Abstract
Functional alterations in irritable bowel syndrome have been associated with defects in bioenergetics and the mitochondrial network. Effects of high fat, adequate-protein, low carbohydrate ketogenic diet (KD) involve oxidative stress, inflammation, mitochondrial function, and biogenesis. The aim was to evaluate the KD efficacy in reducing the effects of stress on gut mitochondria. Newborn Wistar rats were exposed to maternal deprivation to induce IBS in adulthood. Intestinal inflammation (COX-2 and TRL-4); cellular redox status (SOD 1, SOD 2, PrxIII, mtDNA oxidatively modified purines); mitochondrial biogenesis (PPAR-γ, PGC-1α, COX-4, mtDNA content); and autophagy (Beclin-1, LC3 II) were evaluated in the colon of exposed rats fed with KD (IBD-KD) or standard diet (IBS-Std), and in unexposed controls (Ctrl). IBS-Std rats showed dysfunctional mitochondrial biogenesis (PPAR-γ, PGC-1α, COX-4, and mtDNA contents lower than in Ctrl) associated with inflammation and increased oxidative stress (higher levels of COX-2 and TLR-4, SOD 1, SOD 2, PrxIII, and oxidatively modified purines than in Ctrl). Loss of autophagy efficacy appeared from reduced levels of Beclin-1 and LC3 II. Feeding of animals with KD elicited compensatory mechanisms able to reduce inflammation, oxidative stress, restore mitochondrial function, and baseline autophagy, possibly via the upregulation of the PPAR-γ/PGC-1α axis.
1. Introduction
Applying a feasible and straightforward study model to the pathophysiology of functional gastrointestinal disorders (FGIDs) is still challenging since FGIDs result from the complex interplay between several putative factors including genetics, biology, and psychological attitude. Thus, in recent years, to summarize and overcome this complexity, a “biopsychosocial model” has been conceptualized [1].
In this model, an important role is played by the enteric nervous system (ENS) that acts not only as a regulator of digestive and absorptive functions in the gut, but is also tightly connected with the central nervous system (CNS), constituting the brain–gut axis in which stress can modulate or activate several afferent and efferent connections [2].
Among FGIDs, irritable bowel syndrome (IBS) is a highly prevalent disorder affecting an estimated 10–15% of the worldwide population [3]. An inflammatory component has been suggested to contribute to the etiology of this syndrome [4], and miscommunication between the ENS and CNS has been evoked for the onset of several symptoms apart from the classical and major gastrointestinal (GI) ones (e.g., abdominal pain, diarrhea, and constipation). Based on this assumption, migraine, fibromyalgia, cycling vomiting syndrome, and mood disorders have all been considered functional comorbidities for IBS [5].
These functional GI and extra-GI alterations have recently been associated with low cellular energy metabolism, driven by defects in the bioenergetics supply and the intracellular mitochondrial network [6]. Emerging evidence supports a broader role of mitochondria in several GI disorders of both inflammatory and malignant nature [7,8,9]. Mitochondria have also been suggested to play a crucial role in the rescue from the stress-mediated symptoms, as these organelles are the source of the energy supply for adaptive responses [10].
Mitochondrial implications in IBS do not rely only on their role as a cell powerhouse, but also on the extensive crosstalk between these organelles and the intestinal microbiota [11,12], and recent studies have demonstrated that an altered balance of microbial populations in the gut lumen is associated with the IBS pathogenesis and gut–brain connection [13].
There is an increased interest in using low carbohydrate diets like the ketogenic diet (KD) in IBS treatment [14], even if the reports are still few and lack firm conclusions. The low carbohydrate content in KD has been demonstrated to be protective against the proinflammatory effects of saturated fatty acid intake, at least in the contest of a balanced medium-/long-chain triglyceride composition [15].
The high fat, adequate-protein, low carbohydrate KD reprograms metabolism, forcing the body to utilize fat as a source of energy as it occurs during the fasting state. Through ketogenesis, fatty acids are oxidized to ketone bodies in the liver mitochondria and then distributed via the blood to the other organs to be consumed as fuel. It is considered an effective treatment in neurological [16] and mood disorders [17], also given its ability to remodel the gut microbiota and, consequently, impact the brain–gut axis [16].
Pleiotropic effects of KD involve many targets of oxidative stress, inflammation, and mitochondrial function and biogenesis, so it is suggested as a dietary therapeutic approach in the mitochondrial Leber hereditary optic neuropathy [18] and for the impaired mitochondrial functions in the autism spectrum disorder (ASD) [19].
Limited research has explored the effects of chronic psychosocial stress [10] or early-life stress (ELS) on gut mitochondria [20], leading to the conclusion that mitochondrial activity was disturbed [10,20]. On these bases, the present study aimed to evaluate the KD efficacy in reducing the harmful effects of stress on gut mitochondrial functions.
To achieve this goal, newborn Wistar rats were exposed to ELS through maternal deprivation (MD) to induce IBS in adulthood. Different determinants for IBS were taken into account and studied in colon samples of rats exposed to ELS and afterward fed with KD or a standard diet (Std), along with unexposed control rats. In detail, the intestinal inflammation was evaluated by the levels of cyclooxygenase-2 (COX-2) and Toll-like receptor-4 (TRL-4). The antioxidant response and the cellular redox status were investigated by assaying the levels of cytoplasmic and mitochondrial superoxide dismutases (SOD 1 and SOD 2, respectively), the mitochondrial peroxiredoxin III (PrxIII), and the content of oxidatively modified purines in the D-loop region of mtDNA. Mitochondrial biogenesis was studied by assaying the protein levels of the ligand-inducible transcription factor peroxisome proliferator-activated receptors-γ (PPAR-γ) and its co-activator peroxisome proliferator-activated receptor-gamma co-activator-1 alpha (PGC-1α), along with markers of mitochondrial mass such as levels of cytochrome c oxidase, subunit4 (COX-4), and mtDNA relative content. Finally, as markers of autophagy, the levels of Beclin-1 and LC3 II were analyzed.
2. Results
2.1. Evaluation of Markers of Inflammation and Determination of the Redox Status
The etiopathogenetic inflammatory component in the IBS was evaluated by western blot analysis by determining the levels of COX-2 and TLR-4 proteins in the colon tissue of control, IBS-Std, and IBS-KD rats. As shown in Figure 1, the levels of COX-2 (Panel A) and TLR-4 (Panel B) were significantly about 2-fold higher in IBS-Std than the controls. Concerning both proteins, rats treated with KD showed values not significantly different from those in the controls.
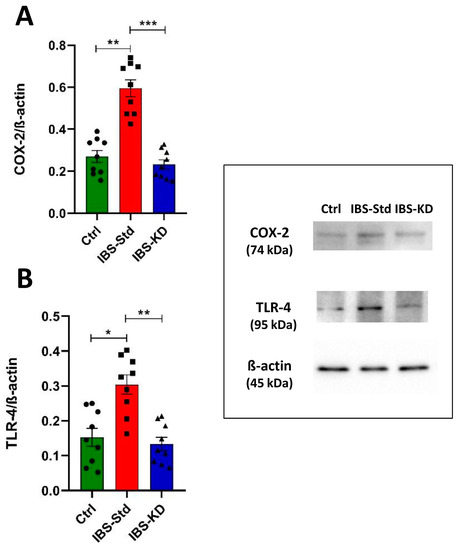
Figure 1.
Western blot analysis of COX-2 (Panel (A)) and TLR-4 (Panel (B)) levels in colon samples of the control, IBS-Std, and IBS-KD rats, with each group consisting of four rats. Data were analyzed by Kruskal–Wallis analysis of variance and Dunn’s Multiple Comparison Test (* p < 0.05, ** p < 0.01, *** p < 0.001).
The possible imbalance in redox status was determined by analyzing the levels of the cellular (SOD 1) and mitochondrial (SOD 2 and PrxIII) antioxidant enzymes, along with the content of oxidatively modified purines in mtDNA.
As in Figure 2 (Panels A and B), the levels of SOD 1 and SOD 2, determined by the ELISA test, were higher, although not significantly, in IBS-Std rats and quite similar to controls in IBS-KD rats.
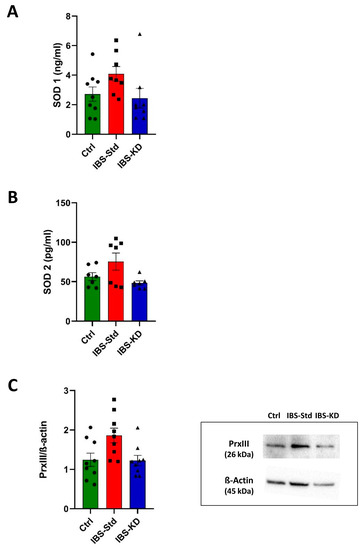
Figure 2.
SOD 1 (Panel (A)) and SOD 2 (Panel (B)) levels, and western blot analysis of PrxIII (Panel (C)) in colon samples of the control, IBS-Std, and IBS-KD rats, with each group consisting of four rats. Data were analyzed by Kruskal–Wallis analysis of variance and Dunn’s Multiple Comparison Test.
The levels of the mitochondrial ROS scavenger PrxIII, evaluated by western blot analysis, showed the same feature with a not significant increase in IBS rats fed with Std and values similar to those of controls in IBS rats fed with KD (Figure 2, Panel C).
The incidence of oxidatively modified purines, mainly 8-oxo-deoxyguanosine (8-oxodG) at the D-loop, the major control region of mtDNA, was determined using the oxidized purines-sensitive enzyme formamidopyrimidine DNA glycosylase (Fpg). The incidence appeared to be approximately two-fold higher in IBS-Std rats than Ctrl rats, and the KD appeared to be able to reduce this content to values similar to those in the controls (Figure 3).
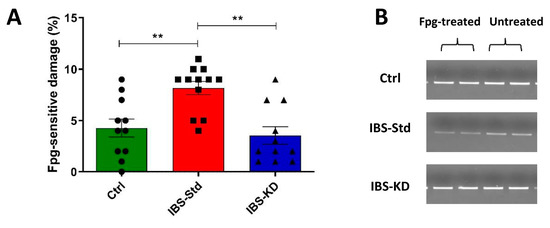
Figure 3.
Incidence of oxidatively modified purines at the D-loop in the colon samples of the control, IBS-Std, and IBS-KD rats, with each group consisting of four rats. Data obtained using the oxidized purines-sensitive enzyme formamidopyrimidine DNA glycosylase (Fpg). Panel (A): the graph represents the ratio between Fpg-treated and untreated band intensities expressed as the complement to 100%. Data were analyzed by Kruskal–Wallis analysis of variance and Dunn’s Multiple Comparison Test (** p < 0.01). Panel (B): Representative gels showing amplicons obtained from Fpg-treated and untreated total DNA.
2.2. Mitochondrial Biogenesis
Levels of PPAR-γ, PGC-1α, COX-4 (evaluated by western blot analysis), and the mtDNA content (evaluated by qPCR) were determined.
All the protein contents resulted in being significantly decreased by approximately two-fold in IBS-Std compared to the controls (Figure 4, Panels A–C).
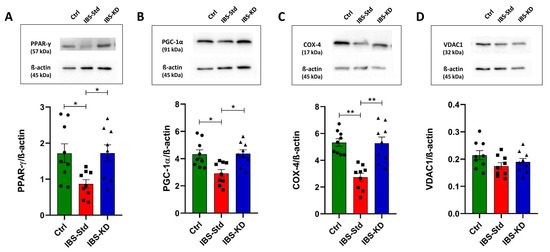
Figure 4.
Western blot analysis of PPAR-γ (Panel (A)), PGC-1α (Panel (B)), COX-4 (Panel (C)), and VDAC1 (Panel (D)) levels in colon samples of the control, IBS-Std, and IBS-KD rats, with each group consisting of four rats. Data were analyzed by Kruskal–Wallis analysis of variance and Dunn’s Multiple Comparison Test (* p < 0.05, ** p < 0.01).
The content of the specific loading marker VDAC1 was analyzed to exclude that the obtained findings could be derived from a different loading of mitochondrial proteins. No statistically significant differences were found among the three experimental groups (Figure 4, Panel D).
The mtDNA also appeared to be significantly reduced (about 2-fold) in IBS-Std than in the controls (Figure 5).
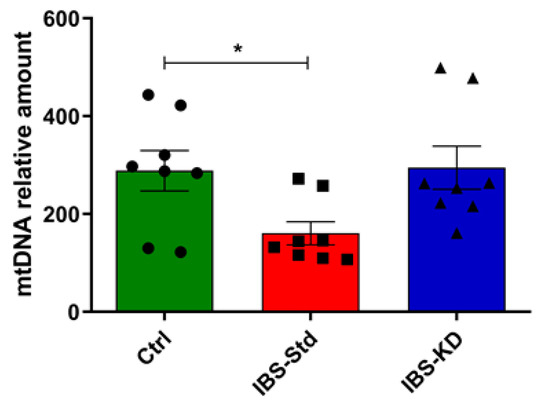
Figure 5.
mtDNA content relative to the β-actin nuclear gene in colon samples of the control, IBS-Std, and IBS-KD rats, with each group consisting of four rats. Data were obtained using qPCR. Data were analyzed by Kruskal–Wallis analysis of variance and Dunn’s Multiple Comparison Test (* p < 0.05).
2.3. Evaluation of Markers of Autophagy
As a measure of autophagy, the protein content of Beclin-1 and LC3 II was determined by western blot analysis.
As shown in Figure 6, significant decreases (1.8 fold) in Beclin-1 (Panel A) and LC3 II (−60%) (Panel B) levels were observed in IBS-Std rats compared with the control ones. The ability of the KD to rescue from these variations and restore values as in the controls was evident.
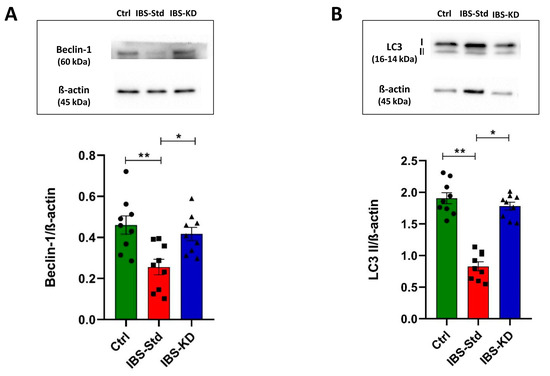
Figure 6.
Western blot analysis of Beclin-1 (Panel (A)) and LC3 II (Panel (B)) levels in colon samples of the control, IBS-Std, and IBS-KD rats, with each group consisting of four rats. Data were analyzed by Kruskal–Wallis analysis of variance and Dunn’s Multiple Comparison Test (* p < 0.05, ** p < 0.01).
3. Discussion
A broad range of intestinal pathologies involves mitochondrial dysfunction [7,8,9,10]. This study aimed to evaluate the KD efficacy in reducing the harmful effects of stress on gut mitochondrial biogenesis using an animal model mimicking the brain–gut axis alterations. The obtained findings support the possible involvement of gut mitochondrial abnormalities in the functional comorbidities associated with IBS [10,20].
One of the more significant results to emerge from this study is that rats exposed to ELS induced by MD and fed with a standard diet had an activated inflammatory response in the colon in adulthood, as shown by the higher levels of COX-2 and TLR-4 than those in the control rats. An abnormally higher expression of COX-2 in the colon has been associated with a worse prognosis in colorectal cancer patients [21], and this protein has been chosen as a pharmacological target to alleviate inflammation of post-inflammatory IBS [22].
Our group has already suggested TLR-4, a leading player in the activation of host immunity being a membrane receptor for LPS, as a useful circulating biomarker for the characterization of IBS subtype with prevalent diarrhea [23]. TLR-4 is also involved in the crosstalk between inflammation and autophagy given its implications in the molecular pathway leading to impaired autophagy, enhanced production of proinflammatory molecules, and oxidative stress in the gut [24].
As recently published elsewhere by our group [25], the histological analysis was performed in the same experimental animals, demonstrating the presence of a normal intestinal mucosa with intact epithelium in the control animals.
In contrast, the IBS-Std animals showed a high inflammatory cell infiltration into both mucosa and submucosa, whereas the IBS-KD animals had mild infiltration of mixed inflammatory cells.
The oxidative hypothesis has been proposed as one of the pathological mechanisms involved in IBS [26]. However, the cytoplasmic antioxidant SOD 1 appeared not to be significantly activated in IBS rats fed with the standard diet. Since mitochondria are the primary cellular ROS source, these organelles are equipped with specific enzymes whose function is to activate an antioxidant response inside them to avoid an oxidative environment. This harmful condition could render the macromolecules therein more vulnerable to substantial damages. In the IBS-Std rats, the not significant increase of the mitochondrial specific scavengers of ROS (SOD 2 and PrxIII), along with the higher presence of oxidatively modified purines in the D-loop region of mtDNA, a hot spot for the damage [27], appeared to be indicative of the inability of these animals to set up an efficacious antioxidant response inside the mitochondria.
Indeed, mitochondria adopt several responses such as the activation of biogenesis, alteration of dynamics, and autophagy to adapt to fluctuating cellular conditions and maintain their proper functions [28]. The expression of nuclear-encoded mitochondrial genes is regulated by a network of transcription factors, co-activators, and co-repressors [29]. In this study, we evaluated the colonic content of the ligand-inducible transcription factor PPAR-γ and its non-enzymatic co-activator PGC-1α, a molecule at the intersection of myriad cellular pathways.
PPAR-γ is primarily expressed in the colon and functions as an endogenous factor in several cellular functions like the metabolism and homeostasis of lipids and carbohydrates, the cell cycle regulation, and the cell differentiation, also being a regulator of bowel inflammation [30]. Decreased PPAR-γ expression has been described in intestinal inflammation [31], so thanks to its role in the interplay between inflammation and the immune system, the pharmacologic use of its agonists has been shown to improve IBS [32].
This transcription factor also regulates the gene coding for PGC-1α, and the PPAR-γ/PGC-1α axis upregulates the mitochondrial biogenesis to promote cellular functional recovery after injury [33,34]. The here shown evidence of lower levels of the transcription factor and its co-activator in IBS rats fed with the standard diet compared to the controls well fits with the observed COX-4 and mtDNA reduced levels.
In line with data reported by other authors [10,20], our findings indicate dysfunctional mitochondrial biogenesis [35] in IBS-Std rats, which appeared to be associated with increased oxidative stress and inflammation in the gut.
The downregulation of PGC-1α within the intestinal epithelium has already been demonstrated in human IBD and experimental murine colitis [36]. Cunningham et al. [36] reported that the reduced levels of PGC-1α were associated with failure in mitochondrial bioenergetics, leading to an impaired intestinal barrier and increased bacterial translocation that contribute to the pathogenesis of colitis. In their animal model, the authors also showed an early increase of PGC-1α, considered as an early adaptive response to oxidative stress that, however, finally failed due to ongoing insult.
Due to the crucial role of mitochondria as a cellular powerhouse, the quality of this organelle pool needs to be continuously controlled to ensure its proper functioning. The clearance of oxidatively modified macromolecules through the induction of autophagy/mitophagy represents an adaptive response when damaged mitochondria generate an excessive amount of ROS, which overwhelms the defenses. In this connection, a new function for the transcriptional co-activator PGC-1α in the adaptive stress response has emerged in the last years [37].
Beyond its function as the master regulator of mitochondrial biogenesis and activator of the antioxidant defense, this protein has a role as coordinator of the autophagic removal of damaged components [37]. The baseline autophagy is crucial for maintaining intestinal homeostasis, as demonstrated by the remarkably rapid mitochondrial turnover (0.7 days in the rat small intestine compared to 24.4 days in the rat brain) [38]. In recent years, autophagy has become a novel issue in GI pathologies [39], providing a further link between stress and the inflammatory status of the GI tract [40]. Actually, the role of autophagy in the inflamed intestinal epithelium is still under debate since available data are conflicting. In IBD patients, Wang et al. [40] showed enhanced intestinal autophagy that appeared to be associated with a worse course of the disease through the modulation of inflammation. More recently, Vincent et al. [41] proposed intestinal autophagy as a homeostatic mechanism to mitigate mitochondrial stress and dysfunction. In the present study, we evaluated the colonic levels of Beclin-1, a protein involved in the starting of autophagy, and the phosphatydilethanolamine-bound form (LC3 II) of the microtubule-associated protein 1 light chain 3 (LC3), which correlates to autophagosome levels. Due to the complexity of the autophagic process, two distinct autophagy markers involved in different steps of the process were evaluated in this study. The reduced levels of Beclin-1 and LC3 II we found in IBS-Std rats compared to the controls indicate the loss of autophagy efficacy in the former group of animals. Although caution is required in interpreting the results since total LC3 levels can change unpredictably [42], the overall agreement of the results obtained by the two markers allowed us to be confident in our interpretation. Considering the role of PGC-1α in the control of the maintenance of a functional mitochondrial pool through the processes of biogenesis, dynamics, and removal of damaged organelles [43], we can speculate that the reduced levels of this protein that we found are not sufficient to coordinate a proper adaptive response [33,34].
The effect of a ketogenic microenvironment, induced in an endogenous or exogenous manner like by the KD, is often studied to understand the pathogenesis of various intestinal inflammatory states. One of the effects resulting from the KD administration is the rise in the circulating ketone bodies. As a rule, the impact of KD consists of a shift toward proteolytic fermentation, leading to a reduction in intestinal mucosa inflammation [44].
Data from this study indicate the ability of KD to rescue from the stress-mediated mitochondrial dysfunction in the gut. In addition to allowing a higher ATP production [45], a complex network links nutritional ketosis and mitochondrial metabolism [46]. Several hypotheses have been proposed on the modes of actions of KD in improving mitochondrial functions. As for the neuroprotective role of KD, some authors have proposed that it could rely on the ability of ketone bodies to cross the blood–brain barrier [47] and activate the fatty acids and ROS sensitive uncoupling protein UCP2, able to reduce ROS production [48]. In liver samples, KD affects mitochondrial protein acetylation [49]. Moore et al. [50] demonstrated that KD, combined with exercise, can suppress de novo lipogenesis and increase mitochondrial biogenesis/content.
In contrast, Newell et al. [19] showed that in the liver of an animal model of ASD, KD increased mitochondrial turnover, thereby decreasing mtDNA. An increase in mitochondrial mass, without a change in oxidative status, has been reported in skeletal muscle tissue [51], whereas in mice defective for the mitochondrial pyruvate carrier, KD improved heart failure [52]. Data concerning the effects of KD in the colon mainly relate to its ability to reshape the architecture of bacterial communities in the gut, whose positive or negative effect is still to be definitively clarified [53].
An interesting result from this study appears to be the rescue of the levels of PPAR-γ in IBS rats fed with KD compared to IBS rats fed with the standard diet. To suggest a possible scheme of the action of KD on gut mitochondria, we can refer to the link existing between PPARs, nutrition, metabolism, and inflammation [54]. It has been reported that a high-fat diet can upregulate PPAR-γ [55] and KD increases fatty acids [56], which are endogenous ligands of this transcription factor [57]. When activated, PPAR-γ exerts its anti-inflammatory and antioxidant roles [30], which in this study was apparent from the values of markers of inflammation and oxidative unbalance that came back to levels similar to those of controls. Furthermore, in this more physiological intracellular environment, mitochondrial functions and baseline autophagy also appear to be restored, possibly via the PPAR-γ/PGC-1α axis, which is able to regulate biogenesis and clearance of damaged cellular components [33,34].
Here, we provide preliminary evidence of the KD efficacy in improving mitochondrial functions in the colon and thus reducing the harmful effects of stress in an animal model of IBS. These data need to be validated by further studies and surely replicated in human clinical trials.
4. Materials and Methods
4.1. Animals and Experimental Design
The study was approved by the Italian Ministry of Health (approval date: 28 November 2018, no. 901/2018-PR) according to European Union guidelines (Directive 2010/63/EU for animal experiments).
The animals were housed at the animal facility of the National Institute of Gastroenterology “S. De Bellis” Research Hospital, Castellana Grotte, Bari, Italy. All the applied procedures followed the International Guidelines for the use of laboratory animals, minimizing animal suffering.
The animal model chosen is the newborn Wistar Rat subjected to ELS through MD to induce IBS in adulthood [58]. Postnatal Day 0 (PND 0) was considered as the birthday. Within PNDs 2 to 14, the puppies experienced MD for 3 h a day.
The experimental design provided that, after weaning, the animals subjected to MD were further divided into two subgroups, one group fed a standard diet (IBS-Std), and one group fed a low carbohydrate, high fat ketogenic diet (IBS-KD). A control group of animals without MD and fed a standard diet was also included (Table 1).

Table 1.
Experimental groups: no irritable bowel syndrome (IBS) rats fed a standard diet (Ctrl); IBS rats fed a standard diet (IBS-Std); IBS rats fed a low carbohydrate, high fat ketogenic diet (IBS-KD). Maternal deprivation (3 h/day from Postnatal Day (PND) 2 to 14). Treatment (for 10 weeks after PND 14).
Diets were supplied as pellets (4RF21 standard diet and KD purchased by Mucedola Srl, Settimo Milanese, Italy) and administered for ten weeks after PND 14 (Table 2).

Table 2.
Composition of standard diet (4RF21) and ketogenic diet.
Rats were checked every day, evaluating different parameters regarding the degree of suffering and stress-induced experimentally (namely, blepharospasm, hollow cheeks, abnormal position of the ears and the whiskers, appetite loss, and liquid stools). Each parameter was recorded, attributing a score from 0 (absent) to 2 (evident) to calculate the possible onset of pain and suffering.
All animals in the study did not show any of the above parameters, except for a slowdown in the growth of puppies with MD, which showed lower weights than the control group puppies.
After treatment, the animals were sacrificed by anesthetic overdose, and colon samples were immediately removed and stored at −80 °C until assayed.
4.2. Western Immunoblotting
Protein extracts were obtained from colon tissue samples of control, IBS-Std, and IBS-KD rats using standard procedure [7]. Aliquots of 50 µg of total protein extracts from each sample were loaded into 4–15% pre-cast polyacrylamide gels (Bio-Rad, Milan, Italy) for western blot analysis. Anti-PrxIII (Ab FRONTIER, Seoul, Korea), anti-PGC-1α (Ab NOVUS, Centennial, CO, USA), anti-COX-2, anti-TLR-4, anti-PPAR-γ, anti-COX-4, anti-Beclin-1, anti-LC3, anti-VDAC1 (Abcam, Cambridge, UK), and anti-β-actin (Cell Signaling, Danvers, MA, USA) were used as primary antibodies. The proteins were detected by chemiluminescence (ECL, Thermo Scientific, Rockford, IL, USA), and the densitometric analysis of each protein-related signal was obtained using the Molecular Imager ChemidocTM (Bio-Rad, Milan, Italy) and normalized against β-actin expression.
4.3. SOD 1 and SOD 2 Levels
The [Cu-Zn] Superoxide dismutase (SOD 1) and the Mitochondrial Superoxide dismutase (SOD 2) levels in colon tissue samples from control and treated Wistar rats were evaluated using the Rat SOD 1 and Rat SOD 2 Enzyme-Linked Immunosorbent Assay (ELISA) Kits (Fine Test, Wuhan, China), respectively, following the manufacturer’s instructions.
4.4. Determination of mtDNA Content
Quantitative real-time polymerase chain reaction (qPCR) was used to determine mtDNA content relative to β-actin nuclear gene, as reported in Chimienti et al. [27]. Briefly, 6 ng total DNA as template and the following primers: mtDNA for 5′GGTTCTTACTTCAGGGCCATCA3′ (nt 15,785-15,806), mtDNA Rev 5′TGATTAGACCCGTTACCATCGA3′ (nt 15,868-15,847) (numbering according to GenBankTM accession number AY172581, Rattus norvegicus, complete mitochondrial genome); β-actin For 5′CCCAGCCATGTACGTAGCCA3′ (nt 2181-2200), β-actin Rev 5′CGTCTCCGGAGTCCATCAC3′ (nt 2266-2248) (GenBankTM accession number V01217.1, Rattus norvegicus, β-actin gene) were used.
4.5. Modified Purines Analysis
Oxidized purines were detected using formamidopyrimidine DNA glycosylase (Fpg) (New England Biolabs, Beverly, MA, USA) digestion of total DNA, as in Chimienti et al. [27]. The PCR amplification of the D-loop mtDNA region was conducted using the following primers: D-loop for 5′TCTGGTCTTGTAAACCAAAAATGA3′ (nt 15,302-15,325), D-loop Rev 5′TGGAATTTTCTGAGGGTAGGC3′ (nt 16,302-16,282) (accession number AY172581, Rattus norvegicus, complete mitochondrial genome) and 7.5 ng of Fpg-treated or untreated total DNA. The Fpg-treated and untreated band intensities ratio was evaluated and expressed as the complement to 100 (%).
4.6. Statistical Analysis
Due to the non-normal distribution of the data, nonparametric tests were performed. Data were analyzed by Kruskal–Wallis analysis of variance and Dunn’s Multiple Comparison Test. Differences were considered significant at p < 0.05. All data represent the results of at least two independent experiments and are expressed as mean ± SEM. A specific statistical package for the exact nonparametric inference package (StataCorp. 2005; Stata Statistical Software: Release 9, College Station, TX, USA) was used.
Author Contributions
Conceptualization, A.O. and G.C.; Methodology, G.C., A.O., B.D., and I.G.; Resources, F.R.; Data curation, A.O. and G.C.; Writing—original draft preparation, A.O., G.C., and F.R.; Writing—review and editing, A.O., G.C., F.R., M.N., A.M.S.L., and V.P.; Visualization, F.R.; Supervision, F.R. and A.O.; Project administration, F.R.; Funding acquisition, F.R. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by RC 2020-2021, Prog. No. 26 (DDG n. 2/2020).
Institutional Review Board Statement
The study was approved by the Italian Ministry of Health (approval date: 28 November 2018, no. 901/2018-PR) according to the European Union guidelines (Directive 2010/63/EU for animal experiments). All the applied procedures followed the International Guidelines for the use of laboratory animals.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Acknowledgments
The authors thank Vito Spilotro for his excellent technical support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Levy, R.L.; Olden, K.W.; Naliboff, B.D.; Bradley, L.A.; Francisconi, C.; Drossman, D.A.; Creed, F. Psychosocial Aspects of the Functional Gastrointestinal Disorders. Gastroenterology 2006, 130, 1447–1458. [Google Scholar] [CrossRef]
- Labanski, A.; Langhorst, J.; Engler, H.; Elsenbruch, S. Stress and the brain-gut axis in functional and chronic-inflammatory gastrointestinal diseases: A transdisciplinary challenge. Psychoneuroendocrinology 2020, 111, 104501. [Google Scholar] [CrossRef] [PubMed]
- Ashburn, T.T.; Gupta, M.S. The IBS market. Nat. Rev. Drug Discov. 2006, 5, 99–100. [Google Scholar] [CrossRef]
- Russo, F.; Chimienti, G.; Clemente, C.; D’Attoma, B.; Linsalata, M.; Orlando, A.; De Carne, M.; Cariola, F.; Semeraro, F.P.; Pepe, G.; et al. Adipokine profile in celiac patients: Differences in comparison with patients suffering from diarrhea-predominant IBS and healthy subjects. Scand. J. Gastroenterol. 2013, 48, 1377–1385. [Google Scholar] [CrossRef] [PubMed]
- O’Malley, D. Immunomodulation of enteric neural function in irritable bowel syndrome. World J. Gastroenterol. 2015, 21, 7362–7366. [Google Scholar] [CrossRef] [PubMed]
- Karabatsiakis, A.; Schonfeldt-Lecuona, C. Depression, mitochondrial bioenergetics, and electroconvulsive therapy: A new approach towards personalized medicine in psychiatric treatment—A short review and current perspective. Transl. Psychiatry 2020, 10, 226. [Google Scholar] [CrossRef] [PubMed]
- Orlando, A.; Chimienti, G.; Pesce, V.; Fracasso, F.; Lezza, A.M.S.; Russo, F. An In Vitro Study on Mitochondrial Compensatory Response Induced by Gliadin Peptides in Caco-2 Cells. Int. J. Mol. Sci. 2019, 20, 1862. [Google Scholar] [CrossRef]
- Picca, A.; Riezzo, G.; Clemente, C.; Pesce, V.; Orlando, A.; Chimienti, G.; Russo, F.; Lezza, A.M.S. Mitochondria and redox balance in coeliac disease: A case-control study. Eur. J. Clin. Investig. 2018, 48, e12877. [Google Scholar] [CrossRef] [PubMed]
- Jackson, D.N.; Theiss, A.L. Gut bacteria signaling to mitochondria in intestinal inflammation and cancer. Gut Microbes 2020, 11, 285–304. [Google Scholar] [CrossRef]
- Vicario, M.; Alonso, C.; Guilarte, M.; Serra, J.; Martínez, C.; González-Castro, A.M.; Lobo, B.; Antolín, M.; Andreu, A.L.; García-Arumí, E.; et al. Chronic psychosocial stress induces reversible mitochondrial damage and corticotropin-releasing factor receptor type-1 upregulation in the rat intestine and IBS-like gut dysfunction. Psychoneuroendocrinology 2012, 37, 65–77. [Google Scholar] [CrossRef]
- Mottawea, W.; Chiang, C.-K.; Mühlbauer, M.; Starr, A.E.; Butcher, J.; Abujamel, T.; Deeke, S.A.; Brandel, A.; Zhou, H.; Shokralla, S.; et al. Altered intestinal microbiota–host mitochondria crosstalk in new onset Crohn’s disease. Nat. Commun. 2016, 7, 13419. [Google Scholar] [CrossRef]
- Galluzzi, L.; Kepp, O.; Kroemer, G. Mitochondria: Master regulators of danger signalling. Nat. Rev. Mol. Cell Biol. 2012, 13, 780–788. [Google Scholar] [CrossRef]
- Sugiyama, T.; Shiotani, A. The Cutting Edge Research of Functional Gastrointestinal Disorders in Japan: Review on JGA Core Symposium 2018–2020. Digestion 2021, 102, 6–11. [Google Scholar] [CrossRef]
- Austin, G.L.; Dalton, C.B.; Hu, Y.; Morris, C.B.; Hankins, J.; Weinland, S.R.; Westman, E.C.; Yancy, W.S.; Drossman, D.A. A Very Low-Carbohydrate Diet Improves Symptoms and Quality of Life in Diarrhea-Predominant Irritable Bowel Syndrome. Clin. Gastroenterol. Hepatol. 2009, 7, 706–708.e1. [Google Scholar] [CrossRef]
- Locker, F.; Leitner, J.; Aminzadeh-Gohari, S.; Weber, D.D.; Sanio, P.; Koller, A.; Feichtinger, R.G.; Weiss, R.; Kofler, B.; Lang, R. The Influence of Ketogenic Diets on Psoriasiform-Like Skin Inflammation. J. Investig. Dermatol. 2020, 140, 707–710.e7. [Google Scholar] [CrossRef] [PubMed]
- Rawat, K.; Singh, N.; Kumari, P.; Saha, L. A review on preventive role of ketogenic diet (KD) in CNS disorders from the gut microbiota perspective. Rev. Neurosci. 2021, 32, 143–157. [Google Scholar] [CrossRef]
- Brietzke, E.; Mansur, R.B.; Subramaniapillai, M.; Balanzá-Martínez, V.; Vinberg, M.; González-Pinto, A.; Rosenblat, J.D.; Ho, R.; McIntyre, R.S. Ketogenic diet as a metabolic therapy for mood disorders: Evidence and developments. Neurosci. Biobehav. Rev. 2018, 94, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Storoni, M.; Robert, M.P.; Plant, G.T. The therapeutic potential of a calorie-restricted ketogenic diet for the management of Leber hereditary optic neuropathy. Nutr. Neurosci. 2017, 22, 156–164. [Google Scholar] [CrossRef]
- Newell, C.; Shutt, T.E.; Ahn, Y.; Hittel, D.S.; Khan, A.; Rho, J.M.; Shearer, J. Tissue Specific Impacts of a Ketogenic Diet on Mitochondrial Dynamics in the BTBR(T+tf/j) Mouse. Front. Physiol. 2016, 7, 654. [Google Scholar] [CrossRef] [PubMed]
- Khorjahani, A.; Peeri, M.; Azarbayjani, M.A. Therapeutic Effect of Exercise on Anxiety and Bowel Oxidative Stress in the Maternal Separation Animal Model. Basic Clin. Neurosci. J. 2019, 11, 69–78. [Google Scholar] [CrossRef]
- Ogino, S.; Kirkner, G.J.; Nosho, K.; Irahara, N.; Kure, S.; Shima, K.; Hazra, A.; Chan, A.T.; Dehari, R.; Giovannucci, E.L.; et al. Cyclooxygenase-2 Expression Is an Independent Predictor of Poor Prognosis in Colon Cancer. Clin. Cancer Res. 2008, 14, 8221–8227. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.Y.; Guo, Y.; Feng, X.J.; Liu, J.J.; Chang, Z.P.; Deng, G.F.; Xu, D.; Gao, J.P.; Hou, R.G. Oridonin Attenuates TNBS-induced Post-inflammatory Irritable Bowel Syndrome via PXR/NF-kappaB Signaling. Inflammation 2020, 44, 645–658. [Google Scholar] [CrossRef]
- Russo, F.; Chimienti, G.; Riezzo, G.; Linsalata, M.; D’Attoma, B.; Clemente, C.; Orlando, A. Adipose Tissue-Derived Biomarkers of Intestinal Barrier Functions for the Characterization of Diarrhoea-Predominant IBS. Dis. Markers 2018, 2018, 1–10. [Google Scholar] [CrossRef]
- Zhou, M.; Xu, W.; Wang, J.; Yan, J.; Shi, Y.; Zhang, C.; Ge, W.; Wu, J.; Du, P.; Chen, Y. Boosting mTOR-dependent autophagy via upstream TLR4-MyD88-MAPK signalling and downstream NF-kappaB pathway quenches intestinal inflammation and oxidative stress injury. EBioMedicine 2018, 35, 345–360. [Google Scholar] [CrossRef]
- Gigante, I.; Tutino, V.; Russo, F.; De Nunzio, V.; Coletta, S.; Armentano, R.; Crovace, A.; Caruso, M.; Orlando, A.; Notarnicola, M. Cannabinoid Receptors Overexpression in a Rat Model of Irritable Bowel Syndrome (IBS) after Treatment with a Ketogenic Diet. Int. J. Mol. Sci. 2021, 22, 2880. [Google Scholar] [CrossRef]
- Balmus, I.-M.; Ilie, O.-D.; Ciobica, A.; Cojocariu, R.; Stanciu, C.; Trifan, A.; Cimpeanu, M.; Cimpeanu, C.; Gorgan, D.L. Irritable Bowel Syndrome between Molecular Approach and Clinical Expertise—Searching for Gap Fillers in the Oxidative Stress Way of Thinking. Medicina 2020, 56, 38. [Google Scholar] [CrossRef] [PubMed]
- Chimienti, G.; Picca, A.; Sirago, G.; Fracasso, F.; Calvani, R.; Bernabei, R.; Russo, F.; Carter, C.S.; Leeuwenburgh, C.; Pesce, V.; et al. Increased TFAM binding to mtDNA damage hot spots is associated with mtDNA loss in aged rat heart. Free Radic. Biol. Med. 2018, 124, 447–453. [Google Scholar] [CrossRef] [PubMed]
- Fu, W.; Liu, Y.; Yin, H. Mitochondrial Dynamics: Biogenesis, Fission, Fusion, and Mitophagy in the Regulation of Stem Cell Behaviors. Stem Cells Int. 2019, 2019, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Maneix, L.; Catic, A. Touch and go: Nuclear proteolysis in the regulation of metabolic genes and cancer. FEBS Lett. 2016, 590, 908–923. [Google Scholar] [CrossRef]
- Vetuschi, A.; Pompili, S.; Gaudio, E.; Latella, G.; Sferra, R. PPAR-gamma with its anti-inflammatory and anti-fibrotic action could be an effective therapeutic target in IBD. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 8839–8848. [Google Scholar] [PubMed]
- Pedersen, G. Development, validation and implementation of an in vitro model for the study of metabolic and immune function in normal and inflamed human colonic epithelium. Dan. Med. J. 2015, 62, 4973. [Google Scholar]
- Roudsari, N.M.; Lashgari, N.A.; Zandi, N.; Pazoki, B.; Momtaz, S.; Sahebkar, A.; Abdolghaffari, A.H. PPAR gamma: A turning point for irritable bowel syndrome treatment. Life Sci. 2020, 257, 118103. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhao, W.; Li, G.; Chen, J.; Guan, X.; Chen, X.; Guan, Z. Neuroprotective Effect and Mechanism of Thiazolidinedione on Dopaminergic Neurons In Vivo and In Vitro in Parkinson’s Disease. PPAR Res. 2017, 2017, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Chidambaram, S.B.; Bhat, A.; Ray, B.; Sugumar, M.; Muthukumar, S.P.; Manivasagam, T.; Justin Thenmozhi, A.; Essa, M.M.; Guillemin, G.J.; Sakharkar, M.K. Cocoa beans improve mitochondrial biogenesis via PPARgamma/PGC1alpha dependent signalling pathway in MPP(+) intoxicated human neuroblastoma cells (SH-SY5Y). Nutr. Neurosci. 2020, 23, 471–480. [Google Scholar] [CrossRef]
- Jornayvaz, F.R.; Shulman, G.I. Regulation of mitochondrial biogenesis. Essays Biochem. 2010, 47, 69–84. [Google Scholar]
- Cunningham, K.E.; Vincent, G.; Sodhi, C.P.; Novak, E.A.; Ranganathan, S.; Egan, C.E.; Stolz, D.B.; Rogers, M.B.; Firek, B.; Morowitz, M.J.; et al. Peroxisome Proliferator-activated Receptor-gamma Coactivator 1-alpha (PGC1alpha) Protects against Experimental Murine Colitis. J. Biol. Chem. 2016, 291, 10184–10200. [Google Scholar] [CrossRef]
- Halling, J.F.; Ringholm, S.; Nielsen, M.M.; Overby, P.; Pilegaard, H. PGC-1α promotes exercise-induced autophagy in mouse skeletal muscle. Physiol. Rep. 2016, 4, e12698. [Google Scholar] [CrossRef]
- Menzies, R.A.; Gold, P.H. The turnover of mitochondria in a variety of tissues of young adult and aged rats. J. Biol. Chem. 1971, 246, 2425–2429. [Google Scholar] [CrossRef]
- Wang, T.; Liu, K.; Wen, L.; Yang, Y.; Yin, X.; Liu, K.; Chen, Y.; He, Y.; Yang, M.; Wei, Y.; et al. Autophagy and Gastrointestinal Diseases. Adv. Exp. Med. Biol. 2020, 1207, 529–556. [Google Scholar]
- Wang, S.-L.; Shao, B.-Z.; Zhao, S.-B.; Chang, X.; Wang, P.; Miao, C.-Y.; Li, Z.-S.; Bai, Y. Intestinal autophagy links psychosocial stress with gut microbiota to promote inflammatory bowel disease. Cell Death Dis. 2019, 10, 1–17. [Google Scholar] [CrossRef]
- Vincent, G.; Novak, E.A.; Siow, V.S.; Cunningham, K.E.; Griffith, B.D.; Iv, T.E.C.; Mentrup, H.L.; Stolz, D.B.; Loughran, P.; Ranganathan, S.; et al. Nix-Mediated Mitophagy Modulates Mitochondrial Damage During Intestinal Inflammation. Antioxidants Redox Signal. 2020, 33, 1–19. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Abdel-Aziz, A.K.; Abdelfatah, S.; Abdellatif, M.; Abdoli, A.; Abel, S.; Abeliovich, H.; Abildgaard, M.H.; Abudu, Y.P.; Acevedo-Arozena, A.; et al. Guidelines for the Use and Interpretation of Assays for Monitoring Autophagy (4th edition). Autophagy 2021, 8, 1–382. [Google Scholar] [CrossRef]
- Halling, J.F.; Pilegaard, H. PGC-1α-mediated regulation of mitochondrial function and physiological implications. Appl. Physiol. Nutr. Metab. 2020, 45, 1–10. [Google Scholar] [CrossRef]
- Ang, Q.Y.; Alexander, M.; Newman, J.C.; Tian, Y.; Cai, J.; Upadhyay, V.; Turnbaugh, J.A.; Verdin, E.; Hall, K.D.; Leibel, R.L.; et al. Ketogenic Diets Alter the Gut Microbiome Resulting in Decreased Intestinal Th17 Cells. Cell 2020, 181, 1263–1275.e16. [Google Scholar] [CrossRef]
- Vidali, S.; Aminzadeh, S.; Lambert, B.; Rutherford, T.; Sperl, W.; Kofler, B.; Feichtinger, R.G. Mitochondria: The ketogenic diet—A metabolism-based therapy. Int. J. Biochem. Cell Biol. 2015, 63, 55–59. [Google Scholar] [CrossRef]
- Miller, V.J.; Villamena, F.A.; Volek, J.S. Nutritional Ketosis and Mitohormesis: Potential Implications for Mitochondrial Function and Human Health. J. Nutr. Metab. 2018, 2018, 1–27. [Google Scholar] [CrossRef]
- Halestrap, A.P.; Meredith, D. The SLC16 gene family?from monocarboxylate transporters (MCTs) to aromatic amino acid transporters and beyond. Pflügers Arch. 2004, 447, 619–628. [Google Scholar] [CrossRef]
- Hasan-Olive, M.; Lauritzen, K.H.; Ali, M.; Rasmussen, L.J.; Storm-Mathisen, J.; Bergersen, L.H. A Ketogenic Diet Improves Mitochondrial Biogenesis and Bioenergetics via the PGC1α-SIRT3-UCP2 Axis. Neurochem. Res. 2018, 44, 22–37. [Google Scholar] [CrossRef] [PubMed]
- Hutfles, L.J.; Wilkins, H.M.; Koppel, S.J.; Weidling, I.W.; Selfridge, J.E.; Tan, E.; Thyfault, J.P.; Slawson, C.; Fenton, A.W.; Zhu, H.; et al. A bioenergetics systems evaluation of ketogenic diet liver effects. Appl. Physiol. Nutr. Metab. 2017, 42, 955–962. [Google Scholar] [CrossRef] [PubMed]
- Moore, M.P.; Cunningham, R.P.; Kelty, T.J.; Boccardi, L.R.; Nguyen, N.; Booth, F.W.; Rector, R.S. Ketogenic diet in combination with voluntary exercise impacts markers of hepatic metabolism and oxidative stress in male and female Wistar rats. Appl. Physiol. Nutr. Metab. 2020, 45, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Parry, H.A.; Kephart, W.C.; Mumford, P.W.; Romero, M.A.; Mobley, C.B.; Zhang, Y.; Roberts, M.D.; Kavazis, A.N. Ketogenic diet increases mitochondria volume in the liver and skeletal muscle without altering oxidative stress markers in rats. Heliyon 2018, 4, e00975. [Google Scholar] [CrossRef]
- McCommis, K.S.; Kovacs, A.; Weinheimer, C.J.; Shew, T.M.; Koves, T.R.; Ilkayeva, O.R.; Kamm, D.R.; Pyles, K.D.; King, M.T.; Veech, R.L.; et al. Nutritional modulation of heart failure in mitochondrial pyruvate carrier–deficient mice. Nat. Metab. 2020, 2, 1232–1247. [Google Scholar] [CrossRef]
- Paoli, A.; Mancin, L.; Bianco, A.; Thomas, E.; Mota, J.F.; Piccini, F. Ketogenic Diet and Microbiota: Friends or Enemies? Genes 2019, 10, 534. [Google Scholar] [CrossRef]
- Duszka, K.; Gregor, A.; Guillou, H.; König, J.; Wahli, W. Peroxisome Proliferator-Activated Receptors and Caloric Restriction—Common Pathways Affecting Metabolism, Health, and Longevity. Cells 2020, 9, 1708. [Google Scholar] [CrossRef] [PubMed]
- Sikder, K.; Shukla, S.K.; Patel, N.; Singh, H.; Rafiq, K. High Fat Diet Up-regulates Fatty Acid Oxidation and Ketogenesis via Intervention of PPAR-gamma. Cell Physiol. Biochem. 2018, 48, 1317–1331. [Google Scholar] [CrossRef] [PubMed]
- Fraser, D.D.; Whiting, S.; Andrew, R.D.; Macdonald, E.A.; Musa-Veloso, K.; Cunnane, S.C. Elevated polyunsaturated fatty acids in blood serum obtained from children on the ketogenic diet. Neurology 2003, 60, 1026–1029. [Google Scholar] [CrossRef] [PubMed]
- Simeone, T.A. Ketogenic Diet and PPAR Gamma. In Ketogenic Diet and Metabolic Therapies: Expanded Roles in Health and Disease; Masino, S.A., Ed.; Oxford University Press: New York, NY, USA, 2017; pp. 167–185. [Google Scholar]
- Vannucchi, M.G.; Evangelista, S. Experimental Models of Irritable Bowel Syndrome and the Role of the Enteric Neurotransmission. J. Clin. Med. 2018, 7, 4. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).