Structural Basis for the Function of the C-Terminal Proton Release Pathway in the Calcium Pump
Abstract
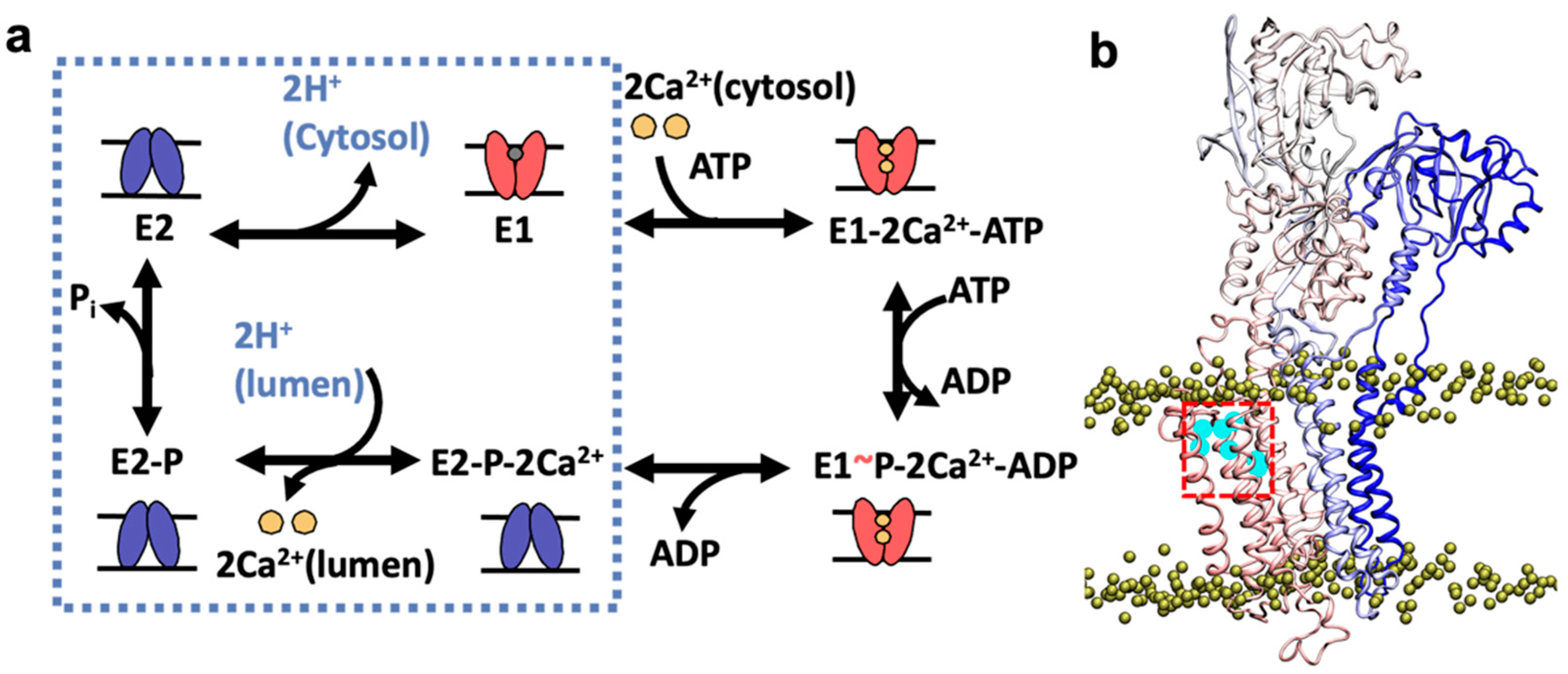
1. Introduction
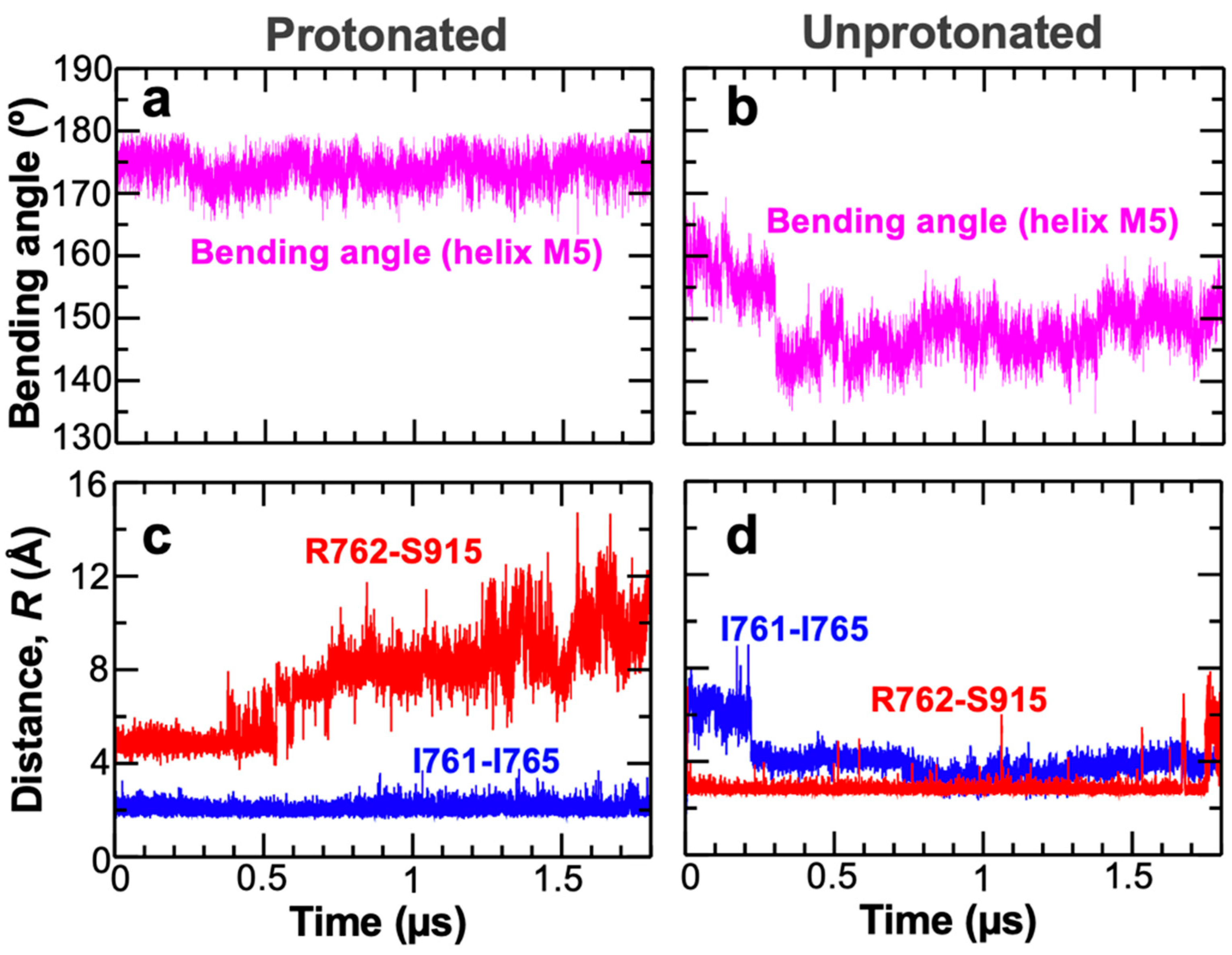
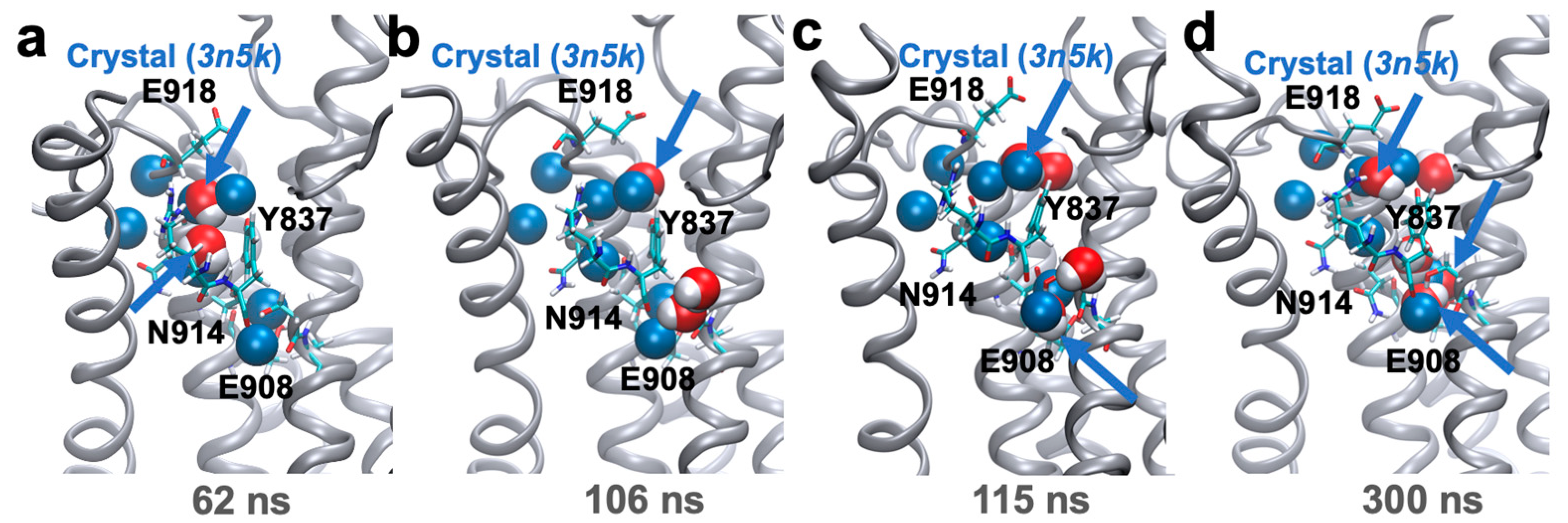
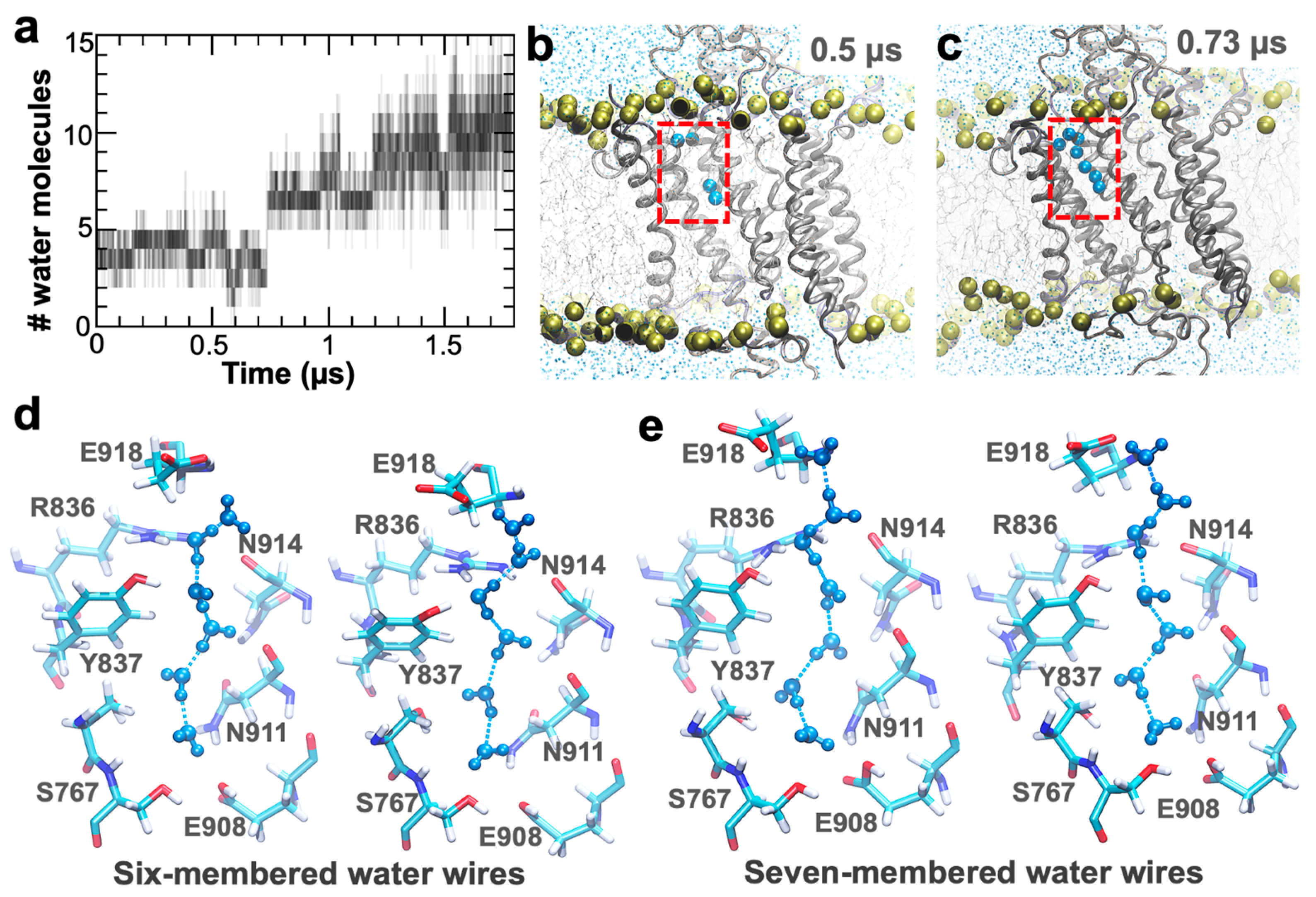
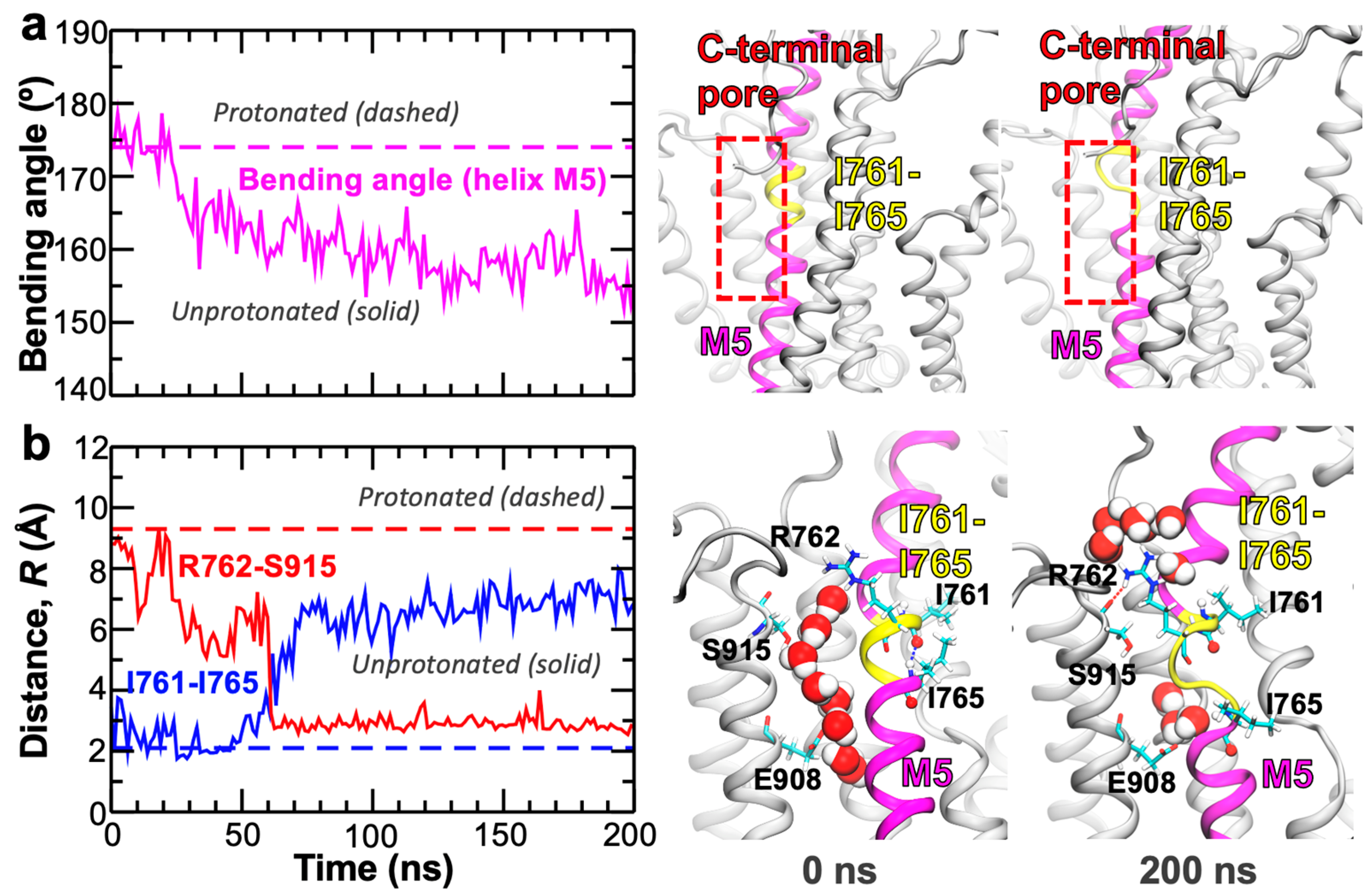
2. Results and Discussion
3. Conclusions
4. Materials and Methods
4.1. Simulation of the Protonated E2 State of SERCA for 300 ns
4.2. Simulation of the Protonated E2 State to Establish the Presence of Water Wires in the C-Terminal Pathway
4.3. Simulation of Proton Release from Residue Glu908 in the E2 State of SERCA
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
References
- Møller, J.V.; Olesen, C.; Winther, A.-M.L.; Nissen, P. What Can Be Learned About the Function of a Single Protein from Its Various X-Ray Structures: The Example of the Sarcoplasmic Calcium Pump. Adv. Struct. Saf. Stud. 2010, 654, 119–140. [Google Scholar] [CrossRef]
- Palmgren, M.G.; Nissen, P. P-Type ATPases. Annu. Rev. Biophys. 2011, 40, 243–266. [Google Scholar] [CrossRef] [PubMed]
- Periasamy, M.; Kalyanasundaram, A. SERCA pump isoforms: Their role in calcium transport and disease. Muscle Nerve 2007, 35, 430–442. [Google Scholar] [CrossRef] [PubMed]
- Obara, K.; Miyashita, N.; Xu, C.; Toyoshima, I.; Sugita, Y.; Inesi, G.; Toyoshima, C. Structural role of countertransport revealed in Ca2+ pump crystal structure in the absence of Ca2+. Proc. Natl. Acad. Sci. USA 2005, 102, 14489–14496. [Google Scholar] [CrossRef]
- Meissner, G.; Young, R. Proton permeability of sarcoplasmic reticulum vesicles. J. Biol. Chem. 1980, 255, 6814–6819. [Google Scholar] [CrossRef]
- Bublitz, M.; Poulsen, H.; Morth, J.P.; Nissen, P. In and out of the cation pumps: P-Type ATPase structure revisited. Curr. Opin. Struct. Biol. 2010, 20, 431–439. [Google Scholar] [CrossRef]
- Toyoshima, C. How Ca2+-ATPase pumps ions across the sarcoplasmic reticulum membrane. Biochim. Biophys. Acta (BBA) Bioenerg. 2009, 1793, 941–946. [Google Scholar] [CrossRef] [PubMed]
- Aguayo-Ortiz, R.; Espinoza-Fonseca, L.M. Linking Biochemical and Structural States of SERCA: Achievements, Challenges, and New Opportunities. Int. J. Mol. Sci. 2020, 21, 21. [Google Scholar] [CrossRef]
- Toyoshima, C.; Mizutani, T. Crystal structure of the calcium pump with a bound ATP analogue. Nat. Cell Biol. 2004, 430, 529–535. [Google Scholar] [CrossRef]
- Hauser, K.; Barth, A. Side-Chain Protonation and Mobility in the Sarcoplasmic Reticulum Ca2+-ATPase: Implications for Proton Countertransport and Ca2+ Release. Biophys. J. 2007, 93, 3259–3270. [Google Scholar] [CrossRef]
- Musgaard, M.; Thøgersen, L.; Schiøtt, B. Protonation States of Important Acidic Residues in the Central Ca2+Ion Binding Sites of the Ca2+-ATPase: A Molecular Modeling Study. Biochem. 2011, 50, 11109–11120. [Google Scholar] [CrossRef]
- Toyoshima, C.; Cornelius, F. New crystal structures of PII-type ATPases: excitement continues. Curr. Opin. Struct. Biol. 2013, 23, 507–514. [Google Scholar] [CrossRef]
- Toyoshima, C.; Nomura, H. Structural changes in the calcium pump accompanying the dissociation of calcium. Nat. Cell Biol. 2002, 418, 605–611. [Google Scholar] [CrossRef]
- Olesen, C.; Sørensen, T.L.-M.; Nielsen, R.C.; Møller, J.V.; Nissen, P. Dephosphorylation of the Calcium Pump Coupled to Counterion Occlusion. Science 2004, 306, 2251–2255. [Google Scholar] [CrossRef] [PubMed]
- Andersson, J.; Hauser, K.; Karjalainen, E.-L.; Barth, A. Protonation and Hydrogen Bonding of Ca2+ Site Residues in the E2P Phosphoenzyme Intermediate of Sarcoplasmic Reticulum Ca2+-ATPase Studied by a Combination of Infrared Spectroscopy and Electrostatic Calculations. Biophys. J. 2008, 94, 600–611. [Google Scholar] [CrossRef] [PubMed]
- Inesi, G.; Lewis, D.; Toyoshima, C.; Hirata, A.; De Meis, L. Conformational Fluctuations of the Ca2+-ATPase in the Native Membrane Environment. J. Biol. Chem. 2008, 283, 1189–1196. [Google Scholar] [CrossRef] [PubMed]
- Tsunekawa, N.; Ogawa, H.; Tsueda, J.; Akiba, T.; Toyoshima, C. Mechanism of the E2 to E1 transition in Ca2+ pump revealed by crystal structures of gating residue mutants. Proc. Natl. Acad. Sci. USA 2018, 115, 12722–12727. [Google Scholar] [CrossRef] [PubMed]
- Bublitz, M.; Musgaard, M.; Poulsen, H.; Thøgersen, L.; Olesen, C.; Schiøtt, B.; Morth, J.; Møller, J.V.; Nissen, P. Ion Pathways in the Sarcoplasmic Reticulum Ca2+-ATPase. J. Biol. Chem. 2013, 288, 10759–10765. [Google Scholar] [CrossRef]
- Ramírez-Salinas, G.L.; Espinoza-Fonseca, L.M. Atomistic Characterization of the First Step of Calcium Pump Activation Associated with Proton Countertransport. Biochemistry 2015, 54, 5235–5241. [Google Scholar] [CrossRef]
- Toyoshima, C.; Iwasawa, S.; Ogawa, H.; Hirata, A.; Tsueda, J.; Inesi, G. Crystal structures of the calcium pump and sarcolipin in the Mg2+-bound E1 state. Nat. Cell Biol. 2013, 495, 260–264. [Google Scholar] [CrossRef]
- Swanson, J.M.J.; Maupin, C.M.; Chen, H.; Petersen, M.K.; Xu, J.; Wu, Y.; Voth, G.A. Proton Solvation and Transport in Aqueous and Biomolecular Systems: Insights from Computer Simulations. J. Phys. Chem. B 2007, 111, 4300–4314. [Google Scholar] [CrossRef] [PubMed]
- Wraight, C.A. Chance and design—Proton transfer in water, channels and bioenergetic proteins. Biochim. Biophys. Acta (BBA) Bioenerg. 2006, 1757, 886–912. [Google Scholar] [CrossRef]
- Clausen, J.D.; Bublitz, M.; Arnou, B.; Montigny, C.; Jaxel, C.; Møller, J.V.; Nissen, P.; Andersen, J.P.; Le Maire, M. SERCA mutant E309Q binds two Ca2+ions but adopts a catalytically incompetent conformation. EMBO J. 2013, 32, 3231–3243. [Google Scholar] [CrossRef]
- Nielsen, G.; Malmendal, A.; Meissner, A.; Møller, J.V.; Nielsen, N.C. NMR studies of the fifth transmembrane segment of sarcoplasmic reticulum Ca2+-ATPase reveals a hinge close to the Ca2+-ligating residues. FEBS Lett. 2003, 544, 50–56. [Google Scholar] [CrossRef]
- Espinoza-Fonseca, L.M.; Ramírez-Salinas, G.L. Microsecond Molecular Simulations Reveal a Transient Proton Pathway in the Calcium Pump. J. Am. Chem. Soc. 2015, 137, 7055–7058. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Yue, Z.; Espinoza-Fonseca, L.M.; Voth, G.A. Multiscale Simulation Reveals Passive Proton Transport Through SERCA on the Microsecond Timescale. Biophys. J. 2020, 119, 1033–1040. [Google Scholar] [CrossRef] [PubMed]
- Sørensen, T.L.-M.; Andersen, J.P. Importance of Stalk Segment S5 for Intramolecular Communication in the Sarcoplasmic Reticulum Ca2+-ATPase. J. Biol. Chem. 2000, 275, 28954–28961. [Google Scholar] [CrossRef] [PubMed]
- Killian, J.A.; Salemink, I.; De Planque, M.R.R.; Lindblom, G.; Koeppe, R.E.; Greathouse, D.V. Induction of Nonbilayer Structures in Diacylphosphatidylcholine Model Membranes by Transmembrane α-Helical Peptides: Importance of Hydrophobic Mismatch and Proposed Role of Tryptophans. Biochemistry 1996, 35, 1037–1045. [Google Scholar] [CrossRef] [PubMed]
- Rice, W.J.; MacLennan, D.H. Scanning Mutagenesis Reveals a Similar Pattern of Mutation Sensitivity in Transmembrane Sequences M4, M5, and M6, but Not in M8, of the Ca2+-ATPase of Sarcoplasmic Reticulum (SERCA1a). J. Biol. Chem. 1996, 271, 31412–31419. [Google Scholar] [CrossRef] [PubMed]
- Asahi, M.; Kimura, Y.; Kurzydlowski, K.; Tada, M.; MacLennan, D.H. Transmembrane Helix M6 in Sarco(endo)plasmic Reticulum Ca2+-ATPase Forms a Functional Interaction Site with Phospholamban. J. Biol. Chem. 1999, 274, 32855–32862. [Google Scholar] [CrossRef]
- Buoninsegni, F.T.; Bartolommei, G.; Moncelli, M.R.; Inesi, G.; Guidelli, R. Time-Resolved Charge Translocation by Sarcoplasmic Reticulum Ca-ATPase Measured on a Solid Supported Membrane. Biophys. J. 2004, 86, 3671–3686. [Google Scholar] [CrossRef]
- Tadini-Buoninsegni, F.; Bartolommei, G.; Moncelli, M.R.; Guidelli, R.; Inesi, G. Pre-steady State Electrogenic Events of Ca2+/H+ Exchange and Transport by the Ca2+-ATPase. J. Biol. Chem. 2006, 281, 37720–37727. [Google Scholar] [CrossRef] [PubMed]
- Espinoza-Fonseca, L.M.; Autry, J.M.; Ramírez-Salinas, G.L.; Thomas, D.D. Atomic-Level Mechanisms for Phospholamban Regulation of the Calcium Pump. Biophys. J. 2015, 108, 1697–1708. [Google Scholar] [CrossRef] [PubMed]
- Bas, D.C.; Rogers, D.M.; Jensen, J.H. Very fast prediction and rationalization of pKa values for protein-ligand complexes. Proteins Struct. Funct. Bioinform. 2008, 73, 765–783. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Robertson, A.D.; Jensen, J.H. Very fast empirical prediction and rationalization of protein pKa values. Proteins Struct. Funct. Bioinform. 2005, 61, 704–721. [Google Scholar] [CrossRef]
- Olsson, M.H.M.; Søndergaard, C.R.; Rostkowski, M.; Jensen, J.H. PROPKA3: Consistent Treatment of Internal and Surface Residues in Empirical pKa Predictions. J. Chem. Theory Comput. 2010, 7, 525–537. [Google Scholar] [CrossRef]
- Søndergaard, C.R.; Olsson, M.H.M.; Rostkowski, M.; Jensen, J.H. Improved Treatment of Ligands and Coupling Effects in Empirical Calculation and Rationalization of pKa Values. J. Chem. Theory Comput. 2011, 7, 2284–2295. [Google Scholar] [CrossRef]
- Pan, J.W.; Hamm, J.R.; Rothman, D.L.; Shulman, R.G. Intracellular pH in human skeletal muscle by 1H NMR. Proc. Natl. Acad. Sci. USA 1988, 85, 7836–7839. [Google Scholar] [CrossRef]
- Steenbergen, C.; DeLeeuw, G.; Rich, T.; Williamson, J.R. Effects of acidosis and ischemia on contractility and intracellular pH of rat heart. Circ. Res. 1977, 41, 849–858. [Google Scholar] [CrossRef]
- Best, R.B.; Zhu, X.; Shim, J.; Lopes, P.E.M.; Mittal, J.; Feig, M.; MacKerell, J.A.D. Optimization of the Additive CHARMM All-Atom Protein Force Field Targeting Improved Sampling of the Backbone ϕ, ψ and Side-Chain χ1 and χ2 Dihedral Angles. J. Chem. Theory Comput. 2012, 8, 3257–3273. [Google Scholar] [CrossRef]
- Klauda, J.B.; Venable, R.M.; Freites, J.A.; O’Connor, J.W.; Tobias, D.J.; Mondragon-Ramirez, C.; Vorobyov, I.; MacKerell, A.D.; Pastor, R.W. Update of the CHARMM All-Atom Additive Force Field for Lipids: Validation on Six Lipid Types. J. Phys. Chem. B 2010, 114, 7830–7843. [Google Scholar] [CrossRef] [PubMed]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kalé, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef]
- Weber, W.; Hünenberger, P.H.; McCammon, J.A. Molecular Dynamics Simulations of a Polyalanine Octapeptide under Ewald Boundary Conditions: Influence of Artificial Periodicity on Peptide Conformation. J. Phys. Chem. B 2000, 104, 3668–3675. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Espinoza-Fonseca, L.M.; Autry, J.M.; Thomas, D.D. Microsecond Molecular Dynamics Simulations of Mg2+- and K+- Bound E1 Intermediate States of the Calcium Pump. PLoS ONE 2014, 9, e95979. [Google Scholar] [CrossRef]
- Sun, B.; Stewart, B.D.; Kucharski, A.N.; Kekenes-Huskey, P.M. Thermodynamics of Cation Binding to the Sarcoendoplasmic Reticulum Calcium ATPase Pump and Impacts on Enzyme Function. J. Chem. Theory Comput. 2019, 15, 2692–2705. [Google Scholar] [CrossRef] [PubMed]
- Inesi, G.; Tadini-Buoninsegni, F. Ca2+/H+ exchange, lumenal Ca2+ release and Ca2+/ATP coupling ratios in the sarcoplasmic reticulum ATPase. J. Cell Commun. Signal. 2013, 8, 5–11. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Espinoza-Fonseca, L.M. Structural Basis for the Function of the C-Terminal Proton Release Pathway in the Calcium Pump. Int. J. Mol. Sci. 2021, 22, 3507. https://doi.org/10.3390/ijms22073507
Espinoza-Fonseca LM. Structural Basis for the Function of the C-Terminal Proton Release Pathway in the Calcium Pump. International Journal of Molecular Sciences. 2021; 22(7):3507. https://doi.org/10.3390/ijms22073507
Chicago/Turabian StyleEspinoza-Fonseca, L. Michel. 2021. "Structural Basis for the Function of the C-Terminal Proton Release Pathway in the Calcium Pump" International Journal of Molecular Sciences 22, no. 7: 3507. https://doi.org/10.3390/ijms22073507
APA StyleEspinoza-Fonseca, L. M. (2021). Structural Basis for the Function of the C-Terminal Proton Release Pathway in the Calcium Pump. International Journal of Molecular Sciences, 22(7), 3507. https://doi.org/10.3390/ijms22073507




