein set in the amino acid composition of the amino-terminal region. This subset of proteins is of particular interest because enzymes for N-degron-dependent protein turnover reside in the cytoplasm and the nucleus.
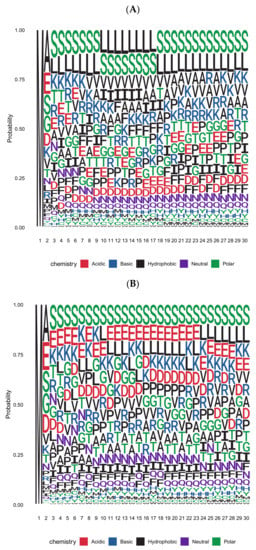
Figure 1.
Position-specific amino acid probability in the first thirty residues of Arabidopsis proteins represented as a sequence logo (in single letter code). (A) Sequence logo based on the complete set of nuclear-encoded proteins with one representative per gene. (B) Sequence logo based on the subset of proteins with predicted localization exclusively in cytoplasmic or nuclear compartments. Predictions were obtained from Araport11.

Table 2.
Abundance of amino termini in the predicted proteome of Arabidopsis 1.
As shown in the graphic summary of Figure 1, Leu and Ser dominate positions 3 to 23 when all proteins are considered. This is consistent with the fact that Leu and Ser are the most frequent amino acids in proteins (with ca. 9.5 and 9.1%, respectively; Table 2). In cytoplasmic/nuclear proteins, however, Leu is much less abundant. Amino acids with polar, charged, or small side chains dominate instead. The sequence following residue 23 is more similar between the two protein sets, and Leu is the second most abundant amino acid in both. One may argue that this section is already part of compactly folded domains, so the differences between organelle-targeted and cytoplasmic proteins disappear. Conversely, amino termini of up to 23 residues may be frequently exposed, making them amenable to processing events. In cases where this region is already part of a compact fold, processing has to be initiated on the nascent polypeptide. In the following, we first discuss the cleavage of amino-terminal methionine (Met) as a processing event followed by a number of other proteases with known roles in the generation of N-degrons.
2. Methionine Aminopeptidase as a Processing Enzyme towards Protein Destabilization
Methionine aminopeptidases (Met APs) are among the first enzymes to reach nascent amino termini at the ribosome. Several isoforms exist and are conserved among eukaryotes. Although isoforms may have distinct substrate preferences [7,8,9], they share the established cleavage specificity that the amino-terminal Met will be removed if the second residue has a small side chain, but it will not if the second residue has a large side chain.
Proteins with Cys as a second residue (small side chain) are processed in vivo by Met cleavage. A prominent group of plant regulatory proteins belong to the set: ERF-VII class transcription factors, ZPR2, and VRN2 [10,11,12,13]. After a Met AP removes the start methionine, there is another set of enzymes that uses amino-terminal Cys as substrate—the plant cysteine oxidases (PCOs [14]). These enzymes use molecular oxygen to convert Cys into cysteic acid. Once a side chain with negative charge is generated, enzymes of the Arg tRNA protein transferase (ATE) class add an Arg residue onto the modified amino terminus. ATE requires an acidic residue on substrate amino termini—i.e., its activity depends on the oxidation of Cys. The Arg-Cys(ox) terminus is recognized by ubiquitin ligases with a UBR domain [15]. All the characterized UBR domains bind amino termini with basic non-acetylated residues, such as Arg. If binding can be followed by ubiquitylation, which requires a spatially accessible internal Lys residue, the result is the turnover of the substrate by the ubiquitin proteasome system (UPS).
The Cys oxidation pathway has one important physiological input: PCOs have a relatively low affinity for molecular oxygen. This property converts them into oxygen sensors [16]. In tissues with low oxygen pressure, or under conditions of low oxygen—e.g., upon root submergence, amino-terminal Cys is not oxidized. As a consequence, the ensuing proteins remain metabolically stable [10,11]. Interestingly, a functional homolog of PCOs was recently described in humans [17], and this latter enzyme also confers oxygen dependence on substrate amino-terminal Cys oxidation. In plants, as well as in animals, there is another messenger input: the absence of nitric monoxide (NO) also prevents Met-Cys protein turnover [18,19]. The gaseous plant hormone ethylene, in turn, impacts NO levels in plants, allowing the modulation of the pathway by additional physiological and developmental inputs [20].
Amino-terminal Pro can be generated by Met cleavage via Met APs from proteins that start with Met-Pro. In budding yeast, a pathway was discovered that quickly removes several gluconeogenesis enzymes upon the addition of glucose to the medium [21]. The substrate recognition subunit (GID4) of the responsible multi-subunit GID ubiquitin ligase has a binding pocket for amino-terminal Pro. Ligase binding depends on additional residues in the amino-terminal region of potential substrates (amino acids 1–7 of yeast fructose bisphosphatase can act as transferrable N-degron). The ligase is conserved in animals and in plants (putative GID4 homolog of Arabidopsis: At2g37680 according to DIOPT 8.0 [22]). Interestingly, the gluconeogenetic enzymes that led to the discovery of the pathway in S. cerevisiae do not start with Pro in most other organisms, including the distantly related fungus K. lactis, but the preference of the ligase for amino-terminal Pro is conserved [23]. This implies that additional, possibly conserved, substrates remain to be discovered.
N-terminal Gly can also be generated via Met AP cleavage. A turnover pathway was recently described that may serve as backup to the N-terminal myristoylation of certain Gly amino termini. In case that linkage of the fatty acyl group to such amino termini fails, which would prevent membrane targeting, the protein is removed from the cell [24].
In insects as well as in mammals, a set of polypeptides with pro-apoptotic function has a conserved amino-terminal motif starting with Ala [25]. This motif can act as N-degron. Whereas the insect proteins are cytoplasmic proteins and processed via Met-AP, the mammalian proteins reside in mitochondria, and their N-degrons are generated by mitochondrial processing proteases. The activity and turnover route of these polypeptides are functionally linked to caspase activation and are discussed below (Section 4).
3. Processing by Met Aminopeptidases: Exceptions from the Rule
As indicated in Table 1, amino termini with small side chains are not the most prominent N-degrons, and no such case is known in plants so far. Of note, however, Knop and coworkers made a systematic analysis of amino-terminal processing in budding yeast [26]. They found that, if Met is followed by Asn, cleavage can occur by Met aminopeptidase. Similarly, Hwang and coworkers [27] identified Met AP-generated substrates resulting from the Met cleavage of Met-Asn and Met-Gln amino termini. It is likely that cleavage before Asn or Gln depends on the sequence context and is blocked by Met acetylation [26], so that only a fraction of the Met-Asn proteome loses the initiator Met. It is currently unclear whether this cleavage affects a potential substrate quantitatively, or whether only a fraction of a particular substrate gets cleaved. While no example from plants has been published so far, there is a general belief that Met AP specificities are conserved across kingdoms, and enzymes that convert Asn into Asp or Gln into Glu, as well as Arg transferase enzymes that add Arg to an acidic amino-terminal residue, are present in plants. Substrates for this pathway in plants may include proteins linked to biotic defense because Asn deamidase mutants display alterations in pathogen response [28].
4. Cleavage of Signal Peptides Can Be Followed by Escape of Proteins from the ER
Proteases that cleave off endoplasmic reticulum (ER) signal sequences frequently expose amino-terminal residues with large side chains. However, because N-degron binding ubiquitin ligases reside in the cytoplasm, this maturation step has no impact on protein stability. The ER can retro-translocate un- or misfolded proteins into the cytoplasm by a pathway called ER-associated degradation (ERAD [29,30]). Retro-translocated proteins are decorated with ubiquitin at the exit channel of retro-translocation (a SEC61 containing membrane protein complex) by dedicated membrane-associated ligases, so amino-terminal recognition may only play a minor role, or no role at all, in marking these proteins with ubiquitin.
A recent finding in mammalian cells, however, generates another link to N-degron recognition in the cytoplasm. Certain forms of stress result in transfer of the (normally) ER-resident chaperone immunoglobulin binding protein (BiP) into the cytoplasm. The protein is apparently functional (implying escape/extrusion from the ER in folded state), but has an amino-terminal Glu residue, emanating from the ER-resident signal peptide cleavage [31]. It is currently unclear how BiP and other proteins escape from the ER, but the exit path for unfolded proteins of ERAD is apparently not involved [32]. Once in the cytoplasm, BiP is arginylated at its amino terminus by Arg transferase ATE. While this amino-terminal processing generates a degron for a UBR domain containing ubiquitin ligases, there is a previously unexpected competition to this process. Two prominent autophagy receptors, p62 and NBR1, have so-called ZZ domains with a high affinity for amino-terminal Arg [33]. One may speculate that under cellular stress, the chaperone BiP is recruited from the ER to bring (cytoplasmic or even ER-generated) misfolded clients. Thereafter, by the binding of BiP to ZZ domains of autophagy receptors, the clients are channeled into autophagy. Interestingly, the occupation of the ZZ domain of p62 promotes its association into larger p62 complexes, which are known to trigger autophagic vesicle formation. Occupancy of these sites by cytoplasmic N-degron proteins can thereby induce ER-phagy [34]. One may speculate that induction of autophagy by accumulation of N-degron substrates, as may occur in the case of suboptimal turnover via the UPS, is not restricted to ER-phagy. While the relevance of the ER protein-initiated turnover route for plants remains to be demonstrated, it should be emphasized that the plant functional homologs of p62 and NBR1, called AtNBR1 in Arabidopsis [35,36] and Joka2 in tobacco [37], contain a ZZ domain and are thus potential receptors for N-degrons. Plant signal peptidases can generate amino-terminal residues that are destabilizing when exposed in the cytoplasm (for instance, Arabidosis BiP2 (identifier At5g42020) is processed in the ER to start with residue Lys 28 according to the prediction, SignalP-5.0 http://www.cbs.dtu.dk/services/SignalP/ (accessed on 26 February 2021) [38], and experimental data, Plant PTM Viewer https://www.psb.ugent.be/webtools/ptm-viewer/ (accessed on 10 March 2021) [39]).
5. Caspases, Metacaspases, and Other Endoproteases Related to Cell Death Programs
In mammals and insects, the cell death program apoptosis is, in several ways, interdigitated with N-degron-based turnover. The central elements of the process are proteases called caspases. Their activity in relation to a broad range of substrates leads to cell death. Caspases require proteolytic processing for activation. As the inactive precursors reside in the cytoplasm, several layers of activity containment exist and ensure that cell death is not initiated inappropriately. In particular, the ubiquitin ligases of the inhibitor of apoptosis (IAP) class can associate with caspases to accomplish their degradation. However, at later stages of caspase activation, caspases cleave IAPs to generate N-degrons [40], resulting in IAP degradation. This activity removes one of the breaks on the cell death program [41]. Another layer of regulation is exerted on IAPs. A number of polypeptides of Drosophila (reaper, grim, and hid, in particular) and of mammals (called Smac and Diablo) have a conserved N-terminal motif starting with Ala [25] that binds to so-called Baculovirus IAP Repeat (BIR) domains. IAPs contain BIR domains, and the binding of the mentioned polypeptides prevents association with (and thereby the inactivation of) caspases. It is possible that these polypeptides are eventually ubiquitylated by IAPs and thereby channeled into degradation, but this latter process is probably slow, so that prevention of caspase binding is the dominating effect. As a consequence, grim and related factors are pro-apoptotic elements. It is shown in Drosophila that another BIR domain containing protein called Bruce binds the Ala N-degron and initiates turnover of grim, reaper, and related pro-apoptotic proteins [42]. Bruce is an unusual ubiquitin ligase because it does not contain a RING domain, but a ubiquitin-conjugating enzyme domain instead.
Caspases contain a Cys active site and generally cleave at the carboxyl-terminal end of internal Asp residues. Depending on the family member, substrates are cleaved at the carboxyl end of sequence motifs such as DEVD or LEHD (single letter code [43]). The newly generated amino terminus of the C-terminal fragment (P1’ position; the peptide bond between the so-called P1 and P1’ positions is cleaved, resulting in a fragment with C-terminal P1 residue, and another one with N-terminal P1’ residue) is more variable and thus often encompasses a destabilizing residue. For this reason, these proteases increase the cellular load of N-degron-containing proteins. There is evidence that the UPS, via N-degrons, removes critical caspase substrates at early stages of caspase activation and thereby antagonizes cell death [44,45]. However, it is currently not known whether N-degron-dependent turnover plays a role during the later stages of programmed cell death processes. This is because, in mammals, the proteasome is also a substrate for caspase cleavage, and this leads to proteasome inactivation [46]. Thus, although the amount of N-degron bearing proteins further increases with ongoing apoptosis, other routes of protein degradation may take over for cell clearance.
In plants, close homologs of caspases do not exist, but metacaspases have a related role [47,48]. Plant metacaspases do not have the Asp specificity for the P1 site, but rather cleave C-terminal to Arg or Lys. Tsiatsiani et al. [49] found, however, a preference for Asp and Glu in the P1´ site, indicating that plant metacaspases also generate N-degrons. Interestingly, some plant proteases with a Ser active site apparently share a preference for Asp or Asn in the P1 position, and may therefore have a cleavage specificity that is more similar to animal caspases. For instance, the vacuolar processing enzyme (VPE) class (also called legumains), recognizes cleavage sites with Asn or Asp in P1 position [50,51]. One critical feature of these enzymes is that they normally reside in the extracellular space (a Ser protease called phytaspase [52]) or in the vacuole (VPEs), so protein fragments generated at the normal locale of these enzymes are sequestered from the cytoplasm with its active N-degron pathway enzymes. However, both types of proteases have been implicated in cell death processes, whereby these enzymes do reach the cytoplasm [52,53]. Finally, the PBA1 subunit of the proteasome, a Thr protease, was also reported to have a caspase-like cleavage specificity in vitro, and its absence impacts on bacterially induced plant cell death [54]. In the intact proteasome, this activity is shielded from the cytoplasm, and PBA1 is produced as an inactive precursor prior to assembly into proteasomes. Therefore, this enzyme also needs to be released into the cytoplasm by a currently unknown mechanism before it can generate N-degron bearing proteins.
Similarly, the plant-specific cell death, senescence, has been linked to endoproteases such as those discussed above. With many indications of varying cellular localization, one may assume that the abundance and/or localization, but not the cleavage specificity, of these proteases depends on the biological context. It is interesting to mention, in this regard, that a mutation in the Arg transferase ATE1, which converts Asp and Glu amino termini into Arg-Asp and Arg-Glu termini, has a delayed senescence phenotype [55]. However, Arg transfer to termini generated by senescence-associated endopeptidases has not been demonstrated so far.
6. Bacterial Effectors
A number of bacterial effectors that are injected into the plant cytoplasm via a type three secretion system contain protease activity. Some of them show activity as de-SUMOylating proteases that release SUMO from conjugates (XopD [56]), but many effectors with protease activity may be active against a broader range of plant proteins, often with key roles in plant defense. A well-known example is AvrRpt2. This protease targets an important component of basal immunity, RIN4 [57,58]. Released fragments have the destabilizing neo-N-termini Asn and Asp [59] (see also Table 1). While these RIN fragments are metabolically unstable, they are apparently not stabilized in the prt6 N-degron ubiquitin ligase mutant. However, AvrRpt2 also cleaves related proteins, such as other members of the NOI domain family to which RIN4 belongs, and some of the resulting fragments accumulate in N-degron turnover mutants such as prt6 [59]. It is therefore possible that RIN 4 and some of its NOI domain protein relatives contain additional cryptic degradation signals that are exposed by endoproteolytic cleavage.
7. Other Specialized Endoproteases
All eukaryotes contain separases, which are necessary to cleave cohesins at the end of mitosis, but they may have other substrates, as well [60]. It has been shown that failure of N-degron-based removal of cohesin fragments after separase cleavage decreases the precision of cell cycle events [61].
In mammals, the calcium-activated proteases, calpains, can also generate N-degrons [62]. Plants have a single copy gene related to calpains—defective kernel 1 (DEK1). DEK1 has an essential function in growth coordination between epidermis and inner leaf cell layers [63]. DEK1 is a fusion of a calpain domain to a transmembrane transporter that may be a touch-sensitive ion channel. DEK1 is functionally coupled to a Ca transporter, but Ca transport activity may reside in a separate polypeptide [64,65]. Activated DEK1 undergoes auto-cleavage, but the released protease may still remain associated with membranes. Unfortunately, the cleavage specificity of DEK1 has not been elucidated yet, and no substrates are known so far.
8. No Proteolytic Processing of Polypeptides Emerging from the Ribosome: Can Met Be a Destabilizing Residue If Followed by an Amino Acid with Hydrophobic Side Chain?
The prototypic N-degron ubiquitin ligase, Ubr1 of S. cerevisiae, has a binding pocket for hydrophobic amino termini, such as Leu, called ClpS homology domain [66]. Animal homologs have a similar domain called N domain [67]. This domain is apparently not conserved in plant homologs of UBR1 type ubiquitin ligases, and binding proteins for aliphatic hydrophobic residues remain to be discovered. For Ubr1, however, protein substrates binding to the ClpS homology domain have been described that start with Met, followed by a hydrophobic amino acid [68]. Another yeast ubiquitin ligase, Doa10, also seems to be involved in Met-Φ (Φ symbolizes a bulky hydrophobic amino acid) substrate turnover [26]. Because more than ten percent of the Arabidopsis ORFs start with Met-Φ (Table 2), it is highly likely that only a fraction of them are substrates for rapid turnover. The additional selection criteria for this class of substrates remain to be determined. All in all, we should be open to the possibility that the selection process for hydrophobic amino termini as N-degrons in plants may also include a fraction of the Met-Φ proteome.
9. Blocking the Alpha-Amino Group Can Generate a Distinct Set of N-Degrons
All the abovementioned amino termini carry a free, positively charged alpha amino group. In contrast, modification by acetylation or by formylation abrogates this positive charge, lowering affinity for the above-mentioned binding pockets on ubiquitin ligases or autophagy substrate adaptors. Nonetheless, N-degrons with a blocked first residue have been discovered, fueling dedicated turnover pathways.
In baker´s yeast, ubiquitin ligases Doa10 and Not4 mediate the degradation of certain proteins dependent on an acetylated first residue, making these ligases key components in the recognition of the specific class of Ac/N-degrons [69]. In plants as in yeast, amino-terminal acetylation does not destabilize the bulk of cytoplasmic proteins [70]. However, this does not rule out the existence of specific plant proteins with Ac/N-degrons because both Doa10 and Not4 [71], in particular, have homologs in plants.
Similarly, proteins in yeast can be modified upon stress or starvation on N-terminal Met residues by formylation, and ubiquitin ligase Psh1 is reported to mediate the removal of formylated proteins from the cytoplasm [72]. Here, again, plants encode proteins related to the recognin (ORTH/VIM proteins [73]), but functional connections to fMet N-degron generation or degradation remain to be demonstrated in plants. The further discussion of these two pathways is beyond the scope of this review.
In conclusion, the elucidation of cleavage specificities of currently uncharacterized proteases would reveal a potential role for protein stability. Even for established enzymes such as Met APs, it would be important to obtain additional, quantitative information regarding cleavage preferences (e.g., in the context of the cleavages described in Section 3).
Furthermore, progress is expected to come from the proteomic identification of protein fragments [6]. Once the occurrence of a certain cleavage pattern has been established, experiments to identify the processing path can be initiated. Alternatively, the over-expression of particular proteases, followed by proteome analysis, is also a promising strategy [49]. As a third route to pathway elucidation, the further characterization of degradation components, particularly the affinities of N-degron binding pockets in ubiquitin ligases and autophagy receptors for certain amino termini, can also help us to fill in the knowledge gaps.
Author Contributions
A.B. was mainly responsible for text writing, N.W. contributed to text writing, Figure design, corrections, and literature research. M.N. performed bioinformatics searches. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Austrian Science Fund FWF (grants P31114 and F7904-B to A.B.).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data supporting reported results can be found in the quoted references.
Acknowledgments
The authors apologize for quoting only a fraction of interesting publications on the topic. Open Access Funding by the Austrian Science Fund (FWF).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Dissmeyer, N. Conditional protein function via N-degron pathway-mediated proteolysis in stress physiology. Annu. Rev. Plant Biol. 2019, 70, 83–117. [Google Scholar] [CrossRef]
- Millar, A.H.; Heazlewood, J.L.; Giglione, C.; Holdsworth, M.J.; Bachmair, A.; Schulze, W.X. The scope, functions, and dynamics of posttranslational modifications. Annu. Rev. Plant Biol. 2019, 70, 119–151. [Google Scholar] [CrossRef]
- Varshavsky, A. N-degron and C-degron pathways of protein degradation. Proc. Natl. Acad. Sci. USA 2019, 116, 358–366. [Google Scholar] [CrossRef]
- Tasaki, T.; Sriram, S.M.; Park, K.S.; Kwon, Y.T. The N-end rule pathway. Annu. Rev. Biochem. 2012, 81, 261–289. [Google Scholar] [CrossRef]
- Gibbs, D.J.; Bacardit, J.; Bachmair, A.; Holdsworth, M.J. The eukaryotic N-end rule pathway: Conserved mechanisms and diverse functions. Trends Cell Biol. 2014, 24, 603–611. [Google Scholar] [CrossRef]
- Perrar, A.; Dissmeyer, N.; Huesgen, P.F. New beginnings and new ends: Methods for large-scale characterization of protein termini and their use in plant biology. J. Exp. Bot 2019, 70, 2021–2038. [Google Scholar] [CrossRef] [PubMed]
- Bradshaw, R.A.; Brickey, W.W.; Walker, K.W. N-terminal processing: The methionine aminopeptidase and N-alpha-acetyl transferase families. Trends Biochem. Sci. 1998, 23, 263–267. [Google Scholar] [CrossRef]
- Giglione, C.; Boularot, A.; Meinnel, T. Protein N-terminal methionine excision. Cell Mol. Life Sci. 2004, 61, 1455–1474. [Google Scholar] [CrossRef] [PubMed]
- Ross, S.; Giglione, C.; Pierre, M.; Espagne, C.; Meinnel, T. Functional and developmental impact of cytosolic protein N-terminal methionine excision in Arabidopsis. Plant Physiol. 2005, 137, 623–637. [Google Scholar] [CrossRef]
- Gibbs, D.J.; Lee, S.C.; Isa, N.M.; Gramuglia, S.; Fukao, T.; Bassel, G.W.; Correia, C.S.; Corbineau, F.; Theodoulou, F.L.; Bailey-Serres, J.; et al. Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature 2011, 479, 415–418. [Google Scholar] [CrossRef]
- Licausi, F.; Kosmacz, M.; Weits, D.A.; Giuntoli, B.; Giorgi, F.M.; Voesenek, L.A.; Perata, P.; van Dongen, J.T. Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature 2011, 479, 419–422. [Google Scholar] [CrossRef]
- Gibbs, D.J.; Tedds, H.M.; Labandera, A.M.; Bailey, M.; White, M.D.; Hartman, S.; Sprigg, C.; Mogg, S.L.; Osborne, R.; Dambire, C.; et al. Oxygen-dependent proteolysis regulates the stability of angiosperm polycomb repressive complex 2 subunit VERNALIZATION 2. Nat. Commun. 2018, 9, 5438. [Google Scholar] [CrossRef]
- Weits, D.A.; Kunkowska, A.B.; Kamps, N.C.W.; Portz, K.M.S.; Packbier, N.K.; Nemec Venza, Z.; Gaillochet, C.; Lohmann, J.U.; Pedersen, O.; van Dongen, J.T.; et al. An apical hypoxic niche sets the pace of shoot meristem activity. Nature 2019, 569, 714–717. [Google Scholar] [CrossRef] [PubMed]
- Weits, D.A.; Giuntoli, B.; Kosmacz, M.; Parlanti, S.; Hubberten, H.M.; Riegler, H.; Hoefgen, R.; Perata, P.; van Dongen, J.T.; Licausi, F. Plant cysteine oxidases control the oxygen-dependent branch of the N-end-rule pathway. Nat. Commun. 2014, 5, 3425. [Google Scholar] [CrossRef] [PubMed]
- Choi, W.S.; Jeong, B.C.; Joo, Y.J.; Lee, M.R.; Kim, J.; Eck, M.J.; Song, H.K. Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases. Nat. Struct. Mol. Biol. 2010, 17, 1175–1181. [Google Scholar] [CrossRef] [PubMed]
- White, M.D.; Kamps, J.J.A.G.; East, S.; Taylor Kearney, L.J.; Flashman, E. The plant cysteine oxidases from Arabidopsis thaliana are kinetically tailored to act as oxygen sensors. J. Biol. Chem. 2018, 293, 11786–11795. [Google Scholar] [CrossRef] [PubMed]
- Masson, N.; Keeley, T.P.; Giuntoli, B.; White, M.D.; Puerta, M.L.; Perata, P.; Hopkinson, R.J.; Flashman, E.; Licausi, F.; Ratcliffe, P.J. Conserved N-terminal cysteine dioxygenases transduce responses to hypoxia in animals and plants. Science 2019, 365, 65–69. [Google Scholar] [CrossRef]
- Hu, R.G.; Sheng, J.; Qi, X.; Xu, Z.; Takahashi, T.T.; Varshavsky, A. The N-end rule pathway as a nitric oxide sensor controlling the levels of multiple regulators. Nature 2005, 437, 981–986. [Google Scholar] [CrossRef]
- Gibbs, D.J.; Md Isa, N.; Movahedi, M.; Lozano-Juste, J.; Mendiondo, G.M.; Berckhan, S.; Marin-de la Rosa, N.; Vicente Conde, J.; Sousa Correia, C.; Pearce, S.P.; et al. Nitric Oxide Sensing in Plants Is Mediated by Proteolytic Control of Group VII ERF Transcription Factors. Mol. Cell 2014, 53, 369–379. [Google Scholar] [CrossRef]
- Hartman, S.; Liu, Z.; van Veen, H.; Vicente, J.; Reinen, E.; Martopawiro, S.; Zhang, H.; van Dongen, N.; Bosman, F.; Bassel, G.W.; et al. Ethylene-mediated nitric oxide depletion pre-adapts plants to hypoxia stress. Nat. Commun. 2019, 10, 4020. [Google Scholar] [CrossRef]
- Chen, S.-J.; Wu, X.; Wadas, B.; Oh, J.-H.; Varshavsky, A. An N-end rule pathway that recognizes proline and destroys gluconeogenic enzymes. Science 2017, 355, eaal3655. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Flockhart, I.; Vinayagam, A.; Bergwitz, C.; Berger, B.; Perrimon, N.; Mohr, S.E. An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinform. 2011, 12, 357. [Google Scholar] [CrossRef]
- Chen, S.J.; Melnykov, A.; Varshavsky, A. Evolution of Substrates and Components of the Pro/N-Degron Pathway. Biochemistry 2020, 59, 582–593. [Google Scholar] [CrossRef] [PubMed]
- Timms, R.T.; Zhang, Z.; Rhee, D.Y.; Harper, J.W.; Koren, I.; Elledge, S.J. A glycine-specific N-degron pathway mediates the quality control of protein N-myristoylation. Science 2019, 365, eaaw4912. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y. A conserved motif: Potentiating apoptosis through IAP-binding. Cell Death Differ. 2002, 9, 93–95. [Google Scholar] [CrossRef] [PubMed]
- Kats, I.; Khmelinskii, A.; Kschonsak, M.; Huber, F.; Knieß, R.A.; Bartosik, A.; Knop, M. Mapping Degradation Signals and Pathways in a Eukaryotic N-terminome. Mol. Cell 2018, 70, 488–501. [Google Scholar] [CrossRef]
- Nguyen, K.T.; Kim, J.M.; Park, S.E.; Hwang, C.S. N-terminal methionine excision of proteins creates tertiary destabilizing N-degrons of the Arg/N-end rule pathway. J. Biol. Chem. 2019, 294, 4464–4476. [Google Scholar] [CrossRef]
- Vicente, J.; Mendiondo, G.M.; Pauwels, J.; Pastor, V.; Izquierdo, Y.; Naumann, C.; Movahedi, M.; Rooney, D.; Gibbs, D.J.; Smart, K.; et al. Distinct branches of the N-end rule pathway modulate the plant immune response. New Phytol. 2018, 221, 988–1000. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Yu, F.; Xie, Q. Insights into endoplasmic reticulum-associated degradation in plants. New Phytol. 2020, 226, 345–350. [Google Scholar] [CrossRef]
- Hwang, J.; Qi, L. Quality control in the endoplasmic reticulum: Crosstalk between ERAD and UPR pathways. Trends Biochem. Sci. 2018, 43, 593–605. [Google Scholar] [CrossRef]
- Cha-Molstad, H.; Sung, K.S.; Hwang, J.; Kim, K.A.; Yu, J.E.; Yoo, Y.D.; Jang, J.M.; Han, D.H.; Molstad, M.; Kim, J.G.; et al. Amino-terminal arginylation targets endoplasmic reticulum chaperone BiP for autophagy through p62 binding. Nat. Cell Biol. 2015, 17, 917–929. [Google Scholar] [CrossRef]
- Shim, S.M.; Choi, H.R.; Sung, K.W.; Lee, Y.J.; Kim, S.T.; Kim, D.; Mun, S.R.; Hwang, J.; Cha-Molstad, H.; Ciechanover, A.; et al. The endoplasmic reticulum-residing chaperone BiP is short-lived and metabolized through N-terminal arginylation. Sci. Signal. 2018, 11, eaan0630. [Google Scholar] [CrossRef] [PubMed]
- Kwon, D.H.; Park, O.H.; Kim, L.; Jung, Y.O.; Park, Y.; Jeong, H.; Hyun, J.; Kim, Y.K.; Song, H.K. Insights into degradation mechanism of N-end rule substrates by p62/SQSTM1 autophagy adapter. Nat. Commun. 2018, 9, 3291. [Google Scholar] [CrossRef] [PubMed]
- Ji, C.H.; Kim, H.Y.; Heo, A.J.; Lee, S.H.; Lee, M.J.; Kim, S.B.; Srinivasrao, G.; Mun, S.R.; Cha-Molstad, H.; Ciechanover, A.; et al. The N-Degron Pathway Mediates ER-phagy. Mol. Cell 2019, 75, 1058–1072. [Google Scholar] [CrossRef] [PubMed]
- Ji, C.; Zhou, J.; Guo, R.; Lin, Y.; Kung, C.-H.; Hu, S.; Ng, W.Y.; Zhuang, X.; Jiang, L. AtNBR1 Is a Selective Autophagic Receptor for AtExo70E2 in Arabidopsis. Plant Physiol. 2020, 184, 777–791. [Google Scholar] [CrossRef]
- Svenning, S.; Lamark, T.; Krause, K.; Johansen, T. Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy 2011, 7, 993–1010. [Google Scholar] [CrossRef]
- Zientara-Rytter, K.; Sirko, A. Significant role of PB1 and UBA domains in multimerization of Joka2, a slelctive autophagy cargo receptor from tobacco. Front. Plant Sci. 2014, 5, 13. [Google Scholar] [CrossRef]
- Almagro-Armenteros, J.J.; Sonderby, C.K.; Petersen, K.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Willems, P.; Horne, A.; Van Parys, T.; Goormachtig, S.; De Smet, I.; Botzki, A.; Van Breusegem, F.; Gevaert, K. The Plant PTM Viewer, a central resource for exploring plant protein modifications. Plant J. 2019, 99, 752–762. [Google Scholar] [CrossRef] [PubMed]
- Ditzel, M.; Wilson, R.; Tenev, T.; Zachariou, A.; Paul, A.; Deas, E.; Meier, P. Degradation of DIAP1 by the N-end rule pathway is essential for regulating apoptosis. Nat. Cell Biol. 2003, 5, 467–473. [Google Scholar] [CrossRef] [PubMed]
- Vasudevan, D.; Ryoo, H.D. Regulation of Cell Death by IAPs and Their Antagonists. Curr. Top. Dev. Biol. 2015, 114, 185–208. [Google Scholar]
- Domingues, C.; Ryoo, H.D. Drosophila BRUCE inhibits apoptosis through non-lysine ubiquitination of the IAP-antagonist REAPER. Cell Death Differ. 2012, 19, 470–477. [Google Scholar] [CrossRef]
- Kumar, S.; van Raam, B.J.; Salvesen, G.S.; Cieplak, P. Caspase cleavage sites of the human proteome: CaspDB, a database of predicted substrates. PLoS ONE 2014, 9, e110539. [Google Scholar] [CrossRef] [PubMed]
- Piatkov, K.I.; Brower, C.S.; Varshavsky, A. The N-end rule pathway counteracts cell death by destroying proapoptotic protein fragments. Proc. Natl. Acad. Sci. USA 2012, 109, E1839–E1847. [Google Scholar] [CrossRef] [PubMed]
- Tenev, T.; Ditzel, M.; Zachariou, A.; Meier, P. The antiapoptotic activity of insect IAPs requires activation by an evolutionarily conserved mechanism. Cell Death Differ. 2007, 14, 1191–1201. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.-M.; Butterwirth, M.; MacFarlane, M.; Dubiel, W.; Ciechanover, A.; Cohen, G.M. Caspase activation inhibits proteasome function during apoptosis. Mol. Cell 2004, 14, 81–93. [Google Scholar] [CrossRef]
- Coll, N.S.; Vercammen, D.; Smidler, A.; Clover, C.; Van Breusegem, F.; Dangl, J.L.; Epple, P. Arabidopsis type I metacaspases control cell death. Science 2010, 330, 1393–1397. [Google Scholar] [CrossRef]
- Salguero-Linares, J.; Coll, N.S. Plant proteases in the control of the hypersensitive response. J. Exp. Bot. 2019, 70, 2087–2095. [Google Scholar] [CrossRef]
- Tsiatsiani, L.; Timmerman, E.; De Bock, P.J.; Vercammen, D.; Stael, S.; van de Cotte, B.; Staes, A.; Goethals, M.; Beunens, T.; Van Damme, P.; et al. The Arabidopsis Metacaspase9 Degradome. Plant Cell 2013, 25, 2831–2847. [Google Scholar] [CrossRef]
- Yamada, K.; Basak, A.K.; Goto-Yamada, S.; Tarnawska-Glatt, K.; Hara-Nishimura, I. Vacuolar processing enzymes in the plant life cyle. New Phytol. 2020, 226, 21–31. [Google Scholar] [CrossRef]
- Vorster, B.J.; Culli, C.A.; Kunert, K.J. Plant vacuolar processing enzymes. Front. Plant Sci. 2019, 10, 479. [Google Scholar] [CrossRef] [PubMed]
- Chichkova, N.V.; Shaw, J.; Galiullina, R.A.; Drury, G.E.; Tuzhikov, A.I.; Kim, S.H.; Kalkum, M.; Hong, T.B.; Goshikova, E.N.; Torrance, L.; et al. Phytaspase, a relocalisable cell death promoting plant protase with caspase specificity. EMBO J. 2010, 29, 1149–1161. [Google Scholar] [CrossRef] [PubMed]
- Hara-Nishimura, I.; Hatsugai, N. The role of vacuole in plant cell death. Cell Death Differ. 2011, 18, 1298–1304. [Google Scholar] [CrossRef]
- Hatsugai, N.; Iwasaki, S.; Tamura, K.; Kondo, M.; Fuji, K.; Ogasawara, K.; Nishimura, M.; Hara-Nishimura, I. A novel membrane fusion-mediated plant immunity against bacterial pathogens. Genes Dev. 2009, 23, 2496–2506. [Google Scholar] [CrossRef]
- Yoshida, S.; Ito, M.; Callis, J.; Nishida, I.; Watanabe, A. A delayed leaf senescence mutant is defective in arginyl-tRNA: Protein arginyltransferase, a component of the N-end rule pathway in Arabidopsis. Plant J. 2002, 32, 129–137. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-G.; Taylor, K.W.; Mudgett, M.B. Comparative analysis of the XopD type III secretion (T3S) effector family in plant pathogenic bacteria. Mol. Plant Pathol. 2011, 12, 715–730. [Google Scholar] [CrossRef] [PubMed]
- Mackey, D.; Belkhadir, Y.; Alonso, J.M.; Ecker, J.R.; Dangl, J.L. Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 2003, 112, 379–389. [Google Scholar] [CrossRef]
- Takemoto, D.; Jones, D.A. Membrane release and destabilization of Arabidopsis RIN4 following cleavage by Pseudomonas syringae AvrRpt2. Mol. Plant Microbe Interact. 2005, 18, 1258–1268. [Google Scholar] [CrossRef]
- Goslin, K.; Eschen-Lippold, L.; Naumann, C.; Linster, E.; Sorel, M.; Klecker, M.; de Marchi, R.; Kind, A.; Wirtz, M.; Lee, J.; et al. Differential N-end rule degradation of RIN4/NOI fragments generated by the AvrRpt2 effector protease. Plant Physiol. 2019, 180, 2272–2289. [Google Scholar] [CrossRef]
- Kumar, R. Separase: Function beyond cohesin cleavage and an emerging oncogene. J. Cell Biochem. 2017, 118, 1283–1299. [Google Scholar] [CrossRef] [PubMed]
- Rao, H.; Uhlmann, F.; Nasmyth, K.; Varshavsky, A. Degradation of a cohesin subunit by the N-end rule pathway is essential for chromosome stability. Nature 2001, 410, 955–959. [Google Scholar] [CrossRef]
- Piatkov, K.I.; Oh, J.H.; Liu, Y.; Varshavsky, A. Calpain-generated natural protein fragments as short-lived substrates of the N-end rule pathway. Proc. Natl. Acad. Sci. USA 2014, 111, E817–E826. [Google Scholar] [CrossRef]
- Johnson, K.L.; Faulkner, C.; Jeffree, C.E.; Ingram, G.C. The phytocalpain defective kernel 1 is a novel Arabidopsis growth regulator whose activity is regulated by proteolytic processing. Plant Cell 2008, 20, 2619–2630. [Google Scholar] [CrossRef] [PubMed]
- Tran, D.; Galletti, R.; Neumann, E.D.; Dubois, A.; Sharif-Naeini, R.; Geitmann, A.; Frachisse, J.M.; Hamant, O.; Ingram, G.C. A mechanosensitive Ca2+ channel activity is dependent on the developmental regulator DEK1. Nat. Commun. 2017, 8, 1009. [Google Scholar] [CrossRef] [PubMed]
- Guerringue, Y.; Thomine, S.; Frachisse, J.M. Sensing and transducing forces in plants with MSL10 and DEK1 mechanosensors. FEBS Lett. 2018, 592, 1968–1979. [Google Scholar] [CrossRef]
- Mogk, A.; Schmidt, R.; Bukau, B. The N-end rule pathway for regulated proteolysis: Prokaryotic and eukaryotic strategies. Trends Cell Biol. 2007, 17, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Tasaki, T.; Zakrzewska, A.; Dudgeon, D.D.; Jiang, Y.; Lazo, J.S.; Kwon, Y.T. The substrate recognition domains of the N-end rule pathway. J. Biol. Chem. 2009, 284, 1884–1895. [Google Scholar] [CrossRef]
- Kim, H.K.; Kim, R.R.; Oh, J.H.; Cho, H.; Varshavsky, A.; Hwang, C.S. The N-Terminal Methionine of Cellular Proteins as a Degradation Signal. Cell 2014, 156, 158–169. [Google Scholar] [CrossRef]
- Hwang, C.S.; Shemorry, A.; Varshavsky, A. N-terminal acetylation of cellular proteins creates specific degradation signals. Science 2010, 327, 973–977. [Google Scholar] [CrossRef] [PubMed]
- Linster, E.; Stephan, I.; Bienvenut, W.V.; Maple-Grodem, J.; Myklebust, L.M.; Huber, M.; Reichelt, M.; Sticht, C.; Geir Moller, S.; Meinnel, T.; et al. Downregulation of N-terminal acetylation triggers ABA-mediated drought responses in Arabidopsis. Nat. Commun. 2015, 6, 7640. [Google Scholar] [CrossRef]
- Bailey, M.; Ivanauskaite, A.; Grimmer, J.; Akintewe, O.; Payne, A.C.; Osborne, R.; Labandera, A.M.; Etherington, R.D.; Rantala, M.; Baginsky, S.; et al. The Arabidopsis NOT4A E3 ligase promotes PGR3 expression and regulates chloroplast translation. Nat. Commun. 2021, 12, 251. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-M.; Seok, O.-H.; Ju, S.; Heo, J.-E.; Yeom, J.; Kim, D.-S.; Yoo, J.-Y.; Varshavsky, A.; Lee, C.P.; Hwang, C.-S. Formyl-Methionine as an N-degron of a eukaryotic N-end rule pathway. Science 2018, 362, eaat0174. [Google Scholar] [CrossRef] [PubMed]
- Kraft, E.; Bostick, M.; Jacobsen, S.E.; Callis, J. ORTH/VIM proteins that regulate DNA methylation are functional ubiquitin E3 ligases. Plant J. 2008, 56, 704–715. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).