Association between HOX Transcript Antisense RNA Single-Nucleotide Variants and Recurrent Implantation Failure
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Genotype Analysis
2.3. Assessment of Blood Coagulation Status
2.4. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Polanski, L.T.; Baumgarten, M.N.; Quenby, S.; Brosens, J.; Campbell, B.K.; Raine-Fenning, N.J. What exactly do we mean by ‘recurrent implantation failure’? A systematic review and opinion. Reprod. Biomed. Online 2014, 28, 409–423. [Google Scholar] [CrossRef]
- Margalioth, E.J.; Ben-Chetrit, A.; Gal, M.; Eldar-Geva, T. Investigation and treatment of repeated implantation failure following IVF-ET. Hum. Reprod. 2006, 21, 3036–3043. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.K.; Vandekerckhove, P.; Kennedy, R.; Keay, S.D. Investigation and current management of recurrent IVF treatment failure in the UK. BJOG 2005, 112, 773–780. [Google Scholar] [CrossRef]
- Yarmishyn, A.A.; Kurochkin, I. V Long noncoding RNAs: A potential novel class of cancer biomarkers. Front. Genet. 2015, 6, 145. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Fu, F.; Chen, Y.; Qiu, W.; Lin, S.; Yang, P.; Huang, M.; Wang, C. Genetic variants in long noncoding RNA H19 contribute to the risk of breast cancer in a southeast China Han population. Onco Targets. Ther. 2017, 10, 4369–4378. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Ge, Y.; Wang, Y.; Ma, G.; Wang, X.; Liu, H.; Wang, M.; Zhang, Z.; Chu, H. The association of rs710886 in lncRNA PCAT1 with bladder cancer risk in a Chinese population. Gene 2017, 627, 226–232. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Yang, C.; Tong, S.; Ding, Y.; Deng, W.; Song, D.; Xiao, K. Genetic variation of long non-coding RNA TINCR contribute to the susceptibility and progression of colorectal cancer. Oncotarget 2017, 8, 33536–33543. [Google Scholar] [CrossRef]
- Zheng, D.; Hou, Y.; Li, Y.; Bian, Y.; Khan, M.; Li, F.; Huang, L.; Qiao, C. Long Non-coding RNA Gas5 Is Associated with Preeclampsia and Regulates Biological Behaviors of Trophoblast via MicroRNA-21. Front. Genet. 2020, 11, 188. [Google Scholar] [CrossRef]
- Huang, J.; Qian, Y.; Cheng, Q.; Yang, J.; Ding, H.; Jia, R. Over expression of long non-coding RNA uc.187 induces preeclampsia-like symptoms in pregnancy rats. Am. J. Hypertens. 2020. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, H.-Z.; Liu, Y.; Wang, H.-J.; Pang, W.-W.; Zhang, J.-J. Downregulated MALAT1 relates to recurrent pregnancy loss via sponging miRNAs. Kaohsiung J. Med. Sci. 2018, 34, 503–510. [Google Scholar] [CrossRef]
- Bhan, A.; Mandal, S.S. LncRNA HOTAIR: A master regulator of chromatin dynamics and cancer. Biochim. Biophys. Acta 2015, 1856, 151–164. [Google Scholar] [CrossRef]
- Tang, Q.; Hann, S.S. HOTAIR: An oncogenic long non-coding RNA in human cancer. Cell. Physiol. Biochem. 2018, 47, 893–913. [Google Scholar] [CrossRef] [PubMed]
- Milevskiy, M.J.G.; Al-Ejeh, F.; Saunus, J.M.; Northwood, K.S.; Bailey, P.J.; Betts, J.A.; McCart Reed, A.E.; Nephew, K.P.; Stone, A.; Gee, J.M.W.; et al. Long-range regulators of the lncRNA HOTAIR enhance its prognostic potential in breast cancer. Hum. Mol. Genet. 2016, 25, 3269–3283. [Google Scholar] [CrossRef]
- Rah, H.; Chung, K.W.; Ko, K.H.; Kim, E.S.; Kim, J.O.; Sakong, J.H.; Kim, J.H.; Lee, W.S.; Kim, N.K. miR-27a and miR-449b polymorphisms associated with a risk of idiopathic recurrent pregnancy loss. PLoS ONE 2017, 12, e0177160. [Google Scholar] [CrossRef]
- Park, H.S.; Kim, E.S.; Ahn, E.H.; Kim, J.O.; An, H.J.; Kim, J.H.; Lee, Y.; Lee, W.S.; Kim, Y.R.; Kim, N.K. The microRNA polymorphisms in miR-150 and miR-1179 are associated with risk of idiopathic recurrent pregnancy loss. Reprod. Biomed. Online 2019, 39, 187–195. [Google Scholar] [CrossRef]
- Lee, H.A.; Ahn, E.H.; Kim, J.H.; Kim, J.O.; Ryu, C.S.; Lee, J.Y.; Cho, S.H.; Lee, W.S.; Kim, N.K. Association study of frameshift and splice variant polymorphisms with risk of idiopathic recurrent pregnancy loss. Mol. Med. Rep. 2018, 18, 2417–2426. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.A.; Ahn, E.H.; Jang, H.G.; Kim, J.O.; Kim, J.H.; Lee, Y.B.; Lee, W.S.; Kim, N.K. Association between miR-605A>G, miR-608G>C, miR-631I>D, miR-938C>T, and miR-1302-3C>T Polymorphisms and Risk of Recurrent Implantation Failure. Reprod. Sci. 2019, 26, 469–475. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Ahn, E.H.; Kim, J.O.; Park, H.S.; Ryu, C.S.; Kim, J.H.; Kim, Y.R.; Lee, W.S.; Kim, N.K. Associations between microRNA (miR-25, miR-32, miR-125, and miR-222) polymorphisms and recurrent implantation failure in Korean women. Hum. Genom. 2019, 13, 68. [Google Scholar] [CrossRef]
- Boudjenah, R.; Molina-Gomes, D.; Wainer, R.; de Mazancourt, P.; Selva, J.; Vialard, F. The vascular endothelial growth factor (VEGF)+ 405 G/C polymorphism and its relationship with recurrent implantation failure in women in an IVF programme with ICSI. J. Assist. Reprod. Genet. 2012, 29, 1415–1420. [Google Scholar] [CrossRef]
- Rakhshan, A.; Zarrinpour, N.; Moradi, A.; Ahadi, M.; Omrani, M.D.; Ghafouri-Fard, S.; Taheri, M. A single nucleotide polymorphism within HOX Transcript Antisense RNA (HOTAIR) is associated with risk of psoriasis. Int. J. Immunogenet. 2020, 47, 430–434. [Google Scholar] [CrossRef]
- Mohammadpour-Gharehbagh, A.; Jahantigh, D.; Saravani, M.; Harati-Sadegh, M.; Maruie-Milan, R.; Teimoori, B.; Salimi, S. Impact of HOTAIR variants on preeclampsia susceptibility based on blood and placenta and in silico analysis. IUBMB Life 2019, 71, 1367–1381. [Google Scholar] [CrossRef] [PubMed]
- Mozdarani, H.; Ezzatizadeh, V.; Rahbar Parvaneh, R. The emerging role of the long non-coding RNA HOTAIR in breast cancer development and treatment. J. Transl. Med. 2020, 18, 152. [Google Scholar] [CrossRef]
- Kim, J.O.; Jun, H.H.; Kim, E.J.; Lee, J.Y.; Park, H.S.; Ryu, C.S.; Kim, S.; Oh, D.; Kim, J.W.; Kim, N.K. Genetic Variants of HOTAIR Associated with Colorectal Cancer Susceptibility and Mortality. Front. Oncol. 2020, 10, 72. [Google Scholar] [CrossRef] [PubMed]
- Lockshin, M.D.; Kim, M.; Laskin, C.A.; Guerra, M.; Branch, D.W.; Merrill, J.; Petri, M.; Porter, T.F.; Sammaritano, L.; Stephenson, M.D.; et al. Prediction of adverse pregnancy outcome by the presence of lupus anticoagulant, but not anticardiolipin antibody, in patients with antiphospholipid antibodies. Arthritis Rheum. 2012, 64, 2311–2318. [Google Scholar] [CrossRef]
- Middeldorp, S.; van Hylckama Vlieg, A. Does thrombophilia testing help in the clinical management of patients? Br. J. Haematol. 2008, 143, 321–335. [Google Scholar] [CrossRef]
- Sayad, A.; Badrlou, E.; Ghafouri-Fard, S.; Taheri, M. Association Analysis Between the rs1899663 Polymorphism of HOTAIR and Risk of Psychiatric Conditions in an Iranian Population. J. Mol. Neurosci. 2020, 70, 953–958. [Google Scholar] [CrossRef]
- Taheri, M.; Noroozi, R.; Sadeghpour, S.; Omrani, M.D.; Ghafouri-Fard, S. The rs4759314 SNP within Hotair lncRNA is associated with risk of multiple sclerosis. Mult. Scler. Relat. Disord. 2020, 40, 101986. [Google Scholar] [CrossRef]
- Li, J.; Liu, R.; Tang, S.; Feng, F.; Wang, X.; Qi, L.; Liu, C.; Yao, Y.; Sun, C. The effect of long noncoding RNAs HOX transcript antisense intergenic RNA single-nucleotide polymorphisms on breast cancer, cervical cancer, and ovarian cancer susceptibility: A meta-analysis. J. Cell. Biochem. 2019, 120, 7056–7067. [Google Scholar] [CrossRef]
- Tsai, M.-C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef]
- Basak, T.; Ain, R. Long non-coding RNAs in placental development and disease. Non-coding RNA Investig. 2019, 3, 14. [Google Scholar] [CrossRef]
- Lv, X.-B.; Lian, G.-Y.; Wang, H.-R.; Song, E.; Yao, H.; Wang, M.-H. Long noncoding RNA HOTAIR is a prognostic marker for esophageal squamous cell carcinoma progression and survival. PLoS ONE 2013, 8, e63516. [Google Scholar] [CrossRef]
- Ren, K.; Li, Y.; Lu, H.; Li, Z.; Li, Z.; Wu, K.; Li, Z.; Han, X. Long Noncoding RNA HOTAIR Controls Cell Cycle by Functioning as a Competing Endogenous RNA in Esophageal Squamous Cell Carcinoma. Transl. Oncol. 2016, 9, 489–497. [Google Scholar] [CrossRef]
- Liu, X.-H.; Sun, M.; Nie, F.-Q.; Ge, Y.-B.; Zhang, E.-B.; Yin, D.-D.; Kong, R.; Xia, R.; Lu, K.-H.; Li, J.-H.; et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol. Cancer 2014, 13, 92. [Google Scholar] [CrossRef]
- Wu, L.; Zhang, L.; Zheng, S. Role of the long non-coding RNA HOTAIR in hepatocellular carcinoma. Oncol. Lett. 2017, 14, 1233–1239. [Google Scholar] [CrossRef] [PubMed]
- Okugawa, Y.; Grady, W.M.; Goel, A. Epigenetic Alterations in Colorectal Cancer: Emerging Biomarkers. Gastroenterology 2015, 149, 1204–1225. [Google Scholar] [CrossRef]
- Liu, K.; Mao, X.; Chen, Y.; Li, T.; Ton, H. Regulatory role of long non-coding RNAs during reproductive disease. Am. J. Transl. Res. 2018, 10, 1–12. [Google Scholar]
- Chang, L.; Guo, R.; Yuan, Z.; Shi, H.; Zhang, D. LncRNA HOTAIR Regulates CCND1 and CCND2 Expression by Sponging miR-206 in Ovarian Cancer. Cell. Physiol. Biochem. 2018, 49, 1289–1303. [Google Scholar] [CrossRef] [PubMed]
- Xiong, X.R.; Lan, D.L.; Li, J.; Yin, S.; Xiong, Y.; Zi, X.D. Identification of differential abundances of mRNA transcript in cumulus cells and CCND1 associated with yak oocyte developmental competence. Anim. Reprod. Sci. 2019, 208, 106135. [Google Scholar] [CrossRef] [PubMed]
- Qiu, H.; Liu, Q.; Li, J.; Wang, X.; Wang, Y.; Yuan, Z.; Li, J.; Pei, D.-S. Analysis of the association of HOTAIR single nucleotide polymorphism (rs920778) and risk of cervical cancer. APMIS 2016, 124, 567–573. [Google Scholar] [CrossRef]
- Fagerberg, L.; Hallström, B.M.; Oksvold, P.; Kampf, C.; Djureinovic, D.; Odeberg, J.; Habuka, M.; Tahmasebpoor, S.; Danielsson, A.; Edlund, K.; et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteomics 2014, 13, 397–406. [Google Scholar] [CrossRef]
- Eun Kwon, H.Y.E.; Taylor, H.S. The Role of HOX Genes in Human Implantation. Ann. N. Y. Acad. Sci. 2004, 1034, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Heubach, J.; Monsior, J.; Deenen, R.; Niegisch, G.; Szarvas, T.; Niedworok, C.; Schulz, W.A.; Hoffmann, M.J. The long noncoding RNA HOTAIR has tissue and cell type-dependent effects on HOX gene expression and phenotype of urothelial cancer cells. Mol. Cancer 2015, 14, 108. [Google Scholar] [CrossRef] [PubMed]
- Aranda, S.; Mas, G.; Di Croce, L. Regulation of gene transcription by Polycomb proteins. Sci. Adv. 2015, 1, e1500737. [Google Scholar] [CrossRef]
- Caley, D.P.; Pink, R.C.; Trujillano, D.; Carter, D.R.F. Long noncoding RNAs, chromatin, and development. Sci. World J. 2010, 10, 90–102. [Google Scholar] [CrossRef]
- Chaligné, R.; Heard, E. X-chromosome inactivation in development and cancer. FEBS Lett. 2014, 588, 2514–2522. [Google Scholar] [CrossRef]
- Salama, E.A.; Adbeltawab, R.E.; El Tayebi, H.M. XIST and TSIX: Novel Cancer Immune Biomarkers in PD-L1-Overexpressing Breast Cancer Patients. Front. Oncol. 2020, 9, 1459. [Google Scholar] [CrossRef]
- Wu, K.; Liu, F.; Wu, W.; Chen, Y.; Wu, H.; Zhang, W. Long non-coding RNA HOX transcript antisense RNA (HOTAIR) suppresses the angiogenesis of human placentation by inhibiting vascular endothelial growth factor A expression. Reprod. Fertil. Dev. 2019, 31, 377–385. [Google Scholar] [CrossRef]
- Zou, Y.; Sun, L. Long noncoding RNA HOTAIR modulates the function of trophoblast cells in pre-eclampsia. Sichuan Da Xue Xue Bao Yi Xue Ban 2015, 46, 113–117. [Google Scholar]
- Zhang, Y.; Jin, F.; Li, X.-C.; Shen, F.-J.; Ma, X.-L.; Wu, F.; Zhang, S.-M.; Zeng, W.-H.; Liu, X.-R.; Fan, J.-X.; et al. The YY1-HOTAIR-MMP2 Signaling Axis Controls Trophoblast Invasion at the Maternal-Fetal Interface. Mol. Ther. 2017, 25, 2394–2403. [Google Scholar] [CrossRef]
- Ebert, M.S.; Sharp, P.A. Emerging roles for natural microRNA sponges. Curr. Biol. 2010, 20, R858–R861. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Zhang, J. Long non-coding RNA HOTAIR functions as miRNA sponge to promote the epithelial to mesenchymal transition in esophageal cancer. Biomed. Pharmacother. 2017, 90, 888–896. [Google Scholar] [CrossRef] [PubMed]
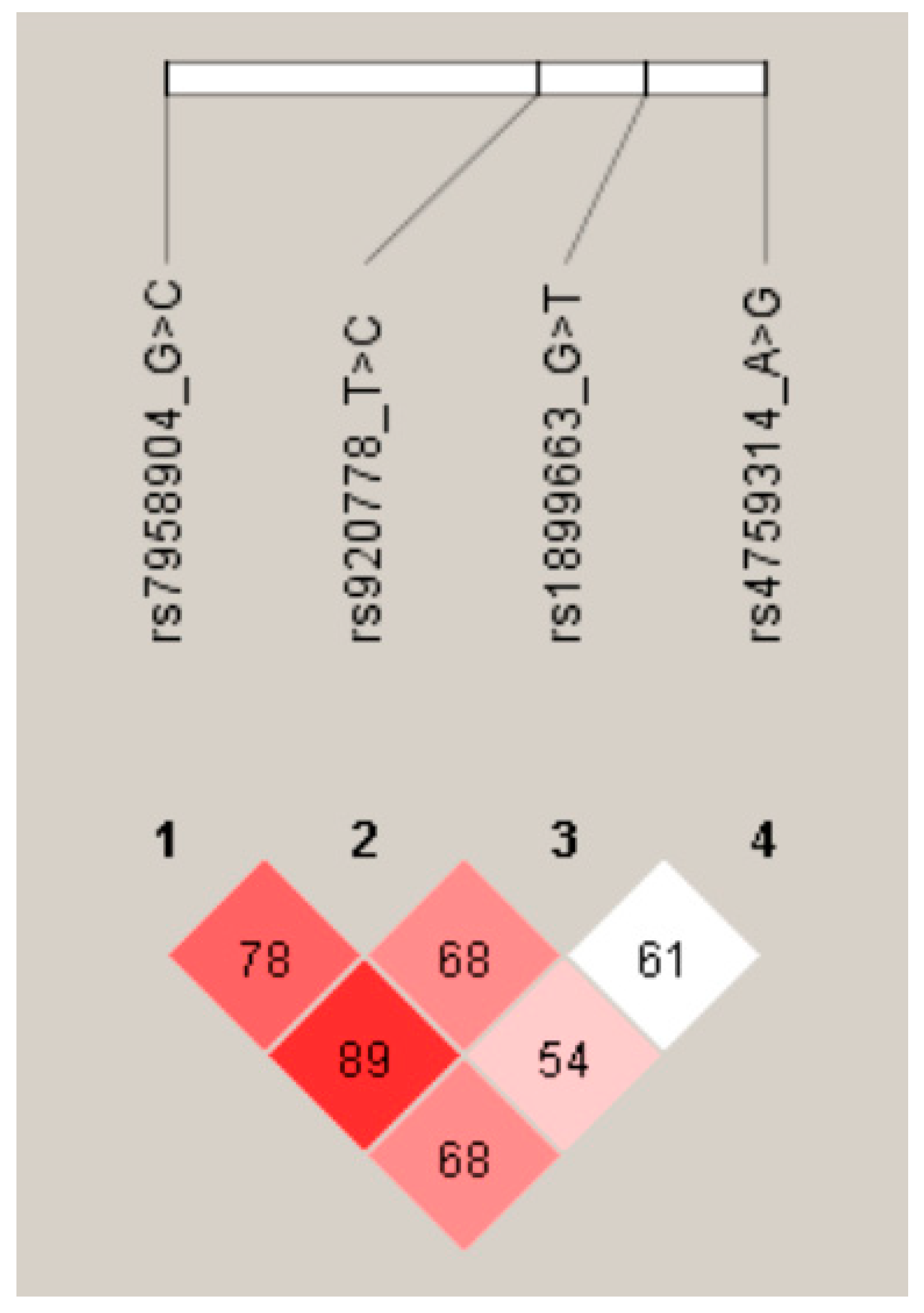
| Characteristics | Controls (n = 330) | RIF (n = 155) | p-Value |
|---|---|---|---|
| Age (years) | 33.69 ± 2.92 | 34.07 ± 3.11 | 0.194 |
| BMI (kg/m²) | 21.79 ± 3.40 | 20.96 ± 2.84 | 0.048 |
| Previous implantation failure (n) | N/A | 4.90 ± 2.12 | |
| Live births (n) | 1.67 ± 0.57 | N/A | |
| PT (sec) | 11.24 ± 3.18 | 10.78 ± 2.27 | 0.332 |
| aPTT (sec) | 30.26 ± 4.48 | 29.37 ± 3.48 | 0.127 |
| PLT (103/µL) | 242.52 ± 60.32 | 237.98 ± 59.63 | 0.895 |
| Homocysteine (µmol/L) | 3.71 ± 4.81 | 6.79 ± 1.48 | <0.0001 |
| Folate (mg/mL) | 13.67 ± 9.26 | 15.5 8 ± 10.19 | 0.617 |
| E2 | 26.27 ±14.72 | 37.88 ± 26.09 | <0.0001 * |
| FSH | 8.16 ± 2.85 | 8.88 ± 5.04 | 0.909 * |
| LH | 3.32 ± 1.76 | 4.84 ± 2.37 | <0.0001 * |
| Hgb | 36.14±4.01 | 12.56±1.44 | <0.0001 * |
| Genotypes | Controls (n = 330) | RIF (n = 155) | COR (95% CI) | p-Value | AOR (95% CI) | p-Value |
|---|---|---|---|---|---|---|
| HOTAIR rs4759314 | ||||||
| AA | 303 (91.8) | 140 (90.3) | 1.000 (reference) | 1.000 (reference) | ||
| AG | 25 (7.6) | 15 (9.7) | 1.299 (0.664–2.540) | 0.445 | 1.299 (0.663–2.544) | 0.445 |
| GG | 2 (0.6) | 0 (0.0) | N/A | 0.996 | N/A | 0.996 |
| Dominant (AA vs. AG + GG) | 1.202 (0.620–2.332 | 0.585 | 1.206 (0.621–2.342) | 0.580 | ||
| Recessive (AA + AG vs. GG) | N/A | 0.996 | N/A | 0.996 | ||
| HWE-P | 0.074 | 0.527 | ||||
| HOTAIR rs920778 | ||||||
| TT | 196 (59.4) | 92 (59.4) | 1.000 (reference) | 1.000 (reference) | ||
| TC | 122 (37.0) | 55 (35.5) | 0.960 (0.641–0.438) | 0.845 | 0.944 (0.629–0.416) | 0.781 |
| CC | 12 (3.6) | 8 (5.2) | 1.420 (0.561–0.594) | 0.459 | 1.475 (0.579–3.751) | 0.415 |
| Dominant (TT vs. TC + CC) | 1.002 (0.679–0.477) | 0.994 | 0.991 (0.671–0.463) | 0.964 | ||
| Recessive (TT + TC vs. CC) | 1.442 (0.577–0.603) | 0.433 | 1.525 (0.606–0.833) | 0.370 | ||
| HWE-P | 0.185 | 0.953 | ||||
| HOTAIR rs1899663 | ||||||
| GG | 188 (57.0) | 104 (67.1) | 1.000 (reference) | 1.000 (reference) | ||
| GT | 125 (37.9) | 45 (29.0) | 0.651 (0.429–0.987) | 0.043 | 0.638 (0.420–0.969) | 0.035 |
| TT | 17 (5.2) | 6 (3.9) | 0.638 (0.244–1.668) | 0.359 | 0.625 (0.239–1.639) | 0.340 |
| Dominant (GG vs. GT + TT) | 0.649 (0.435–0.968) | 0.034 | 0.638 (0.427–0.952) | 0.028 | ||
| Recessive (GG + GT vs. TT) | 0.741 (0.287–1.919) | 0.537 | 0.728 (0.281–1.889) | 0.515 | ||
| HWE-P | 0.517 | 0.684 | ||||
| HOTAIR rs7958904 | ||||||
| GG | 176 (53.3) | 99 (63.9) | 1.000 (reference) | 1.000 (reference) | ||
| GC | 129 (39.1) | 48 (31.0) | 0.662 (0.438–1.000) | 0.050 | 0.654 (0.432–0.989) | 0.044 |
| CC | 25 (7.6) | 8 (5.2) | 0.569 (0.247–1.309) | 0.185 | 0.566 (0.246–1.302) | 0.181 |
| Dominant (GG vs. GC + CC) | 0.647 (0.437–0.957) | 0.029 | 0.640 (0.432–0.948) | 0.026 | ||
| Recessive (GG + GC vs. CC) | 0.664 (0.292–1.508) | 0.328 | 0.655 (0.288–1.490) | 0.313 | ||
| HWE-P | 0.840 | 0.494 |
| Genotypes | Controls (n = 330) | RIF ≥ 3 (n = 130) | AOR (95% CI) | p-Value | RIF ≥ 4 (n = 98) | AOR (95% CI) | p-Value |
|---|---|---|---|---|---|---|---|
| HOTAIR rs4759314 | |||||||
| AA | 303 (91.8) | 119 (91.5) | 1.000 (reference) | 88 (89.8) | 1.000 (reference) | ||
| AG | 25 (7.6) | 11 (8.5) | 1.123 (0.535–2.359) | 0.759 | 10 (10.2) | 1.371 (0.632–2.971) | 0.424 |
| GG | 2 (0.6) | 0 (0.0) | N/A | 0.994 | 0 (0.0) | N/A | 0.994 |
| Dominant (GG vs. GA + AA) | 1.043 (0.501–2.175) | 0.910 | 1.275 (0.593–2.742) | 0.535 | |||
| Recessive (GG + GA vs. AA) | N/A | 0.994 | N/A | 0.994 | |||
| HOTAIR rs920778 | |||||||
| TT | 196 (59.4) | 80 (61.5) | 1.000 (reference) | 59 (60.2) | 1.000 (reference) | ||
| TC | 122 (37.0) | 43 (33.1) | 0.846 (0.547–0.309) | 0.452 | 32 (32.7) | 0.854 (0.524–0.391) | 0.526 |
| CC | 12 (3.6) | 7 (5.4) | 1.500 (0.565–0.974) | 0.415 | 7 (7.1) | 2.086 (0.776–0.604) | 0.145 |
| Dominant (CC vs. CT + TT) | 0.903 (0.594–0.370) | 0.631 | 0.957 (0.603–0.519) | 0.853 | |||
| Recessive (CC + CT vs. TT) | 1.610 (0.614–0.217) | 0.332 | 2.213 (0.837–0.846) | 0.109 | |||
| HOTAIR rs1899663 | |||||||
| GG | 188 (57.0) | 89 (68.5) | 1.000 (reference) | 66 (67.3) | 1.000 (reference) | ||
| GT | 125 (37.9) | 35 (26.9) | 0.576 (0.365–0.907) | 0.017 | 26 (26.5) | 0.578 (0.347–0.963) | 0.035 |
| TT | 17 (5.2) | 6 (4.6) | 0.729 (0.277–1.918) | 0.522 | 6 (6.1) | 0.976 (0.368–2.592) | 0.962 |
| Dominant (GG vs. GT + TT) | 0.596 (0.387–0.917) | 0.019 | 0.628 (0.390–1.013) | 0.056 | |||
| Recessive (GG + GT vs. TT) | 0.873 (0.335-2.272) | 0.781 | 1.174 (0.448–3.076) | 0.744 | |||
| HOTAIR rs7958904 | |||||||
| GG | 176 (53.3) | 86 (66.2) | 1.000 (reference) | 65 (66.3) | 1.000 (reference) | ||
| GC | 129 (39.1) | 37 (28.5) | 0.582 (0.372–0.912) | 0.018 | 26 (26.5) | 0.544 (0.327–0.906) | 0.019 |
| CC | 25 (7.6) | 7 (5.4) | 0.565 (0.235–1.361) | 0.203 | 7 (7.1) | 0.745 (0.306–1.809) | 0.515 |
| Dominant (GG vs. GC + CC) | 0.579 (0.379–0.885) | 0.012 | 0.577 (0.360–0.926) | 0.023 | |||
| Recessive (GG + GC vs. CC) | 0.681 (0.286–1.619) | 0.384 | 0.918 (0.383–2.199) | 0.848 |
| Allele Combination | Controls (2n = 660) | Case (2n = 310) | OR (95% CI) | p-Value |
|---|---|---|---|---|
| HOTAIR rs4759314A>G/rs920778T>C/rs1899663G>T/rs7958904G>C | ||||
| A-T-G-G | 0.6811 (450) | 0.7512 (233) | 1.000 (reference) | |
| A-T-T-C | 0.0678 (45) | 0.0032 (1) | 0.043 (0.005–0.314) | <0.0001 |
| A-C-G-G | 0.0293 (19) | 0.0033 (1) | 0.102 (0.013–0.764) | 0.006 |
| A-C-T-G | 0.006 (4) | 0.0293 (9) | 4.345 (1.324–14.260) | 0.015 |
| HOTAIR rs4759314A>G/rs920778T>C/rs1899663G>T | ||||
| A-T-G | 0.6929 (457) | 0.7612 (236) | 1.000 (reference) | |
| A-T-T | 0.0688 (45) | 0.0032 (1) | 0.043 (0.005–0.314) | <0.0001 |
| A-C-G | 0.0305 (20) | 0.0065 (2) | 0.194 (0.044–0.836) | 0.012 |
| HOTAIR rs4759314A>G/rs920778T>C/rs7958904G>C | ||||
| A-T-G | 0.6831 (451) | 0.7536 (234) | 1.000 (reference) | |
| A-T-C | 0.0768 (51) | 0.0134 (4) | 0.151 (0.053–0.424) | <0.0001 |
| HOTAIR rs4759314A>G/rs1899663G>T/rs7958904G>C | ||||
| A-G-G | 0.7105 (469) | 0.7576 (235) | 1.000 (reference) | |
| A-T-C | 0.2287 (151) | 0.1512 (47) | 0.621 (0.432–0.893) | 0.010 |
| HOTAIR rs920778T>C/rs1899663G>T/rs7958904G>C | ||||
| T-G-G | 0.6868 (453) | 0.7543 (234) | 1.000 (reference) | |
| T-T-C | 0.0696 (46) | 0.0032 (1) | 0.042 (0.005–0.307) | <0.0001 |
| C-G-G | 0.031 (20) | 0.0066 (2) | 0.194 (0.044–0.836) | 0.012 |
| C-T-G | 0.0061 (4) | 0.0293 (9) | 4.356 (1.327–14.300) | 0.015 |
| HOTAIR rs4759314A>G/rs920778T>C | ||||
| A-T | 0.7573 (500) | 0.767 (238) | 1.000 (reference) | |
| G-T | 0.0215 (14) | 0.004 (1) | 0.150 (0.019–1.148) | 0.046 |
| G-C | 0.0194 (13) | 0.0476 (15) | 2.424 (1.135–5.177) | 0.019 |
| HOTAIR rs4759314A>G/rs1899663G>T | ||||
| A-G | 0.718 (474) | 0.7753 (240) | 1.000 (reference) | |
| A-T | 0.2381 (157) | 0.1763 (55) | 0.692 (0.490–0.976) | 0.035 |
| HOTAIR rs4759314A>G/rs7958904G>C | ||||
| A-G | 0.7192 (475) | 0.7859 (244) | 1.000 (reference) | |
| A-C | 0.2369 (156) | 0.1658 (51) | 0.636 (0.448–0.905) | 0.011 |
| HOTAIR rs920778T>C/rs1899663G>T | ||||
| T-G | 0.7017 (463) | 0.7642 (237) | 1.000 (reference) | |
| T-T | 0.077 (51) | 0.0068 (2) | 0.077 (0.018–0.318) | <0.0001 |
| HOTAIR rs920778T>C/rs7958904G>C | ||||
| T-G | 0.691 (456) | 0.7576 (235) | 1.000 (reference) | |
| T-C | 0.0878 (58) | 0.0134 (4) | 0.134 (0.047–0.373) | <0.0001 |
| HOTAIR rs1899663G>T/rs7958904G>C | ||||
| G-G | 0.7177 (474) | 0.7594 (235) | 1.000 (reference) | |
| T-G | 0.0111 (7) | 0.0341 (11) | 3.170 (1.213–8.284) | 0.013 |
| T-C | 0.2298 (152) | 0.1497 (46) | 0.610 (0.424–0.879) | 0.008 |
| Genotypes | Homocysteine (mmol/L) | CD56+ NK Cells (%) | PT (sec) | Uric Acid (mg/dl) | T. Chol (mg/dl) | BUN (mg/dl) | Creatinine (mg/dl) | Hgb (mg/dl) | Estradiol (pg/mL) | FSH (mIU/mL) | LH (mIU/mL) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean ± SD (133) | Mean ± SD (132) | Mean ± SD (164) | Mean ± SD (77) | Mean ± SD (126) | Mean ± SD (152) | Mean ± SD (153) | Mean ± SD (277) | Mean ± SD (220) | Mean ± SD (206) | Mean ± SD (200) | |
| HOTAIR rs4759314 | |||||||||||
| AA | 5.17 ± 4.21 | 18.89 ± 9.56 | 10.88 ± 2.65 | 4.01 ± 1.01 | 190.73 ± 50.34 | 9.82 ± 2.83 | 0.76 ± 0.10 | 27.16 ± 11.99 | 32.63 ± 22.61 | 8.37 ± 3.73 | 4.06 ± 2.16 |
| AG | 4.28 ± 2.51 | 17.39 ± 7.08 | 11.18 ± 0.54 | 3.43 ± 0.83 | 189.18 ± 27.23 | 11.90 ± 2.39 | 0.80 ± 0.09 | 25.97 ± 11.85 | 26.78 ± 11.33 | 9.90 ± 6.43 | 3.78 ± 2.56 |
| GG | 2 | N/A | 8.93 ± 1.59 | N/A | N/A | N/A | N/A | 34.20 ± 3.95 | N/A | N/A | N/A |
| P | 0.422 | 0.572 | 0.480 | 0.181 | 0.920 | 0.015 | 0.220 | 0.633 | 0.308 | 0.540 | 0.606 |
| HOTAIR rs920778 | |||||||||||
| TT | 4.61 ± 3.09 | 19.07 ± 9.98 | 10.74 ± 2.12 | 4.07 ± 1.09 | 188.13 ± 45.24 | 9.90 ± 2.87 | 0.77 ± 0.10 | 26.95 ± 12.08 | 32.09 ± 20.16 | 8.60 ± 4.12 | 4.33 ± 2.22 |
| TC | 6.41 ± 5.67 | 19.15 ± 8.42 | 11.12 ± 3.21 | 3.76 ± 0.82 | 190.61 ± 45.85 | 10.20 ± 2.91 | 0.77 ± 0.10 | 27.52 ± 11.80 | 32.85 ± 25.28 | 8.52 ± 4.07 | 3.58 ± 2.16 |
| CC | 3.24 ± 2.47 | 12.99 ± 5.53 | 11.07 ± 0.59 | 3.80 ± 0.80 | 221.67 ± 96.16 | 9.43 ± 2.01 | 0.73 ± 0.08 | 25.91 ± 11.92 | 26.95 ± 13.24 | 6.77 ± 1.77 | 3.93 ± 1.53 |
| P | 0.027 | 0.199 | 0.641 | 0.434 | 0.269 | 0.745 | 0.752 | 0.875 | 0.769 | 0.462 | 0.071 |
| HOTAIR rs1899663 | |||||||||||
| GG | 4.62 ± 3.04 | 19.32 ± 9.74 | 10.87 ± 1.79 | 3.98 ± 1.11 | 186.94 ± 43.30 | 10.14 ± 2.95 | 0.77 ± 0.11 | 25.59 ± 12.21 | 32.78 ± 19.96 | 8.56 ± 4.14 | 4.31 ± 2.29 |
| GT | 6.04 ± 5.76 | 18.54 ± 8.35 | 10.92 ± 3.66 | 3.96 ± 0.77 | 197.25 ± 59.29 | 9.67 ± 2.74 | 0.76 ± 0.10 | 28.93 ± 11.33 | 32.70 ± 26.08 | 8.22 ± 3.33 | 3.75 ± 2.09 |
| TT | 4.20 ± 3.11 | 11.07 ± 4.38 | 10.80 ± 0.76 | 3.50 ± 0.71 | 203.00 ± 33.18 | 10.08 ± 1.02 | 0.80 ± 0.08 | 31.57 ± 10.82 | 24.20 ± 10.60 | 9.47 ± 6.20 | 3.11 ± 1.46 |
| P | 0.178 | 0.109 | 0.991 | 0.806 | 0.498 | 0.636 | 0.613 | 0.033 | 0.374 | 0.551 | 0.064 |
| HOTAIR rs7958904 | |||||||||||
| GG | 4.80 ± 3.05 | 19.47 ± 9.87 | 10.85 ± 1.85 | 4.00 ± 1.07 | 191.16 ± 49.92 | 9.90 ± 2.91 | 0.77 ± 0.11 | 25.35 ± 12.28 | 33.87 ± 23.50 | 8.75 ± 4.57 | 4.46 ± 2.38 |
| GC | 5.65 ± 5.96 | 18.35 ± 8.34 | 11.13 ± 3.26 | 3.83 ± 0.87 | 190.22 ± 48.77 | 10.17 ± 2.86 | 0.76 ± 0.11 | 29.14 ± 11.27 | 31.65 ± 21.19 | 8.31 ± 3.48 | 3.43 ± 1.86 |
| CC | 4.40 ± 2.73 | 12.88 ± 6.00 | 9.26 ± 4.09 | 4.20 ± 0.57 | 184.00 ±30.31 | 10.08 ± 1.73 | 0.78 ± 0.04 | 30.82 ± 10.46 | 23.63 ± 11.63 | 7.76 ± 2.23 | 3.75 ± 1.72 |
| P | 0.504 | 0.153 | 0.215 | 0.756 | 0.950 | 0.866 | 0.733 | 0.016 | 0.164 | 0.548 | 0.007 |
| Genotypes | Homocysteine (mmol/L) | PLT (103/µL) | aPTT (sec) | PT (sec) | Uric Acid (mg/dl) | BUN (mg/dl) | Creatinine (mg/dl) | Hgb (mg/dl) | Estradiol (pg/mL) | FSH (mIU/mL) | LH (mIU/mL) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean ± SD (57) | Mean ± SD (128) | Mean ± SD (127) | Mean ± SD (127) | Mean ± SD (70) | Mean ± SD (122) | Mean ± SD (123) | Mean ± SD (106) | Mean ± SD(111) | Mean ± SD (97) | Mean ± SD (94) | |
| HOTAIR rs4759314 | |||||||||||
| AA | 6.82 ± 1.51 | 240.15 ± 60.63 | 29.30 ± 3.33 | 10.73 ± 2.38 | 4.05 ± 1.00 | 10.28 ± 2.86 | 0.78 ± 0.10 | 12.48 ± 1.47 | 39.28 ± 27.04 | 8.65 ± 4.52 | 4.91 ± 2.32 |
| AG | 6.54 ± 1.33 | 215.00 ± 43.31 | 29.95 ± 4.76 | 11.27 ± 0.57 | 3.43 ± 0.83 | 12.26 ± 2.13 | 0.81 ± 0.08 | 13.31 ± 0.72 | 27.44 ± 12.13 | 10.71 ± 8.13 | 4.32 ± 2.75 |
| GG | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| P | 0.666 | 0.182 | 0.525 | 0.413 | 0.145 | 0.027 | 0.403 | 0.082 | 0.198 | 0.955 | 0.443 |
| HOTAIR rs920778 | |||||||||||
| TT | 6.53 ± 1.31 | 242.20 ± 63.76 | 29.58 ± 3.58 | 10.72 ± 2.37 | 4.14 ± 1.08 | 10.42 ± 2.93 | 0.79 ± 0.10 | 12.43 ± 1.48 | 37.83 ± 23.07 | 8.92 ± 5.04 | 4.92 ± 2.35 |
| TC | 7.17 ± 1.70 | 233.86 ± 53.68 | 29.14 ± 3.30 | 10.85 ± 2.23 | 3.77 ± 0.79 | 10.58 ± 2.86 | 0.78 ± 0.10 | 12.71 ± 1.35 | 40.86 ± 32.45 | 9.34 ± 5.41 | 4.81 ± 2.56 |
| CC | 8.12 | 207.60 ± 30.57 | 28.22 ± 4.04 | 11.22 ± 0.49 | 3.40 ± 0.57 | 9.88 ± 1.89 | 0.76 ± 0.05 | 13.00 ± 1.65 | 27.48 ± 15.55 | 6.06 ± 0.97 | 4.28 ± 1.62 |
| P | 0.193 | 0.389 | 0.603 | 0.867 | 0.244 | 0.863 | 0.826 | 0.508 | 0.506 | 0.343 | 0.818 |
| HOTAIR rs1899663 | |||||||||||
| GG | 6.52 ± 1.29 | 240.84 ± 63.68 | 29.73 ± 3.77 | 10.90 ± 1.97 | 4.34 ± 1.09 | 10.64 ± 2.95 | 0.79 ± 0.10 | 12.54 ± 1.45 | 37.39 ± 22.13 | 8.77 ± 4.87 | 4.88 ± 2.34 |
| GT | 7.44 ± 1.74 | 233.97 ± 50.45 | 28.69 ± 2.78 | 10.51 ± 2.90 | 3.96 ± 0.72 | 10.12 ± 2.74 | 0.78 ± 0.10 | 12.57 ± 1.38 | 41.05 ± 35.07 | 8.72 ± 4.11 | 4.99 ± 2.50 |
| TT | N/A | 206.00 ± 46.13 | 27.70 ± 0.66 | 10.80 ± 0.76 | 3.50 ± 0.71 | 9.80 ± 1.06 | 0.80 ± 0.10 | 12.90 ± 2.26 | 31.58 ± 6.98 | 11.73 ± 11.77 | 3.34 ± 2.02 |
| P | 0.031 | 0.543 | 0.222 | 0.678 | 0.743 | 0.601 | 0.871 | 0.914 | 0.632 | 0.842 | 0.426 |
| HOTAIR rs7958904 | |||||||||||
| GG | 6.54 ± 1.28 | 245.49 ± 62.17 | 29.62 ± 3.59 | 10.89 ± 2.00 | 4.03 ± 1.06 | 10.42 ± 2.94 | 0.79 ± 0.10 | 12.48 ± 1.45 | 38.93 ± 26.86 | 9.14 ± 5.45 | 4.97 ± 2.51 |
| GC | 7.52 ± 1.87 | 233.42 ± 55.35 | 28.91 ± 3.18 | 10.77 ± 2.42 | 3.89 ± 0.84 | 10.62 ± 2.83 | 0.78 ± 0.10 | 12.57 ± 1.45 | 39.20 ± 26.71 | 8.83 ± 4.42 | 4.63 ± 2.15 |
| CC | 7.10 ± 1.57 | 217.83 ± 12.89 | 28.64 ± 4.15 | 9.10 ± 4.55 | 4.20 ± 0.57 | 9.92 ± 1.88 | 0.78 ± 0.04 | 13.66 ± 0.67 | 26.08 ± 14.87 | 6.27 ± 1.18 | 4.36 ± 1.68 |
| P | 0.099 | 0.122 | 0.526 | 0.232 | 0.847 | 0.858 | 0.737 | 0.206 | 0.508 | 0.413 | 0.743 |
| Genotypes | Homocysteine (mmol/L) | PLT (103/µL) | aPTT (sec) | PT (sec) | T. Chol (mg/dl) | BUN (mg/dl) | Creatinine (mg/dl) | Hgb (mg/dl) | Estradiol (pg/mL) | FSH (mIU/mL) | LH (mIU/mL) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean ± SD (76) | Mean ± SD (175) | Mean ± SD (67) | Mean ± SD (37) | Mean ± SD (9) | Mean ± SD (30) | Mean ± SD (30) | Mean ± SD (171) | Mean ± SD (109) | Mean ± SD (109) | Mean ± SD (106) | |
| HOTAIR rs4759314 | |||||||||||
| AA | 3.90 ± 5.11 | 243.31 ± 59.20 | 30.05 ± 4.49 | 11.44 ± 3.47 | 229.44 ± 90.57 | 8.07 ± 1.86 | 0.68 ± 0.08 | 36.20 ± 3.98 | 26.30 ± 14.97 | 8.13 ± 2.91 | 3.35 ± 1.74 |
| AG | 2.59 ± 1.66 | 234.39 ± 78.94 | 32.32 ± 4.98 | 10.93 ± 0.38 | N/A | 7.90 | 0.7 | 35.72 ± 4.56 | 25.93 ± 11.12 | 8.64 ± 1.97 | 2.91 ± 2.14 |
| GG | 2 | 232.50 ±7.78 | 30.35 ± 1.63 | 8.93 ± 1.59 | N/A | N/A | N/A | 34.20 ± 3.96 | N/A | N/A | N/A |
| P | 0.680 | 0.854 | 0.504 | 0.554 | N/A | 0.931 | 0.806 | 0.726 | 0.949 | 0.648 | 0.530 |
| HOTAIR rs920778 | |||||||||||
| TT | 3.29 ± 3.28 | 239.28 ± 54.86 | 30.18 ± 4.63 | 10.81 ± 0.96 | 183.67 ± 42.30 | 8.09 ± 1.71 | 0.69 ± 0.08 | 36.16 ± 4.19 | 26.07 ± 14.48 | 8.30 ± 3.00 | 3.76 ± 1.94 |
| TC | 5.47 ± 8.30 | 248.08 ± 67.46 | 30.58 ± 4.40 | 12.40 ± 6.00 | 276.50 ± 84.15 | 8.09 ± 2.33 | 0.66 ± 0.07 | 36.22 ± 3.69 | 26.58 ± 15.49 | 7.95 ± 2.71 | 2.77 ± 1.35 |
| CC | 2.63 ± 1.77 | 239.86 ± 74.61 | 29.78 ± 4.05 | 10.82 ± 0.77 | 410.00 | 7.2 | 0.60 | 35.13 ± 4.57 | 25.35 ± 3.32 | 8.90 ± 2.26 | 2.90 ± 0.71 |
| P | 0.216 | 0.654 | 0.928 | 0.415 | 0.015 | 0.899 | 0.438 | 0.791 | 0.981 | 0.772 | 0.015 |
| HOTAIR rs1899663 | |||||||||||
| GG | 3.16± 3.20 | 237.89 ± 56.67 | 30.18 ± 4.59 | 10.76 ± 0.85 | 174.60 ± 40.25 | 7.88 ± 1.55 | 0.68 ± 0.08 | 36.16 ± 4.15 | 26.57 ± 14.63 | 8.29 ± 2.98 | 3.58 ± 2.03 |
| GT | 4.97 ± 7.41 | 248.58 ± 66.51 | 30.52 ± 4.43 | 12.13 ± 5.26 | 298.00 ± 91.85 | 8.10 ± 2.19 | 0.67 ± 0.08 | 35.97 ± 4.06 | 27.01 ± 15.59 | 7.94 ± 2.81 | 3.11 ± 1.50 |
| TT | 4.20 ± 3.11 | 240.90 ± 44.42 | 27.70 | N/A | N/A | 10.9 | 0.80 | 37.17 ± 1.97 | 21.25 ± 10.59 | 8.57 ± 2.48 | 3.02 ± 1.30 |
| P | 0.336 | 0.528 | 0.815 | 0.215 | 0.029 | 0.283 | 0.324 | 0.677 | 0.525 | 0.741 | 0.364 |
| HOTAIR rs7958904 | |||||||||||
| GG | 3.36 ± 3.35 | 241.63 ± 55.35 | 29.56 ± 4.39 | 10.70 ± 0.94 | 216.00 ± 93.86 | 7.79 ± 1.52 | 0.68 ± 0.08 | 36.26 ± 3.94 | 26.43 ± 14.78 | 8.22 ± 3.03 | 3.79 ± 2.02 |
| GC | 4.57 ± 7.25 | 244.65 ± 68.76 | 31.43 ± 4.61 | 11.98 ± 4.70 | 276.50 ± 84.15 | 8.36 ± 2.32 | 0.68 ± 0.08 | 35.90 ± 4.32 | 27.15 ± 15.75 | 8.03 ± 2.85 | 2.80 ± 1.33 |
| CC | 3.10 ± 2.20 | 237.60 ± 46.48 | 29.60 ± 2.69 | 10.05 | N/A | 10.9 | 0.80 | 36.54 ± 2.84 | 22.50 ± 10.30 | 8.45 ± 2.28 | 3.47 ± 1.73 |
| P | 0.594 | 0.902 | 0.262 | 0.466 | 0.442 | 0.218 | 0.327 | 0.789 | 0.602 | 0.878 | 0.022 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, J.Y.; Ahn, E.H.; Park, H.W.; Kim, J.H.; Kim, Y.R.; Lee, W.S.; Kim, N.K. Association between HOX Transcript Antisense RNA Single-Nucleotide Variants and Recurrent Implantation Failure. Int. J. Mol. Sci. 2021, 22, 3021. https://doi.org/10.3390/ijms22063021
Lee JY, Ahn EH, Park HW, Kim JH, Kim YR, Lee WS, Kim NK. Association between HOX Transcript Antisense RNA Single-Nucleotide Variants and Recurrent Implantation Failure. International Journal of Molecular Sciences. 2021; 22(6):3021. https://doi.org/10.3390/ijms22063021
Chicago/Turabian StyleLee, Jeong Yong, Eun Hee Ahn, Hyeon Woo Park, Ji Hyang Kim, Young Ran Kim, Woo Sik Lee, and Nam Keun Kim. 2021. "Association between HOX Transcript Antisense RNA Single-Nucleotide Variants and Recurrent Implantation Failure" International Journal of Molecular Sciences 22, no. 6: 3021. https://doi.org/10.3390/ijms22063021
APA StyleLee, J. Y., Ahn, E. H., Park, H. W., Kim, J. H., Kim, Y. R., Lee, W. S., & Kim, N. K. (2021). Association between HOX Transcript Antisense RNA Single-Nucleotide Variants and Recurrent Implantation Failure. International Journal of Molecular Sciences, 22(6), 3021. https://doi.org/10.3390/ijms22063021






