Long Noncoding RNAs—Crucial Players Organizing the Landscape of the Neuronal Nucleus
Abstract
1. Introduction
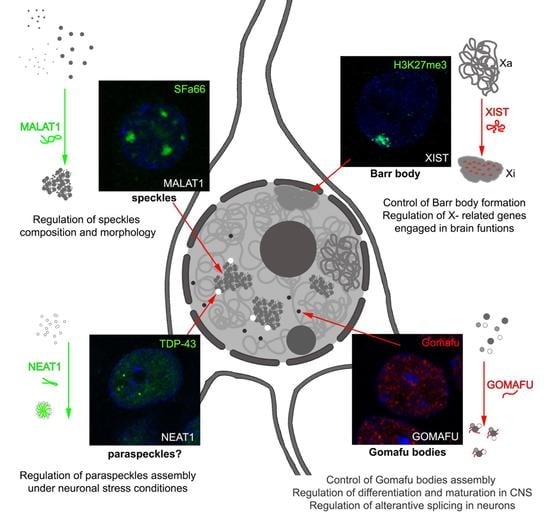
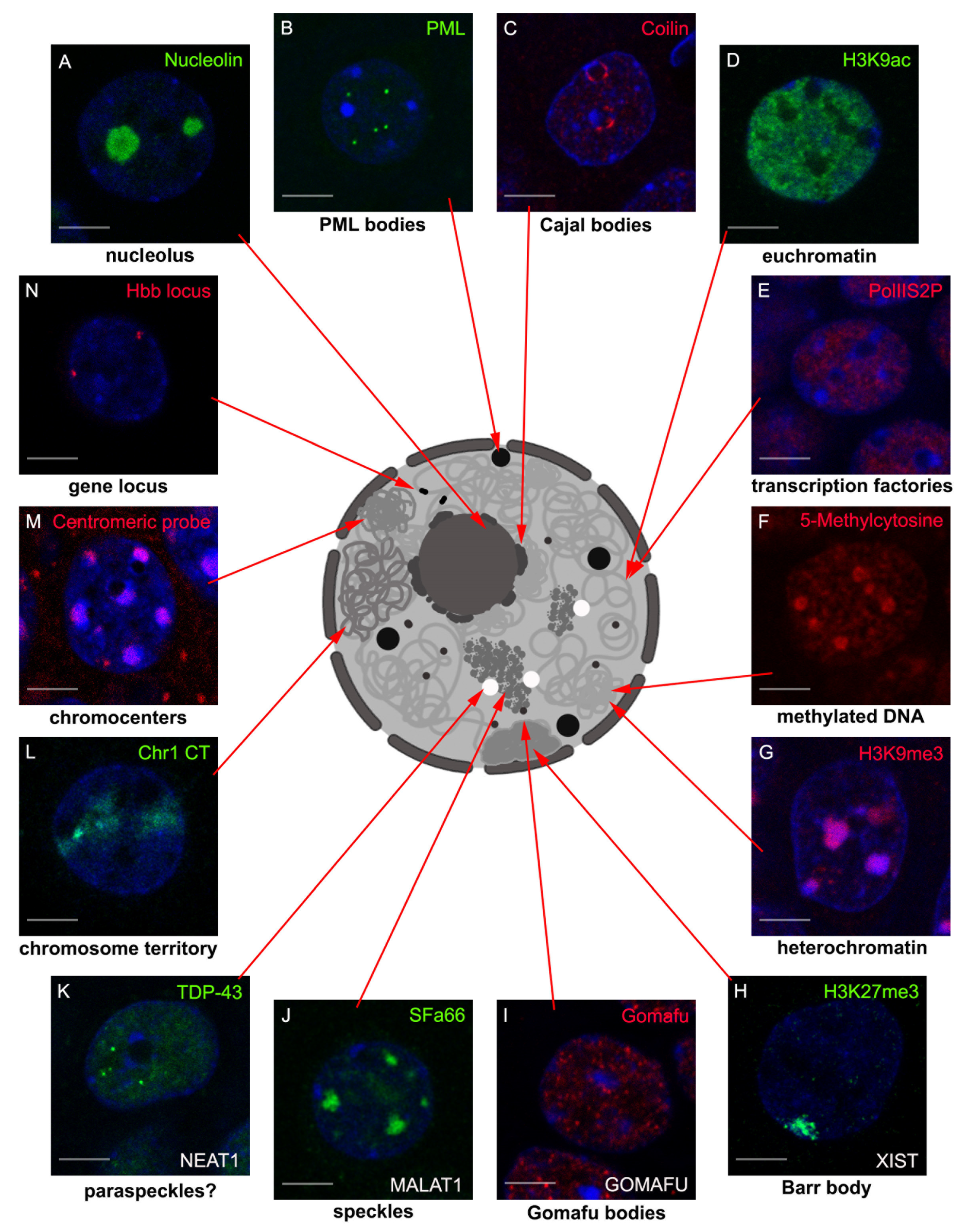
2. LncRNAs Involved in Nuclear Body Formation
2.1. XIST and Barr Body
2.1.1. The X Chromosome Inactivation
2.1.2. Xi Structure and Localization
2.1.3. Genes That Escape from XCI
2.2. NEAT1 and Paraspeckles
2.3. MALAT1 and Nuclear Speckles
2.4. Gomafu and Gomafu Bodies
3. Perspective
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cremer, T.; Cremer, M.; Hübner, B.; Strickfaden, H.; Smeets, D.; Popken, J.; Sterr, M.; Markaki, Y.; Rippe, K.; Cremer, C. The 4D nucleome: Evidence for a dynamic nuclear landscape based on co-aligned active and inactive nuclear compartments. FEBS Lett. 2015, 589, 2931–2943. [Google Scholar] [CrossRef]
- Cremer, T.; Cremer, M.; Hübner, B.; Silahtaroglu, A.; Hendzel, M.; Lanctôt, C.; Strickfaden, H.; Cremer, C. The Interchromatin Compartment Participates in the Structural and Functional Organization of the Cell Nucleus. BioEssays 2020, 42, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Staněk, D.; Fox, A. Nuclear bodies: News insights into structure and function. Curr. Opin. Cell Biol. 2017, 46, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Hall, M.H.; Magalska, A.; Malinowska, M.; Ruszczycki, B.; Czaban, I.; Patel, S.; Ambrożek-Latecka, M.; Zołocińska, E.; Broszkiewicz, H.; Parobczak, K.; et al. Localization and regulation of PML bodies in the adult mouse brain. Brain Struct. Funct. 2016, 221, 2511–2525. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Brangwynne, C.P. Nuclear bodies: The emerging biophysics of nucleoplasmic phases. Curr. Opin. Cell Biol. 2015, 34, 23–30. [Google Scholar] [CrossRef]
- van Steensel, B.; Furlong, E.E.M. The role of transcription in shaping the spatial organization of the genome. Nat. Rev. Mol. Cell Biol. 2019, 20, 327–337. [Google Scholar] [CrossRef]
- Heinz, S.; Texari, L.; Hayes, M.G.B.; Urbanowski, M.; Chang, M.W.; Givarkes, N.; Rialdi, A.; White, K.M.; Albrecht, R.A.; Pache, L.; et al. Transcription Elongation Can Affect Genome 3D Structure. Cell 2018, 174, 1522–1536.e22. [Google Scholar] [CrossRef]
- Weipoltshammer, K.; Schöfer, C. Morphology of nuclear transcription. Histochem. Cell Biol. 2016, 145, 343–358. [Google Scholar] [CrossRef]
- Solovei, I.; Kreysing, M.; Lanctôt, C.; Kösem, S.; Peichl, L.; Cremer, T.; Guck, J.; Joffe, B. Nuclear Architecture of Rod Photoreceptor Cells Adapts to Vision in Mammalian Evolution. Cell 2009, 137, 356–368. [Google Scholar] [CrossRef]
- Benito, E.; Barco, A. The Neuronal Activity-Driven Transcriptome. Mol. Neurobiol. 2015, 51, 1071–1088. [Google Scholar] [CrossRef]
- Yap, E.L.; Greenberg, M.E. Activity-Regulated Transcription: Bridging the Gap between Neural Activity and Behavior. Neuron 2018, 100, 330–348. [Google Scholar] [CrossRef]
- Billia, F.; Baskys, A.; Carlen, P.L.; De Boni, U. Rearrangement of centromeric satellite DNA in hippocampal neurons exhibiting long-term potentiation. Mol. Brain Res. 1992, 14, 101–108. [Google Scholar] [CrossRef]
- Tao-Cheng, J.H. Stimulation-induced structural changes at the nucleus, endoplasmic reticulum and mitochondria of hippocampal neurons. Mol. Brain 2018, 11, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Crepaldi, L.; Policarpi, C.; Coatti, A.; Sherlock, W.T.; Jongbloets, B.C.; Down, T.A.; Riccio, A. Binding of TFIIIC to SINE Elements Controls the Relocation of Activity-Dependent Neuronal Genes to Transcription Factories. PLoS Genet. 2013, 9. [Google Scholar] [CrossRef] [PubMed]
- Walczak, A.; Szczepankiewicz, A.A.; Ruszczycki, B.; Magalska, A.; Zamlynska, K.; Dzwonek, J.; Wilczek, E.; Zybura-Broda, K.; Rylski, M.; Malinowska, M.; et al. Novel higher-order epigenetic regulation of the Bdnf gene upon seizures. J. Neurosci. 2013, 33, 2507–2511. [Google Scholar] [CrossRef] [PubMed]
- Salomoni, P.; Betts-Henderson, J. The role of PML in the nervous system. Mol. Neurobiol. 2011, 43, 114–123. [Google Scholar] [CrossRef]
- Lafarga, M.; Tapia, O.; Romero, A.M.; Berciano, M.T. RNA Biology Cajal bodies in neurons. 2016. [Google Scholar] [CrossRef]
- Yamazaki, T.; Nakagawa, S.; Hirose, T. Architectural RNAs for Membraneless Nuclear Body Formation. Cold Spring Harb. Symp. Quant. Biol. 2019, 84, 227–237. [Google Scholar] [CrossRef]
- Thakur, J.; Henikoff, S. Architectural RNA in chromatin organization. Biochem. Soc. Trans. 2020, 48, 1967–1978. [Google Scholar] [CrossRef] [PubMed]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef]
- Gendron, J.; Colace-Sauty, C.; Beaume, N.; Cartonnet, H.; Guegan, J.; Ulveling, D.; Pardanaud-Glavieux, C.; Moszer, I.; Cheval, H.; Ravassard, P. Long non-coding RNA repertoire and open chromatin regions constitute midbrain dopaminergic neuron - specific molecular signatures. Sci. Rep. 2019, 9, 1409. [Google Scholar] [CrossRef]
- Zimmer-Bensch, G. Emerging Roles of Long Non-Coding RNAs as Drivers of Brain Evolution. Cells 2019, 8, 1399. [Google Scholar] [CrossRef] [PubMed]
- Kadakkuzha, B.M.; Liu, X.-A.; Mccrate, J.; Shankar, G.; Rizzo, V.; Afinogenova, A.; Young, B.; Fallahi, M.; Carvalloza, A.C.; Raveendra, B.; et al. Transcriptome analyses of adult mouse brain reveal enrichment of lncRNAs in specific brain regions and neuronal populations. 2015. [Google Scholar] [CrossRef]
- Ziats, M.N.; Rennert, O.M. Aberrant Expression of Long Noncoding RNAs in Autistic Brain. J. Mol. Neurosci. 2013, 49, 589–593. [Google Scholar] [CrossRef] [PubMed]
- Cogill, S.B.; Srivastava, A.K.; Yang, M.Q.; Wang, L. Co-expression of long non-coding RNAs and autism risk genes in the developing human brain. BMC Syst. Biol. 2018, 12, 91. [Google Scholar] [CrossRef] [PubMed]
- Parikshak, N.N.; Swarup, V.; Belgard, T.G.; Irimia, M.; Ramaswami, G.; Gandal, M.J.; Hartl, C.; Leppa, V.; Ubieta, L.D.L.T.; Huang, J.; et al. Genome-wide changes in lncRNA, splicing, and regional gene expression patterns in autism. Nature 2016, 540, 423–427. [Google Scholar] [CrossRef]
- Petazzi, P.; Sandoval, J.; Szczesna, K.; Jorge, O.C.; Roa, L.; Sayols, S.; Gomez, A.; Huertas, D.; Esteller, M. Dysregulation of the long non-coding RNA transcriptome in a Rett syndrome mouse model. RNA Biol. 2013, 10, 1197–1203. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R. Long non-coding RNAs in Huntington’s disease neurodegeneration. Neurobiol. Dis. 2012, 46, 245–254. [Google Scholar] [CrossRef]
- Idda, M.L.; Munk, R.; Abdelmohsen, K.; Gorospe, M. Noncoding RNAs in Alzheimer’s disease. Wiley Interdiscip. Rev. RNA 2018, 9, e1463. [Google Scholar] [CrossRef]
- Iarovaia, O.V.; Minina, E.P.; Sheval, E.V.; Onichtchouk, D.; Dokudovskaya, S.; Razin, S.V.; Vassetzky, Y.S. Nucleolus: A Central Hub for Nuclear Functions. Trends Cell Biol. 2019, 29, 647–659. [Google Scholar] [CrossRef] [PubMed]
- Joh, R.I.; Palmieri, C.M.; Hill, I.T.; Motamedi, M. Regulation of histone methylation by noncoding RNAs. Biochim. Biophys. Acta Gene Regul. Mech. 2014, 1839, 1385–1394. [Google Scholar] [CrossRef]
- Villaseñor, R.; Baubec, T. Regulatory mechanisms governing chromatin organization and function. Curr. Opin. Cell Biol. 2021, 70, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Sun, H.; Wang, H. Long noncoding RNAs in DNA methylation: New players stepping into the old game. Cell Biosci. 2016, 6, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Yen, Y.P.; Hsieh, W.F.; Tsai, Y.Y.; Lu, Y.L.; Liau, E.S.; Hsu, H.C.; Chen, Y.C.; Liu, T.C.; Chang, M.; Li, J.; et al. Dlk1-Dio3 locus-derived lncRNAs perpetuate postmitotic motor neuron cell fate and subtype identity. Elife 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Mozdarani, H.; Ezzatizadeh, V.; Rahbar Parvaneh, R. The emerging role of the long non-coding RNA HOTAIR in breast cancer development and treatment. J. Transl. Med. 2020, 18, 1–15. [Google Scholar] [CrossRef]
- Arun, G.; Aggarwal, D.; Spector, D.L. MALAT1 Long Non-Coding RNA: Functional Implications. Non-Coding RNA 2020, 6, 22. [Google Scholar] [CrossRef]
- Chujo, T.; Yamazaki, T.; Hirose, T. Architectural RNAs (arcRNAs): A class of long noncoding RNAs that function as the scaffold of nuclear bodies. Biochim. Biophys. Acta Gene Regul. Mech. 2016, 1859, 139–146. [Google Scholar] [CrossRef]
- Bernard, D.; Prasanth, K.V.; Tripathi, V.; Colasse, S.; Nakamura, T.; Xuan, Z.; Zhang, M.Q.; Sedel, F.; Jourdren, L.; Coulpier, F.; et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 2010, 29, 3082–3093. [Google Scholar] [CrossRef]
- Smith, K.P.; Hall, L.L.; Lawrence, J.B. Nuclear hubs built on RNAs and clustered organization of the genome. Curr. Opin. Cell Biol. 2020, 64, 67–76. [Google Scholar] [CrossRef]
- Sone, M.; Hayashi, T.; Tarui, H.; Agata, K.; Takeichi, M.; Nakagawa, S. The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons. J. Cell Sci. 2007, 120, 2498–2506. [Google Scholar] [CrossRef]
- Barr, M.L.; Bertram, E.G. A morphological distinction between neurones of the male and female, and the behaviour of the nucleolar satellite during accelerated nucleoprotein synthesis. Nature 1949, 163, 676–677. [Google Scholar] [CrossRef]
- Ohno, S.; Kaplan, W.D.; Kinosita, R. Formation of the sex chromatin by a single X-chromosome in liver cells of Rattus norvegicus. Exp. Cell Res. 1959, 18, 415–418. [Google Scholar] [CrossRef]
- Lyon, M. Gene Action in the X-chromosome of the Mouse (Mus musculus L.). Nature 1961, 190, 372–373. [Google Scholar] [CrossRef] [PubMed]
- Ohno, S. Sex Chromosomes and Sex-linked Genes. Springer: Berlin/Heidelberg, Germany, 1967; ISBN 9783642881800. [Google Scholar]
- Chen, J.; Wang, M.; He, X.; Yang, J.-R.; Chen, X. The Evolution of Sex Chromosome Dosage Compensation in Animals. J. Genet. Genomics 2020. [Google Scholar] [CrossRef]
- Muller, H.J. Evidence of the precision of genetic adaptation. Harvey Lect Ser. 1948, 43, 165–229. [Google Scholar]
- Lucchesi, J.C.; Kuroda, M.I. Dosage compensation in drosophila. Cold Spring Harb. Perspect. Biol. 2015, 7, 1–21. [Google Scholar] [CrossRef]
- Herzing, L.B.K.; Romer, J.T.; Horn, J.M.; Ashworth, A. Xist has properties of the X-chromosome inactivation centre. Nature 1997, 386, 272–275. [Google Scholar] [CrossRef]
- Lee, J.T.; Lu, N.; Han, Y. Genetic analysis of the mouse X inactivation center defines an 80-kb multifunction domain. Proc. Natl. Acad. Sci. USA 1999, 96, 3836–3841. [Google Scholar] [CrossRef]
- Lee, J.T. Molecular biology: Regulation of X-chromosome counting by Tsix and Xite sequences. Science 2005, 309, 768–771. [Google Scholar] [CrossRef]
- Tian, D.; Sun, S.; Lee, J.T. The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell 2010, 143, 390–403. [Google Scholar] [CrossRef]
- Lee, J.T.; Jaenisch, R. Long-range cis effects of ectopic X-inactivation centres on a mouse autosome. Nature 1997, 386, 275–279. [Google Scholar] [CrossRef] [PubMed]
- Smeets, D.; Markaki, Y.; Schmid, V.J.; Kraus, F.; Tattermusch, A.; Cerase, A.; Sterr, M.; Fiedler, S.; Demmerle, J.; Popken, J.; et al. Three-dimensional super-resolution microscopy of the inactive X chromosome territory reveals a collapse of its active nuclear compartment harboring distinct Xist RNA foci. Epigenet. Chromatin 2014, 7, 1–27. [Google Scholar] [CrossRef]
- Gartler, S.M.; Riggs, A.D. Mammalian X-chromosome inactivation. Annu. Rev. Genet. 1983, 17, 155–190. [Google Scholar] [CrossRef]
- Boyle, A.L.; Ballard, S.G.; Ward, D.C. Differential distribution of long and short interspersed element sequences in the mouse genome: Chromosome karyotyping by fluorescence in situ hybridization. Proc. Natl. Acad. Sci. USA 1990, 87, 7757–7761. [Google Scholar] [CrossRef] [PubMed]
- Lyon, M.F. X-chromosome inactivation: A repeat hypothesis. Cytogenet. Cell Genet. 1998, 80, 133–137. [Google Scholar] [CrossRef] [PubMed]
- Paço, A.; Adega, F.; Chaves, R. LINE-1 retrotransposons: From “parasite” sequences to functional elements. J. Appl. Genet. 2015, 56, 133–145. [Google Scholar] [CrossRef]
- Bailey, J.A.; Carrel, L.; Chakravarti, A.; Eichler, E.E. Molecular evidence for a relationship between LINE-1 elements and X chromosome inactivation: The Lyon repeat hypothesis. Proc. Natl. Acad. Sci. USA 2000, 97, 6634–6639. [Google Scholar] [CrossRef]
- Chow, J.C.; Ciaudo, C.; Fazzari, M.J.; Mise, N.; Servant, N.; Glass, J.L.; Attreed, M.; Avner, P.; Wutz, A.; Barillot, E.; et al. LINE-1 activity in facultative heterochromatin formation during X chromosome inactivation. Cell 2010, 141, 956–969. [Google Scholar] [CrossRef]
- Chaumeil, J.; Okamoto, I.; Guggiari, M.; Heard, E. Integrated kinetics of X chromosome inactivation in differentiating embryonic stem cells. Cytogenet. Genome Res. 2002, 99, 75–84. [Google Scholar] [CrossRef]
- Jeppesen, P.; Turner, B.M. The inactive X chromosome in female mammals is distinguished by a lack of histone H4 acetylation, a cytogenetic marker for gene expression. Cell 1993, 74, 281–289. [Google Scholar] [CrossRef]
- Żylicz, J.J.; Bousard, A.; Žumer, K.; Dossin, F.; Mohammad, E.; da Rocha, S.T.; Schwalb, B.; Syx, L.; Dingli, F.; Loew, D.; et al. The Implication of Early Chromatin Changes in X Chromosome Inactivation. Cell 2019, 176, 182–197.e23. [Google Scholar] [CrossRef]
- Plath, K.; Fang, J.; Mlynarczyk-Evans, S.K.; Cao, R.; Worringer, K.A.; Wang, H.; Zhang, Y. Role of histone H3 lysine 27 methylation in X inactivation. Science 2003, 300, 131–135. [Google Scholar] [CrossRef]
- Heard, E.; Rougeulle, C.; Arnaud, D.; Avner, P.; Allis, C.D.; Spector, D.L. Methylation of histone H3 at Lys-9 Is an early mark on the X chromosome during X inactivation. Cell 2001, 107, 727–738. [Google Scholar] [CrossRef]
- Duncan, C.G.; Grimm, S.A.; Morgan, D.L.; Bushel, P.R.; Bennett, B.D.; Roberts, J.D.; Tyson, F.L.; Merrick, B.A.; Wade, P.A. Dosage compensation and DNA methylation landscape of the X chromosome in mouse liver. Sci. Rep. 2018, 8, 1–17. [Google Scholar] [CrossRef]
- Gdula, M.R.; Nesterova, T.B.; Pintacuda, G.; Godwin, J.; Zhan, Y.; Ozadam, H.; McClellan, M.; Moralli, D.; Krueger, F.; Green, C.M.; et al. The non-canonical SMC protein SmcHD1 antagonises TAD formation and compartmentalisation on the inactive X chromosome. Nat. Commun. 2019, 10, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.K.; Blanco, M.; Jackson, C.; Aznauryan, E.; Ollikainen, N.; Surka, C.; Chow, A.; Cerase, A.; McDonel, P.; Guttman, M. Xist recruits the X chromosome to the nuclear lamina to enable chromosome-wide silencing. Science 2016, 354, 468–472. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.-F.; Huynh, K.D.; Lee, J.T. Perinucleolar targeting of the inactive X during S phase: Evidence for a role in the maintenance of silencing. Cell 2007, 129, 693–706. [Google Scholar] [CrossRef] [PubMed]
- Bourgeois, C.A.; Laquerriere, F.; Hemon, D.; Hubert, J.; Bouteille, M. New data on the in-situ position of the inactive X chromosome in the interphase nucleus of human fibroblasts. Hum. Genet. 1985, 69, 122–129. [Google Scholar] [CrossRef]
- Yang, F.; Deng, X.; Ma, W.; Berletch, J.B.; Rabaia, N.; Wei, G.; Moore, J.M.; Filippova, G.N.; Xu, J.; Liu, Y.; et al. The lncRNA Firre anchors the inactive X chromosome to the nucleolus by binding CTCF and maintains H3K27me3 methylation. Genome Biol. 2015, 16, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Ma, W.; Ramani, V.; Hill, A.; Yang, F.; Ay, F.; Berletch, J.B.; Blau, C.A.; Shendure, J.; Duan, Z.; et al. Bipartite structure of the inactive mouse X chromosome. Genome Biol. 2015, 16, 1–21. [Google Scholar] [CrossRef]
- Monfort, A.; Wutz, A. The B-side of Xist [version 1; peer review: 3 approved]. F1000Research 2020, 9, 1–12. [Google Scholar]
- Fackelmayer, F.; Dahm, K.; Renz, A.; Ramsperger, U.; Richter, A. Nucleic-acid-binding properties of hnRNP-U/SAF-A. Eur. J. Biochem. 1994, 221, 749–757. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, Y.; Brockdorff, N.; Kawano, S.; Tsutui, K.; Tsutui, K.; Nakagawa, S. The matrix protein hnRNP U is required for chromosomal localization of xist RNA. Dev. Cell 2010, 19, 469–476. [Google Scholar] [CrossRef] [PubMed]
- Kipp, M.; Göhring, F.; Ostendorp, T.; van Drunen, C.M.; van Driel, R.; Przybylski, M.; Fackelmayer, F.O. SAF-Box, a Conserved Protein Domain That Specifically Recognizes Scaffold Attachment Region DNA. Mol. Cell. Biol. 2000, 20, 7480–7489. [Google Scholar] [CrossRef]
- Lv, Q.; Yuan, L.; Song, Y.; Sui, T.; Li, Z.; Lai, L. D-repeat in the XIST gene is required for X chromosome inactivation. RNA Biol. 2016, 13, 172–176. [Google Scholar] [CrossRef] [PubMed]
- Hoki, Y.; Kimura, N.; Kanbayashi, M.; Amakawa, Y.; Ohhata, T.; Sasaki, H.; Sado, T. A proximal conserved repeat in the Xist gene is essential as a genomic element for X-inactivation in mouse. Development 2009, 136, 139–146. [Google Scholar] [CrossRef]
- Wutz, A.; Rasmussen, T.P.; Jaenisch, R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat. Genet. 2002, 30, 167–174. [Google Scholar] [CrossRef]
- Monfort, A.; Di Minin, G.; Postlmayr, A.; Freimann, R.; Arieti, F.; Thore, S.; Wutz, A. Identification of Spen as a crucial factor for Xist function through forward genetic screening in haploid embryonic stem cells. Cell Rep. 2015, 12, 554–561. [Google Scholar] [CrossRef]
- Dossin, F.; Pinheiro, I.; Żylicz, J.J.; Roensch, J.; Collombet, S.; Le Saux, A.; Chelmicki, T.; Attia, M.; Kapoor, V.; Zhan, Y.; et al. SPEN integrates transcriptional and epigenetic control of X-inactivation. Nature 2020, 578, 455–460. [Google Scholar] [CrossRef]
- Almeida, M.; Pintacuda, G.; Masui, O.; Koseki, Y.; Gdula, M.; Cerase, A.; Brown, D.; Mould, A.; Innocent, C.; Nakayama, M.; et al. PCGF3/5-PRC1 initiates Polycomb recruitment in X chromosome inactivation. Science 2017, 356, 1081–1084. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Guo, J.K.; Wei, Y.; Dou, D.R.; Zarnegar, B.; Ma, Q.; Li, R.; Zhao, Y.; Liu, F.; Choudhry, H.; et al. Structural modularity of the XIST ribonucleoprotein complex. Nat. Commun. 2020, 11, 1–14. [Google Scholar] [CrossRef]
- Chu, C.; Zhang, Q.C.; da Rocha, S.T.; Flynn, R.A.; Bharadwaj, M.; Calabrese, J.M.; Magnuson, T.; Heard, E.; Chang, H.Y. Systematic discovery of Xist RNA binding proteins. Cell 2015, 161, 404–416. [Google Scholar] [CrossRef]
- Minajigi, A.; Froberg, J.; Wei, C.; Sunwoo, H.; Kesner, B.; Colognori, D.; Lessing, D.; Payer, B.; Boukhali, M.; Haas, W.; et al. Chromosomes. A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science 2015, 349. [Google Scholar] [CrossRef] [PubMed]
- Loda, A.; Heard, E. Xist RNA in action: Past, present, and future. PLoS Genet. 2019, 15, 1–17. [Google Scholar] [CrossRef]
- Jégu, T.; Aeby, E.; Lee, J.T. The X chromosome in space. Nat. Rev. Genet. 2017, 18, 377–389. [Google Scholar] [CrossRef]
- Giorgetti, L.; Lajoie, B.R.; Carter, A.C.; Attia, M.; Zhan, Y.; Xu, J.; Chen, C.J.; Kaplan, N.; Chang, H.Y.; Heard, E.; et al. Structural organization of the inactive X chromosome in the mouse. Nature 2016, 535, 575–579. [Google Scholar] [CrossRef]
- Bischoff, A.; Albers, J.; Kharboush, I.; Stelzer, E.; Cremer, T.; Cremer, C. Differences of size and shape of active and inactive X-chromosome domains in human amniotic fluid cell nuclei. Microsc. Res. Tech. 1993, 25, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Eils, R.; Dietzel, S.; Bertin, E.; Schröck, E.; Speicher, M.R.; Ried, T.; Robert-Nicoud, M.; Cremer, C.; Cremer, T. Three-dimensional reconstruction of painted human interphase chromosomes: Active and inactive X chromosome territories have similar volumes but differ in shape and surface structure. J. Cell Biol. 1996, 135, 1427–1440. [Google Scholar] [CrossRef]
- Teller, K.; Illner, D.; Thamm, S.; Casas-Delucchi, C.S.; Versteeg, R.; Indemans, M.; Cremer, T.; Cremer, M. A top-down analysis of Xa- and Xi-territories reveals differences of higher order structure at ≥20 Mb genomic length scales. Nucleus 2011, 2, 465–477. [Google Scholar] [CrossRef]
- Kelsey, A.D.; Yang, C.; Leung, D.; Minks, J.; Dixon-McDougall, T.; Baldry, S.E.L.; Bogutz, A.B.; Lefebvre, L.; Brown, C.J. Impact of flanking chromosomal sequences on localization and silencing by the human non-coding RNA XIST. Genome Biol. 2015, 16, 1–16. [Google Scholar] [CrossRef] [PubMed]
- BARR, M.L.; BERTRAM, E.G. The behaviour of nuclear structures during depletion and restoration of Nissl material in motor neurons. J. Anat. 1951, 85, 171–17181. [Google Scholar]
- Borden, J.; Manuelidis, L. Movement of the X chromosome in epilepsy. Science 1988, 242, 1686–1691. [Google Scholar] [CrossRef]
- van Steensel, B.; Belmont, A.S. Lamina-Associated Domains: Links with Chromosome Architecture, Heterochromatin, and Gene Repression. Cell 2017, 169, 780–791. [Google Scholar] [CrossRef] [PubMed]
- Berletch, J.B.; Ma, W.; Yang, F.; Shendure, J.; Noble, W.S.; Disteche, C.M.; Deng, X. Escape from X inactivation varies in mouse tissues. PLoS Genet. 2015, 11, e1005079. [Google Scholar] [CrossRef] [PubMed]
- Carrel, L.; Willard, H.F. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature 2005, 434, 400–404. [Google Scholar] [CrossRef]
- Tukiainen, T.; Villani, A.-C.; Yen, A.; Rivas, M.A.; Marshall, J.L.; Satija, R.; Aguirre, M.; Gauthier, L.; Fleharty, M.; Kirby, A.; et al. Landscape of X chromosome inactivation across human tissues. Nature 2017, 550, 244–248. [Google Scholar] [CrossRef] [PubMed]
- Murakami, K.; Ohhira, T.; Oshiro, E.; Qi, D.; Oshimura, M.; Kugoh, H. Identification of the chromatin regions coated by non-coding Xist RNA. Cytogenet. Genome Res. 2009, 125, 19–25. [Google Scholar] [CrossRef]
- Filippova, G.N.; Cheng, M.K.; Moore, J.M.; Truong, J.P.; Hu, Y.J.; Nguyen, D.K.; Tsuchiya, K.D.; Disteche, C.M. Boundaries between chromosomal domains of X inactivation and escape bind CTCF and lack CpG methylation during early development. Dev. Cell 2005, 8, 31–42. [Google Scholar] [CrossRef]
- Chen, C.Y.; Shi, W.; Balaton, B.P.; Matthews, A.M.; Li, Y.; Arenillas, D.J.; Mathelier, A.; Itoh, M.; Kawaji, H.; Lassmann, T.; et al. YY1 binding association with sex-biased transcription revealed through X-linked transcript levels and allelic binding analyses. Sci. Rep. 2016, 6, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Fang, H.; Disteche, C.M.; Berletch, J.B. X Inactivation and Escape: Epigenetic and Structural Features. Front. Cell Dev. Biol. 2019, 7, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Reinius, B.; Shi, C.; Hengshuo, L.; Sandhu, K.S.; Radomska, K.J.; Rosen, G.D.; Lu, L.; Kullander, K.; Williams, R.W.; Jazin, E. Female-biased expression of long non-coding RNAs in domains that escape X-inactivation in mouse. BMC Genomics 2010, 11. [Google Scholar] [CrossRef] [PubMed]
- Neri, G.; Schwartz, C.E.; Lubs, H.A.; Stevenson, R.E. X-linked intellectual disability update 2017. Am. J. Med. Genet. A 2018, 176, 1375–1388. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.K.; Disteche, C.M. Dosage compensation of the active X chromosome in mammals. Nat. Genet. 2006, 38, 47–53. [Google Scholar] [CrossRef]
- Nguyen, D.K.; Disteche, C.M. High expression of the mammalian X chromosome in brain. Brain Res. 2006, 1126, 46–49. [Google Scholar] [CrossRef]
- Scandaglia, M.; Lopez-Atalaya, J.P.; Medrano-Fernandez, A.; Lopez-Cascales, M.T.; del Blanco, B.; Lipinski, M.; Benito, E.; Olivares, R.; Iwase, S.; Shi, Y.; et al. Loss of Kdm5c Causes Spurious Transcription and Prevents the Fine-Tuning of Activity-Regulated Enhancers in Neurons. Cell Rep. 2017, 21, 47–59. [Google Scholar] [CrossRef]
- Jensen, L.R.; Amende, M.; Gurok, U.; Moser, B.; Gimmel, V.; Tzschach, A.; Janecke, A.R.; Tariverdian, G.; Chelly, J.; Fryns, J.P.; et al. Mutations in the JARID1C gene, which is involved in transcriptional regulation and chromatin remodeling, cause X-linked mental retardation. Am. J. Hum. Genet. 2005, 76, 227–236. [Google Scholar] [CrossRef]
- Claes, S.; Devriendt, K.; Van Goethem, G.; Roelen, L.; Meireleire, J.; Raeymaekers, P.; Cassiman, J.J.; Fryns, J.P. Novel syndromic form of X-linked complicated spastic paraplegia. Am. J. Med. Genet. 2000, 94, 1–4. [Google Scholar] [CrossRef]
- Ji, B.; Higa, K.K.; Kelsoe, J.R.; Zhou, X. Over-expression of XIST, the Master Gene for X Chromosome Inactivation, in Females With Major Affective Disorders. EBioMedicine 2015, 2, 909–918. [Google Scholar] [CrossRef]
- Yu, S.; Chen, C.; Pan, Y.; Kurz, M.C.; Datner, E.; Hendry, P.L.; Velilla, M.-A.; Lewandowski, C.; Pearson, C.; Domeier, R.; et al. Genes known to escape X chromosome inactivation predict co-morbid chronic musculoskeletal pain and posttraumatic stress symptom development in women following trauma exposure. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2019, 180, 415–427. [Google Scholar] [CrossRef]
- Hong, D.S.; Reiss, A.L. Cognition and behavior in turner syndrome: A brief review. Pediatr. Endocrinol. Rev. 2012, 9, 710–712. [Google Scholar] [PubMed]
- Clement-Jones, M.; Schiller, S.; Rao, E.; Blaschke, R.J.; Zuniga, A.; Zeller, R.; Robson, S.C.; Binder, G.; Glass, I.; Strachan, T.; et al. The short stature homeobox gene SHOX is involved in skeletal abnormalities in Turner syndrome. Hum. Mol. Genet. 2000, 9, 695–702. [Google Scholar] [CrossRef] [PubMed]
- Trolle, C.; Nielsen, M.M.; Skakkebæk, A.; Lamy, P.; Vang, S.; Hedegaard, J.; Nordentoft, I.; Ørntoft, T.F.; Pedersen, J.S.; Gravholt, C.H. Widespread DNA hypomethylation and differential gene expression in Turner syndrome. Sci. Rep. 2016, 6, 1–14. [Google Scholar] [CrossRef]
- Kuntsi, J.; Skuse, D.; Elgar, K.; Morris, E.; Turner, C. Ring-X chromosomes: Their cognitive and behavioural phenotype. Ann. Hum. Genet. 2000. [Google Scholar] [CrossRef]
- Viana, J.; Pidsley, R.; Troakes, C.; Spiers, H.; Wong, C.C.Y.; Al-Sarraj, S.; Craig, I.; Schalkwyk, L.; Mill, J. Epigenomic and transcriptomic signatures of a Klinefelter syndrome (47,XXY) karyotype in the brain. Epigenetics 2014, 9, 587–599. [Google Scholar] [CrossRef]
- Zhang, X.; Yang, J.; Li, Y.; Ma, X.; Li, R. Sex chromosome abnormalities and psychiatric diseases. Oncotarget 2017, 8, 3969–3979. [Google Scholar] [CrossRef]
- Navarro-Cobos, M.J.; Balaton, B.P.; Brown, C.J. Genes that escape from X-chromosome inactivation: Potential contributors to Klinefelter syndrome. Am. J. Med. Genet. C Semin. Med. Genet. 2020, 184, 226–238. [Google Scholar] [CrossRef]
- Clemson, C.M.; Hutchinson, J.N.; Sara, S.A.; Ensminger, A.W.; Fox, A.H.; Chess, A.; Lawrence, J.B. An Architectural Role for a Nuclear Noncoding RNA: NEAT1 RNA Is Essential for the Structure of Paraspeckles. Mol. Cell 2009, 33, 717–726. [Google Scholar] [CrossRef] [PubMed]
- Naganuma, T.; Nakagawa, S.; Tanigawa, A.; Sasaki, Y.F.; Goshima, N.; Hirose, T. Alternative 3′-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. EMBO J. 2012, 31, 4020–4034. [Google Scholar] [CrossRef]
- Sunwoo, H.; Dinger, M.E.; Wilusz, J.E.; Amaral, P.P.; Mattick, J.S.; Spector, D.L. Men ε/β nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009, 19, 347–359. [Google Scholar] [CrossRef]
- Sasaki, Y.T.F.; Ideue, T.; Sano, M.; Mituyama, T.; Hirose, T. MENε/β noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc. Natl. Acad. Sci. USA 2009, 106, 2525–2530. [Google Scholar] [CrossRef]
- Chen, L.-L.; Carmichael, G.G. Altered Nuclear Retention of mRNAs Containing Inverted Repeats in Human Embryonic Stem Cells: Functional Role of a Nuclear Noncoding RNA. Mol. Cell 2009, 35, 467–478. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, S.; Naganuma, T.; Shioi, G.; Hirose, T. Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J. Cell Biol. 2011, 193, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Chujo, T.; Yamazaki, T.; Kawaguchi, T.; Kurosaka, S.; Takumi, T.; Nakagawa, S.; Hirose, T. Unusual semi-extractability as a hallmark of nuclear body-associated architectural noncoding RNAs. EMBO J. 2017, 36, 1447–1462. [Google Scholar] [CrossRef] [PubMed]
- Knott, G.J.; Bond, C.S.; Fox, A.H. The DBHS proteins SFPQ, NONO and PSPC1: A multipurpose molecular scaffold. Nucleic Acids Res. 2016, 44, 3989–4004. [Google Scholar] [CrossRef] [PubMed]
- Passon, D.M.; Lee, M.; Rackham, O.; Stanley, W.A.; Sadowska, A.; Filipovska, A.; Fox, A.H.; Bond, C.S. Structure of the heterodimer of human NONO and paraspeckle protein component 1 and analysis of its role in subnuclear body formation. Proc. Natl. Acad. Sci. USA 2012, 109, 4846–4850. [Google Scholar] [CrossRef]
- Hennig, S.; Kong, G.; Mannen, T.; Sadowska, A.; Kobelke, S.; Blythe, A.; Knott, G.J.; Iyer, S.S.; Ho, D.; Newcombe, E.A.; et al. Prion-like domains in RNA binding proteins are essential for building subnuclear paraspeckles. J. Cell Biol. 2015, 210, 529–539. [Google Scholar] [CrossRef]
- Maharana, S.; Wang, J.; Papadopoulos, D.K.; Richter, D.; Pozniakovsky, A.; Poser, I.; Bickle, M.; Rizk, S.; Guillén-Boixet, J.; Franzmann, T.M.; et al. RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science 2018, 360, 918–921. [Google Scholar] [CrossRef]
- Mao, Y.S.; Sunwoo, H.; Zhang, B.; Spector, D.L. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat. Cell Biol. 2011, 13, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Li, K.; Huang, W. Long non-coding RNA NEAT1-centric gene regulation. Cell. Mol. Life Sci. 2020, 77, 3769–3779. [Google Scholar] [CrossRef]
- Yamazaki, T.; Souquere, S.; Chujo, T.; Kobelke, S.; Chong, Y.S.; Fox, A.H.; Bond, C.S.; Nakagawa, S.; Pierron, G.; Hirose, T. Functional Domains of NEAT1 Architectural lncRNA Induce Paraspeckle Assembly through Phase Separation. Mol. Cell 2018, 70, 1038–1053.e7. [Google Scholar] [CrossRef]
- West, J.A.; Mito, M.; Kurosaka, S.; Takumi, T.; Tanegashima, C.; Chujo, T.; Yanaka, K.; Kingston, R.E.; Hirose, T.; Bond, C.; et al. Structural, super-resolution microscopy analysis of paraspeckle nuclear body organization. J. Cell Biol. 2016, 214, 817–830. [Google Scholar] [CrossRef]
- Kawaguchi, T.; Tanigawa, A.; Naganuma, T.; Ohkawa, Y.; Souquere, S.; Pierron, G.; Hirose, T. SWI/SNF chromatin-remodeling complexes function in noncoding RNA-dependent assembly of nuclear bodies. Proc. Natl. Acad. Sci. USA 2015, 112, 4304–4309. [Google Scholar] [CrossRef] [PubMed]
- Hirose, T.; Virnicchi, G.; Tanigawa, A.; Naganuma, T.; Li, R.; Kimura, H.; Yokoi, T.; Nakagawa, S.; Bénard, M.; Fox, A.H.; et al. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol. Biol. Cell 2014, 25, 169–183. [Google Scholar] [CrossRef]
- West, J.A.; Davis, C.P.; Sunwoo, H.; Simon, M.D.; Sadreyev, R.I.; Wang, P.I.; Tolstorukov, M.Y.; Kingston, R.E. The Long Noncoding RNAs NEAT1 and MALAT1 Bind Active Chromatin Sites. Mol. Cell 2014, 55, 791–802. [Google Scholar] [CrossRef]
- Chakravarty, D.; Sboner, A.; Nair, S.S.; Giannopoulou, E.; Li, R.; Hennig, S.; Mosquera, J.M.; Pauwels, J.; Park, K.; Kossai, M.; et al. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Grosch, M.; Ittermann, S.; Rusha, E.; Greisle, T.; Ori, C.; Truong, D.J.J.; O’Neill, A.C.; Pertek, A.; Westmeyer, G.G.; Drukker, M. Nucleus size and DNA accessibility are linked to the regulation of paraspeckle formation in cellular differentiation. BMC Biol. 2020, 18, 42. [Google Scholar] [CrossRef]
- Imamura, K.; Imamachi, N.; Akizuki, G.; Kumakura, M.; Kawaguchi, A.; Nagata, K.; Kato, A.; Kawaguchi, Y.; Sato, H.; Yoneda, M.; et al. Long Noncoding RNA NEAT1-Dependent SFPQ Relocation from Promoter Region to Paraspeckle Mediates IL8 Expression upon Immune Stimuli. Mol. Cell 2014, 53, 393–406. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; He, L.; Yin, L. lncRNA NEAT1 Binds to MiR-339-5p to Increase HOXA1 and Alleviate Ischemic Brain Damage in Neonatal Mice. Mol. Ther. Nucleic Acid 2020, 20, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Li, J.-W.; Ren, S.-H.; Ren, J.-R.; Zhen, Z.-G.; Li, L.-R.; Hao, X.-D.; Ji, H.-M. Nimodipine Improves Cognitive Impairment After Subarachnoid Hemorrhage in Rats Through IncRNA NEAT1/miR-27a/MAPT Axis. Drug Des. Dev. Ther. 2020, 14, 2295–2306. [Google Scholar] [CrossRef]
- Jiang, L.; Shao, C.; Wu, Q.J.; Chen, G.; Zhou, J.; Yang, B.; Li, H.; Gou, L.T.; Zhang, Y.; Wang, Y.; et al. NEAT1 scaffolds RNA-binding proteins and the Microprocessor to globally enhance pri-miRNA processing. Nat. Struct. Mol. Biol. 2017, 24, 816–824. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Fan, P.; Zhao, Y.; Zhang, S.; Lu, J.; Xie, W.; Jiang, Y.; Lei, F.; Xu, N.; Zhang, Y. NEAT1 modulates herpes simplex virus-1 replication by regulating viral gene transcription. Cell. Mol. Life Sci. 2017, 74, 1117–1131. [Google Scholar] [CrossRef] [PubMed]
- Pisani, G.; Baron, B. Nuclear paraspeckles function in mediating gene regulatory and apoptotic pathways. Non-coding RNA Res. 2019, 4, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Nishimoto, Y.; Nakagawa, S.; Hirose, T.; Okano, H.J.; Takao, M.; Shibata, S.; Suyama, S.; Kuwako, K.I.; Imai, T.; Murayama, S.; et al. The long non-coding RNA nuclear-enriched abundant transcript 1-2 induces paraspeckle formation in the motor neuron during the early phase of amyotrophic lateral sclerosis. Mol. Brain 2013, 6, 1–18. [Google Scholar] [CrossRef]
- Butler, A.A.; Johnston, D.R.; Kaur, S.; Lubin, F.D. Long noncoding RNA NEAT1 mediates neuronal histone methylation and age-related memory impairment. Sci. Signal. 2019, 12. [Google Scholar] [CrossRef]
- Kukharsky, M.S.; Ninkina, N.N.; An, H.; Telezhkin, V.; Wei, W.; de Meritens, C.R.; Cooper-knock, J.; Nakagawa, S.; Hirose, T.; Buchman, V.L.; et al. Long non-coding RNA Neat1 regulates adaptive behavioural response to stress in mice. Transl. Psychiatry 2020, 10, 1–19. [Google Scholar] [CrossRef]
- Nakagawa, S.; Shimada, M.; Yanaka, K.; Mito, M.; Arai, T.; Takahashi, E.; Fujita, Y.; Fujimori, T.; Standaert, L.; Marine, J.C.; et al. The lncRNA Neat1 is required for corpus luteum formation and the establishment of pregnancy in a subpopulation of mice. Development 2014, 141, 4618–4627. [Google Scholar] [CrossRef]
- Cheng, C.; Spengler, R.M.; Keiser, M.S.; Monteys, A.M.; Rieders, J.M.; Ramachandran, S.; Davidson, B.L. The long non-coding RNA NEAT1 is elevated in polyglutamine repeat expansion diseases and protects from disease gene-dependent toxicities. Hum. Mol. Genet. 2018, 27, 4303–4314. [Google Scholar] [CrossRef]
- Lipovich, L.; Dachet, F.; Cai, J.; Bagla, S.; Balan, K.; Jia, H.; Loeb, J.A. Activity-dependent human brain coding/noncoding gene regulatory networks. Genetics 2012, 192, 1133–1148. [Google Scholar] [CrossRef]
- Barry, G.; Briggs, J.A.; Hwang, D.W.; Nayler, S.P.; Fortuna, P.R.J.; Jonkhout, N.; Dachet, F.; Maag, J.L.V.; Mestdagh, P.; Singh, E.M.; et al. The long non-coding RNA NEAT1 is responsive to neuronal activity and is associated with hyperexcitability states. Sci. Rep. 2017, 7, 40127. [Google Scholar] [CrossRef]
- Pongs, O. Regulation of excitability by potassium channels. Results Probl. Cell Differ. 2007, 44, 145–161. [Google Scholar] [CrossRef]
- Pereira Fernandes, D.; Bitar, M.; Jacobs, F.; Barry, G. Long Non-Coding RNAs in Neuronal Aging. Non-Coding RNA 2018, 4, 12. [Google Scholar] [CrossRef]
- Barry, G.; Guennewig, B.; Fung, S.; Kaczorowski, D.; Weickert, C.S. Long non-coding RNA expression during aging in the human subependymal zone. Front. Neurol. 2015, 6, 1–10. [Google Scholar] [CrossRef]
- Suzuki, H.; Shibagaki, Y.; Hattori, S.; Matsuoka, M. C9-ALS/FTD-linked proline–arginine dipeptide repeat protein associates with paraspeckle components and increases paraspeckle formation. Cell Death Dis. 2019, 10, 746. [Google Scholar] [CrossRef] [PubMed]
- Harley, J.; Patani, R. Stress-Specific Spatiotemporal Responses of RNA-Binding Proteins in Human Stem Cell-Derived Motor Neurons. Int. J. Mol. Sci. 2020, 21, 8346. [Google Scholar] [CrossRef] [PubMed]
- Koza, P.; Beroun, A.; Konopka, A.; Górkiewicz, T.; Bijoch, L.; Torres, J.C.; Bulska, E.; Knapska, E.; Kaczmarek, L.; Konopka, W. Neuronal TDP-43 depletion affects activity-dependent plasticity. Neurobiol. Dis. 2019, 130, 104499. [Google Scholar] [CrossRef] [PubMed]
- Tollervey, J.R.; Curk, T.; Rogelj, B.; Briese, M.; Cereda, M.; Kayikci, M.; Hortobágyi, T.; Nishimura, A.L.; Župunski, V.; Patani, R.; et al. Characterising the RNA targets and position-dependent splicing regulation by TDP-43; implications for neurodegenerative diseases Europe PMC Funders Group. Nat. Neurosci. 2011, 14, 452–458. [Google Scholar] [CrossRef]
- Wang, Z.; Zhao, Y.; Xu, N.; Zhang, S.; Wang, S.; Mao, Y.; Zhu, Y.; Li, B.; Jiang, Y.; Tan, Y.; et al. NEAT1 regulates neuroglial cell mediating Aβ clearance via the epigenetic regulation of endocytosis-related genes expression. Cell. Mol. Life Sci. 2019, 76, 3005–3018. [Google Scholar] [CrossRef]
- Shelkovnikova, T.A.; Kukharsky, M.S.; An, H.; Dimasi, P.; Alexeeva, S.; Shabir, O.; Heath, P.R.; Buchman, V.L. Protective paraspeckle hyper-assembly downstream of TDP-43 loss of function in amyotrophic lateral sclerosis. Mol. Neurodegener. 2018, 13, 1–17. [Google Scholar] [CrossRef]
- Ji, P.; Diederichs, S.; Wang, W.; Boïng, S.; Metzger, R.; Schneider, P.M.; Tidow, N.; Brandt, B.; Buerger, H.; Bulk, E.; et al. MALAT-1, a novel noncoding RNA, and thymosin b4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 2003, 22, 8031–8041. [Google Scholar] [CrossRef]
- Johnsson, P.; Lipovich, L.; Grandér, D.; Morris, K.V. Evolutionary conservation of long non-coding RNAs; Sequence, structure, function. Biochim. Biophys. Acta Gen. Subj. 2014, 1840, 1063–1071. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, V.; Ellis, J.D.; Shen, Z.; Song, D.Y.; Pan, Q.; Watt, A.T.; Freier, S.M.; Bennett, C.F.; Sharma, A.; Bubulya, P.A.; et al. The Nuclear-Retained Noncoding RNA MALAT1 Regulates Alternative Splicing by Modulating SR Splicing Factor Phosphorylation. Mol. Cell 2010, 39, 925–938. [Google Scholar] [CrossRef]
- Lamond, A.I.; Spector, D.L. Nuclear speckles: A model for nuclear organelles. Nat. Rev. Mol. Cell Biol. 2003, 4, 605–612. [Google Scholar] [CrossRef]
- O’Keefe, R.T.; Henderson, S.C.; Spector, D.L. Dynamic organization of DNA replication in mammalian cell nuclei: Spatially and temporally defined replication of chromosome-specific α- satellite DNA sequences. J. Cell Biol. 1992, 116, 1095–1110. [Google Scholar] [CrossRef] [PubMed]
- Spector, D.L.; Fu, X.D.; Maniatis, T. Associations between distinct pre-mRNA splicing components and the cell nucleus. EMBO J. 1991, 10, 3467–3481. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Han, K.Y.; Khanna, N.; Ha, T.; Belmont, A.S. Nuclear speckle fusion via long-range directional motion regulates speckle morphology after transcriptional inhibition. J. Cell Sci. 2019, 132. [Google Scholar] [CrossRef] [PubMed]
- Galganski, L.; Urbanek, M.O.; Krzyzosiak, W.J. Nuclear speckles: Molecular organization, biological function and role in disease. Nucleic Acids Res. 2017, 45, 10350–10368. [Google Scholar] [CrossRef]
- Shopland, L.S.; Johnson, C.V.; Lawrence, J.B. Evidence that all SC-35 domains contain mRNAs and that transcripts can be structurally constrained within these domains. J. Struct. Biol. 2002, 140, 131–139. [Google Scholar] [CrossRef]
- Carter, K.C.; Taneja, K.L.; Lawrence, J.B. Discrete nuclear domains of poly(A) RNA and their relationship to the functional organization of the nucleus. J. Cell Biol. 1991, 115, 1191–1202. [Google Scholar] [CrossRef] [PubMed]
- Fei, J.; Jadaliha, M.; Harmon, T.S.; Li, I.T.S.; Hua, B.; Hao, Q.; Holehouse, A.S.; Reyer, M.; Sun, Q.; Freier, S.M.; et al. Quantitative analysis of multilayer organization of proteins and RNA in nuclear speckles at super resolution. J. Cell Sci. 2017, 130, 4180–4192. [Google Scholar] [CrossRef]
- Rieder, D.; Ploner, C.; Krogsdam, A.M.; Stocker, G.; Fischer, M.; Scheideler, M.; Dani, C.; Amri, E.-Z.; Müller, W.G.; Mcnally, J.G.; et al. Co-expressed genes prepositioned in spatial neighborhoods stochastically associate with SC35 speckles and RNA polymerase II factories. Cell. Mol. Life Sci. 2014, 71, 1741–1759. [Google Scholar] [CrossRef]
- Xing, Y.; Johnson, C.V.; Moen, P.T.; McNeil, J.A.; Lawrence, J.B. Nonrandom gene organization: Structural arrangements of specific pre-mRNA transcription and splicing with SC-35 domains. J. Cell Biol. 1995, 131, 1635–1647. [Google Scholar] [CrossRef] [PubMed]
- Szczerbal, I.; Bridger, J.M. Association of adipogenic genes with SC-35 domains during porcine adipogenesis. Chromosome Res. 2010, 18, 887–895. [Google Scholar] [CrossRef]
- Jolly, C.; Vourc’h, C.; Robert-Nicoud, M.; Morimoto, R.I. Intron-independent association of splicing factors with active genes. J. Cell Biol. 1999, 145, 1133–1143. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.P.; Moen, P.T.; Wydner, K.L.; Coleman, J.R.; Lawrence, J.B. Processing of endogenous pre-mRNAs in association with SC-35 domains is gene specific. J. Cell Biol. 1999, 144, 617–629. [Google Scholar] [CrossRef] [PubMed]
- Moen, P.T.; Johnson, C.V.; Byron, M.; Shopland, L.S.; De La Serna, I.L.; Imbalzano, A.N.; Lawrence, J.B. Repositioning of Muscle-specific Genes Relative to the Periphery of SC-35 Domains during Skeletal Myogenesis. Mol. Biol. Cell 2004, 15, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.M.; Leach, J.; Reittie, J.E.; Atzberger, A.; Lee-Prudhoe, J.; Wood, W.G.; Higgs, D.R.; Iborra, F.J.; Buckle, V.J. Coregulated human globin genes are frequently in spatial proximity when active. J. Cell Biol. 2006, 172, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.M.; Green, J.; Das Neves, R.P.; Wallace, H.A.C.; Smith, A.J.H.; Hughes, J.; Gray, N.; Taylor, S.; Wood, W.G.; Higgs, D.R.; et al. Association between active genes occurs at nuclear speckles and is modulated by chromatin environment. J. Cell Biol. 2008, 182, 1083–1097. [Google Scholar] [CrossRef]
- Nielsen, J.A.; Hudson, L.D.; Armstrong, R.C. Nuclear organization in differentiating oligodendrocytes. J. Cell Sci. 2002, 115, 4071–4079. [Google Scholar] [CrossRef]
- Kim, J.; Venkata, N.C.; Hernandez Gonzalez, G.A.; Khanna, N.; Belmont, A.S. Gene expression amplification by nuclear speckle association. J. Cell Biol. 2019, 219, e201904046. [Google Scholar] [CrossRef]
- Quinodoz, S.A.; Ollikainen, N.; Tabak, B.; Palla, A.; Schmidt, J.M.; Detmar, E.; Lai, M.M.; Shishkin, A.A.; Bhat, P.; Takei, Y.; et al. Higher-Order Inter-chromosomal Hubs Shape 3D Genome Organization in the Nucleus. Cell 2018, 174, 744–757.e24. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, Y.; Wang, Y.; Zhang, L.; Brinkman, E.K.; Adam, S.A.; Goldman, R.; Van Steensel, B.; Ma, J.; Belmont, A.S. Mapping 3D genome organization relative to nuclear compartments using TSA-Seq as a cytological ruler. J. Cell Biol. 2018, 217, 4025–4048. [Google Scholar] [CrossRef]
- Romero-Barrios, N.; Legascue, M.F.; Benhamed, M.; Ariel, F.; Crespi, M. Splicing regulation by long noncoding RNAs. Nucleic Acids Res. 2018, 46, 2169–2184. [Google Scholar] [CrossRef]
- Kataoka, N.; Fujita, M.; Ohno, M. Functional Association of the Microprocessor Complex with the Spliceosome. Mol. Cell. Biol. 2009, 29, 3243–3254. [Google Scholar] [CrossRef]
- Nakagawa, S.; Ip, J.Y.; Shioi, G.; Tripathi, V.; Zong, X.; Hirose, T.; Prasanth, K. V Malat1 is not an essential component of nuclear speckles in mice. RNA 2012, 18, 1487–1499. [Google Scholar] [CrossRef]
- Zhang, B.; Arun, G.; Mao, Y.S.; Lazar, Z.; Hung, G.; Bhattacharjee, G.; Xiao, X.; Booth, C.J.; Wu, J.; Zhang, C.; et al. The lncRNA Malat1 Is Dispensable for Mouse Development but Its Transcription Plays a cis-Regulatory Role in the Adult. Cell Rep. 2012, 2, 111–123. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.M.; Kabotyanski, E.B.; Reineke, L.C.; Shao, J.; Xiong, F.; Lee, J.-H.; Dubrulle, J.; Johnson, H.; Stossi, F.; Tsoi, P.S.; et al. The SINEB1 element in the long non-coding RNA Malat1 is necessary for TDP-43 proteostasis. Nucleic Acids Res. 2019, 48, 2621–2642. [Google Scholar] [CrossRef] [PubMed]
- Tani, H.; Okuda, S.; Nakamura, K.; Aoki, M.; Umemura, T. Short-lived long non-coding RNAs as surrogate indicators for chemical exposure and LINC00152 and MALAT1 modulate their neighboring genes. PLoS One 2017, 12. [Google Scholar] [CrossRef]
- Yang, L.; Lin, C.; Liu, W.; Zhang, J.; Ohgi, K.A.; Grinstein, J.D.; Dorrestein, P.C.; Rosenfeld, M.G. NcRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell 2011, 147, 773–788. [Google Scholar] [CrossRef] [PubMed]
- Medrano-Fernández, A.; Barco, A. Nuclear organization and 3D chromatin architecture in cognition and neuropsychiatric disorders. Mol. Brain 2016, 9, 1–12. [Google Scholar] [CrossRef]
- Navascues, J.; Casafont, I.; Villagra, N.T.; Lafarga, M.; Berciano, M.T. Reorganization of nuclear compartments of type A neurons of trigeminal ganglia in response to inflammatory injury of peripheral nerve endings. J. Neurocytol. 2004, 33, 393–405. [Google Scholar] [CrossRef] [PubMed]
- Meng, C.; Yang, X.; Liu, Y.; Zhou, Y.; Rui, J.; Li, S.; Xu, C.; Zhuang, Y.; Lao, J.; Zhao, X. Decreased expression of lncRNA Malat1 in rat spinal cord contributes to neuropathic pain by increasing neuron excitability after brachial plexus avulsion. J. Pain Res. 2019, 12, 1297–1310. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Cai, Z.; Ji, Z.; Zou, J.; Liang, Z.; Zhang, G.; Liang, Y.; Lin, H.; Tan, M. The lncRNA MALAT1/miR-30/Spastin Axis Regulates Hippocampal Neurite Outgrowth. Front. Cell. Neurosci. 2020, 14. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.; Ma, J.; Yan, L.; Li, T.; Li, Z.; Han, X.; Shui, S. Down-Regulation of Lncrna MALAT1 Attenuates Neuronal Cell Death Through Suppressing Beclin1-Dependent Autophagy by Regulating Mir-30a in Cerebral Ischemic Stroke. Cell. Physiol. Biochem. 2017, 43, 182–194. [Google Scholar] [CrossRef]
- Wu, Q.; Yi, X. Down-regulation of Long Noncoding RNA MALAT1 Protects Hippocampal Neurons Against Excessive Autophagy and Apoptosis via the PI3K/Akt Signaling Pathway in Rats with Epilepsy. J. Mol. Neurosci. 2018, 65, 234–245. [Google Scholar] [CrossRef]
- Liu, W.; Zhang, Q.; Zhang, J.; Pan, W.; Zhao, J.; Xu, Y. Long non-coding RNA MALAT1 contributes to cell apoptosis by sponging miR-124 in Parkinson disease. Cell Biosci. 2017, 7, 19. [Google Scholar] [CrossRef]
- Li, L.; Xu, Y.; Zhao, M.; Gao, Z. Neuro-protective roles of long non-coding RNA MALAT1 in Alzheimer’s disease with the involvement of the microRNA-30b/CNR1 network and the following PI3K/AKT activation. Exp. Mol. Pathol. 2020, 117. [Google Scholar] [CrossRef] [PubMed]
- Ma, P.; Li, Y.; Zhang, W.; Fang, F.; Sun, J.; Liu, M.; Li, K.; Dong, L. Long Non-coding RNA MALAT1 Inhibits Neuron Apoptosis and Neuroinflammation While Stimulates Neurite Outgrowth and Its Correlation With MiR-125b Mediates PTGS2, CDK5 and FOXQ1 in Alzheimer’s Disease. Curr. Alzheimer Res. 2019, 16, 596–612. [Google Scholar] [CrossRef]
- Rapicavoli, N.A.; Poth, E.M.; Blackshaw, S. The long noncoding RNA RNCR2 directs mouse retinal cell specification. BMC Dev. Biol. 2010, 10. [Google Scholar] [CrossRef]
- Mercer, T.R.; Qureshi, I.A.; Gokhan, S.; Dinger, M.E.; Li, G.; Mattick, J.S.; Mehler, M.F. Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. 2010, 11, 14. [Google Scholar] [CrossRef]
- Mercer, T.R.; Dinger, M.E.; Sunkin, S.M.; Mehler, M.F.; Mattick, J.S. Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl. Acad. Sci. USA 2008, 105, 716–721. [Google Scholar] [CrossRef] [PubMed]
- Aprea, J.; Prenninger, S.; Dori, M.; Ghosh, T.; Monasor, L.S.; Wessendorf, E.; Zocher, S.; Massalini, S.; Alexopoulou, D.; Lesche, M.; et al. Transcriptome sequencing during mouse brain development identifies long non-coding RNAs functionally involved in neurogenic commitment. EMBO J. 2013, 32, 3145–3160. [Google Scholar] [CrossRef]
- Stilling, R.M.; Benito, E.; Gertig, M.; Barth, J.; Capece, V.; Burkhardt, S.; Bonn, S.; Fischer, A. De-regulation of gene expression and alternative splicing affects distinct cellular pathways in the aging hippocampus. Front. Cell. Neurosci. 2014, 8, 1–15. [Google Scholar] [CrossRef]
- Michelhaugh, S.K.; Lipovich, L.; Blythe, J.; Jia, H.; Kapatos, G.; Bannon, M.J. Mining Affymetrix microarray data for long non-coding RNAs: Altered expression in the nucleus accumbens of heroin abusers. J. Neurochem. 2011, 116, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Ip, J.Y.; Sone, M.; Nashiki, C.; Pan, Q.; Kitaichi, K.; Yanaka, K.; Abe, T.; Takao, K.; Miyakawa, T.; Blencowe, B.J.; et al. Gomafu lncRNA knockout mice exhibit mild hyperactivity with enhanced responsiveness to the psychostimulant methamphetamine. Sci. Rep. 2016, 6, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Barry, G.; Briggs, J.A.; Vanichkina, D.P.; Poth, E.M.; Beveridge, N.J.; Ratnu, V.S.; Nayler, S.P.; Nones, K.; Hu, J.; Bredy, T.W.; et al. The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol. Psychiatry 2014, 19, 486–494. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sas-Nowosielska, H.; Magalska, A. Long Noncoding RNAs—Crucial Players Organizing the Landscape of the Neuronal Nucleus. Int. J. Mol. Sci. 2021, 22, 3478. https://doi.org/10.3390/ijms22073478
Sas-Nowosielska H, Magalska A. Long Noncoding RNAs—Crucial Players Organizing the Landscape of the Neuronal Nucleus. International Journal of Molecular Sciences. 2021; 22(7):3478. https://doi.org/10.3390/ijms22073478
Chicago/Turabian StyleSas-Nowosielska, Hanna, and Adriana Magalska. 2021. "Long Noncoding RNAs—Crucial Players Organizing the Landscape of the Neuronal Nucleus" International Journal of Molecular Sciences 22, no. 7: 3478. https://doi.org/10.3390/ijms22073478
APA StyleSas-Nowosielska, H., & Magalska, A. (2021). Long Noncoding RNAs—Crucial Players Organizing the Landscape of the Neuronal Nucleus. International Journal of Molecular Sciences, 22(7), 3478. https://doi.org/10.3390/ijms22073478