Cardiac Oxidative Signaling and Physiological Hypertrophy in the Na/K-ATPase α1s/sα2s/s Mouse Model of High Affinity for Cardiotonic Steroids
Abstract
1. Introduction
2. Results
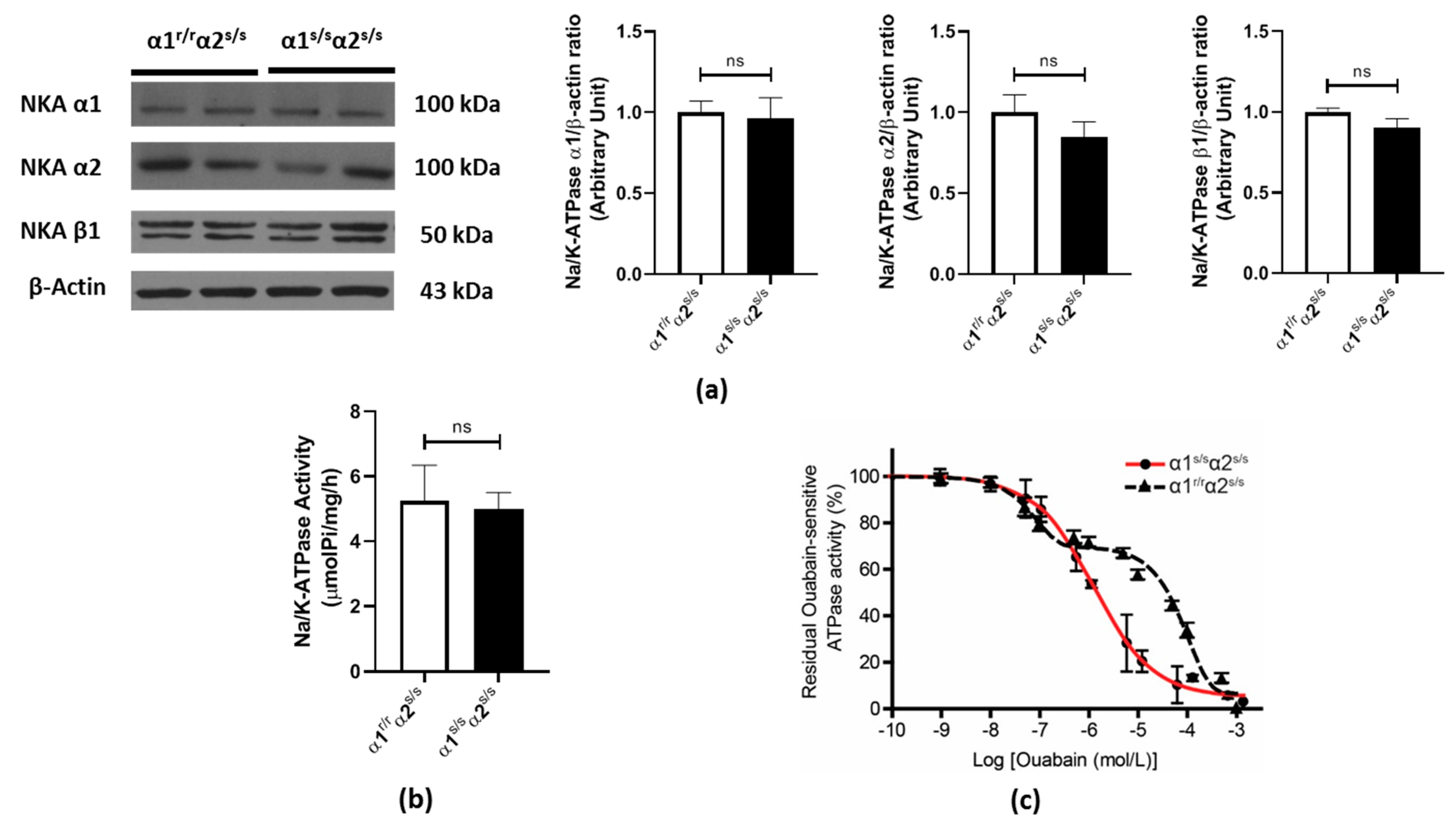
2.1. Cardiac Na+/K+-ATPase in the α1s/sα2s/s Mouse
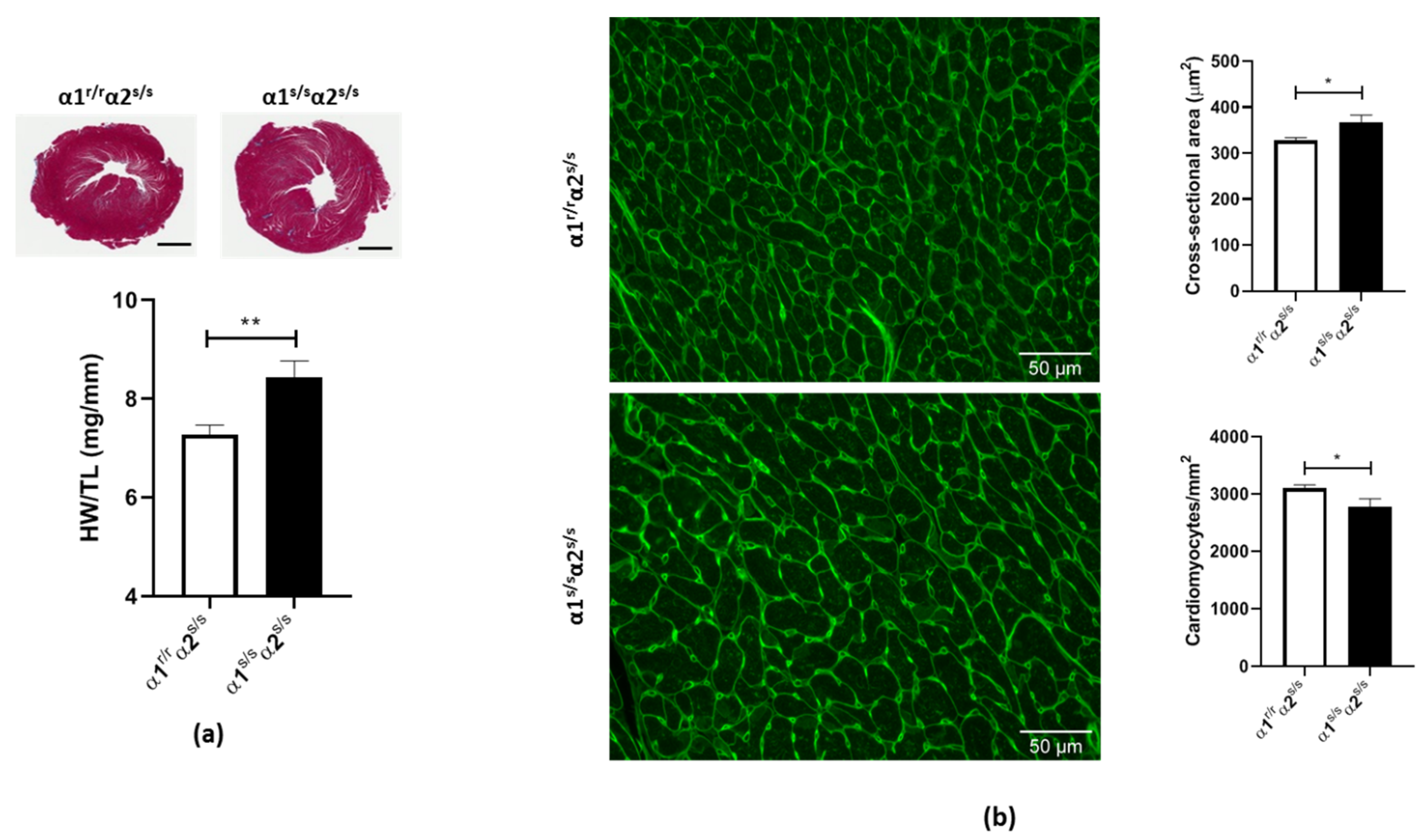
2.2. Cardiac Phenotype of the α1s/sα2s/s Mouse
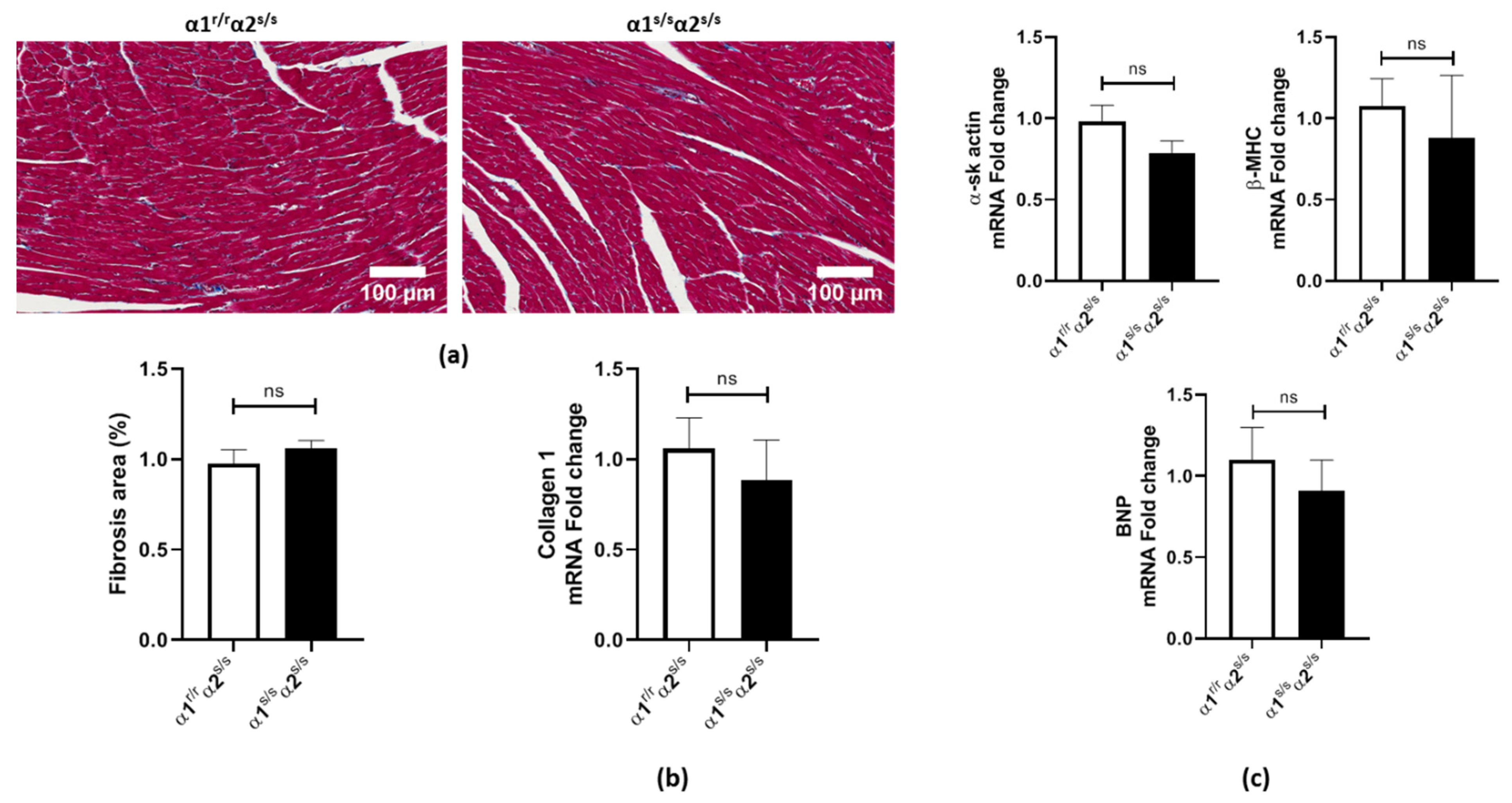
2.3. Absence of Adverse Cardiac Remodeling in α1s/sα2s/s Mice
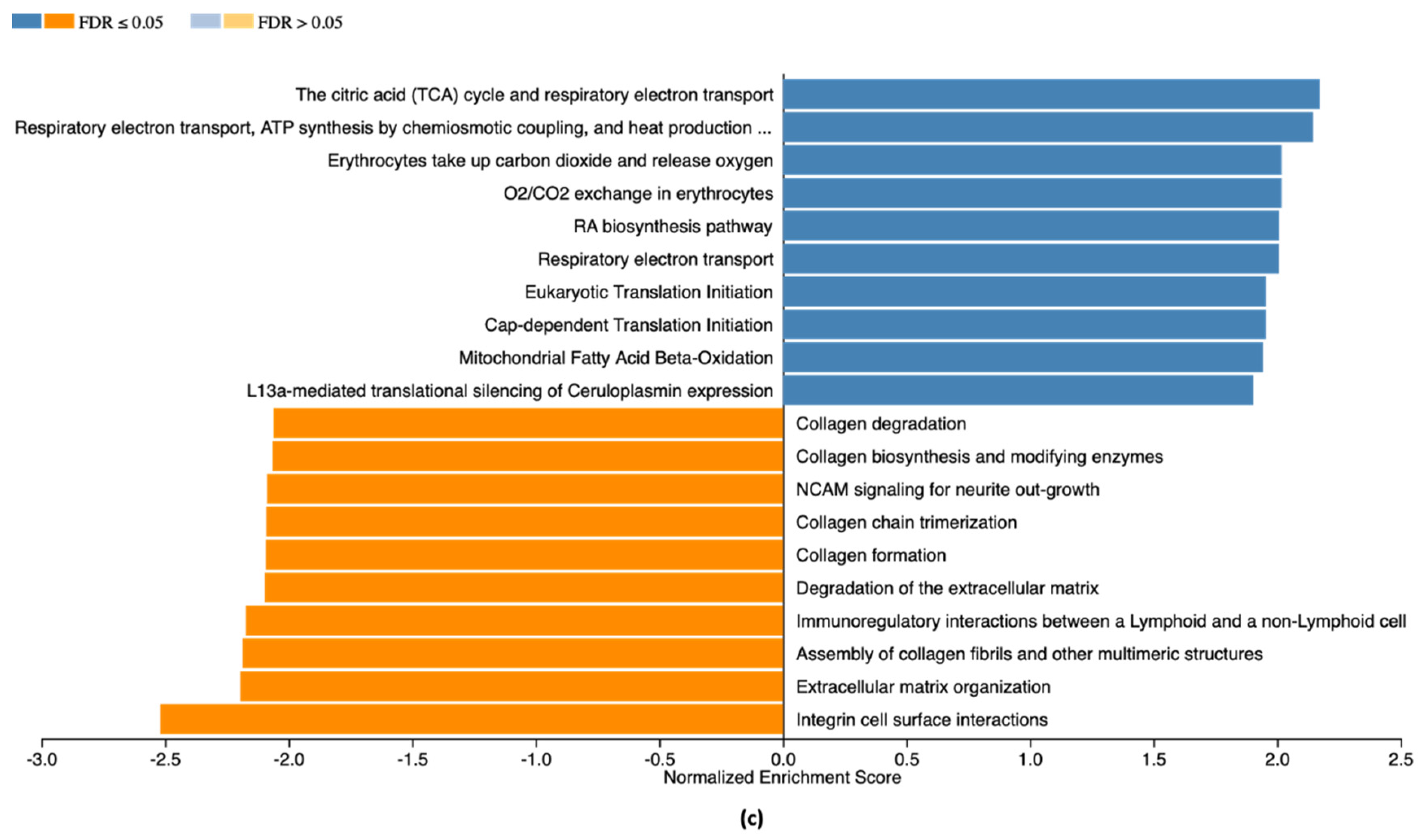
2.4. RNA-Seq Analysis
2.5. Evidence of Activation of the Na/K-ATPase Oxidant Amplification Loop in α1s/sα2s/s Mice
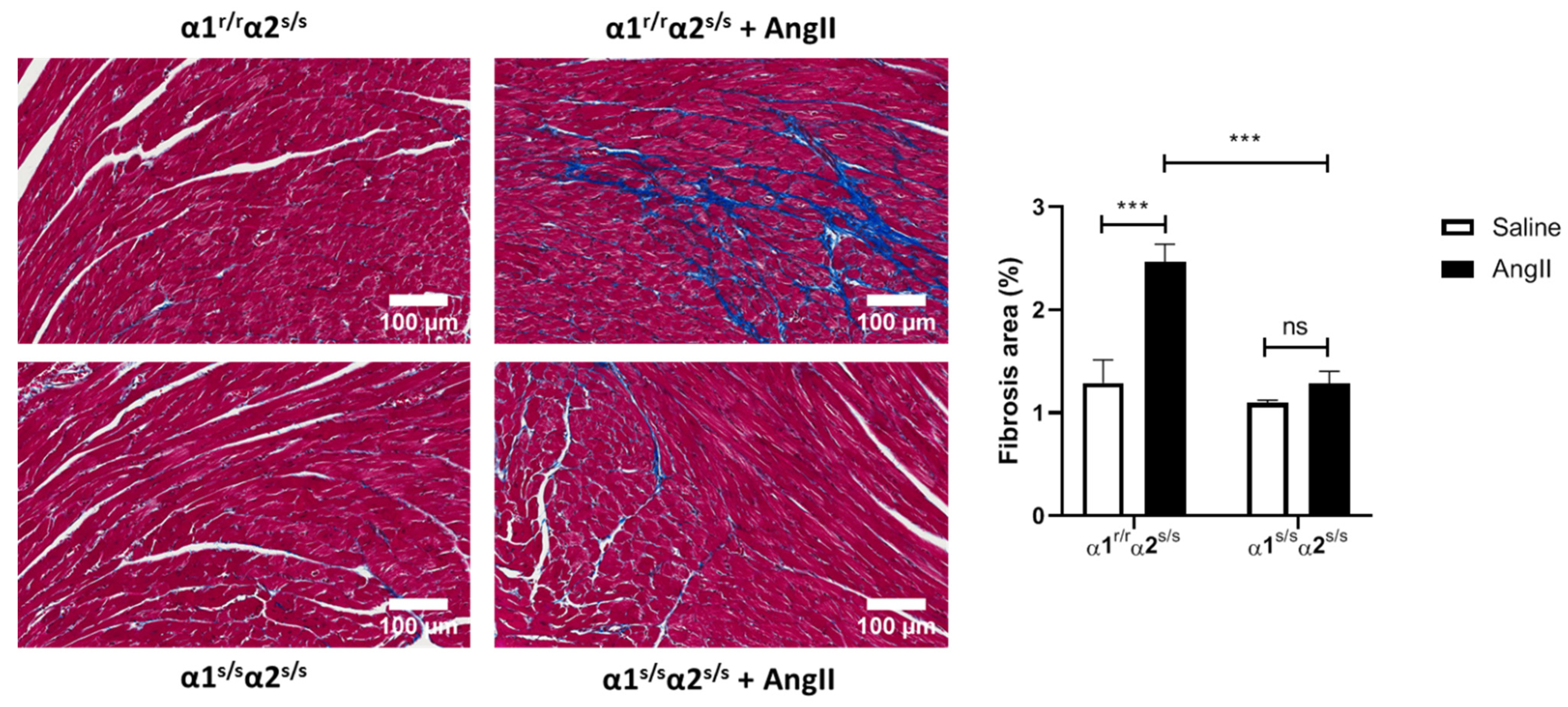
2.6. Angiotensin-II Does Not Induce Cardiac Fibrosis in α1s/sα2s/s Mice
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Generation of the NKA α1 Sensitive Mouse Model
4.3. Western Blot Analysis
4.4. Na/K-ATPase Activity and Ouabain Dose-Response in Heart Lysates
4.5. Echocardiography
4.6. Histology
4.7. Determination of mRNA Levels
4.8. Angiotensin-II Treatment
4.9. RNA Sequencing (RNA-Seq) Analysis
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bauer, N.; Müller-Ehmsen, J.; Krämer, U.; Hambarchian, N.; Zobel, C.; Schwinger, R.H.; Neu, H.; Kirch, U.; Grünbaum, E.G.; Schoner, W. Ouabain-like compound changes rapidly on physical exercise in humans and dogs: Effects of beta-blockade and angiotensin-converting enzyme inhibition. Hypertension 2005, 45, 1024–1028. [Google Scholar] [CrossRef]
- Bagrov, A.Y.; Shapiro, J.I.; Fedorova, O.V. Endogenous cardiotonic steroids: Physiology, pharmacology, and novel therapeutic targets. Pharmacol. Rev. 2009, 61, 9–38. [Google Scholar] [CrossRef]
- Ishkaraeva-Yakovleva, V.V.; Fedorova, O.V.; Solodovnikova, N.G.; Frolova, E.V.; Bzhelyansky, A.M.; Emelyanov, I.V.; Adair, C.D.; Zazerskaya, I.E.; Bagrov, A.Y. DigiFab interacts with endogenous cardiotonic steroids and reverses preeclampsia-induced Na/K-ATPase inhibition. Reprod. Sci. 2012, 19, 1260–1267. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Xie, Z.J. The sodium pump and cardiotonic steroids-induced signal transduction protein kinases and calcium-signaling microdomain in regulation of transporter trafficking. Biochim. Biophys. Acta 2010, 1802, 1237–1245. [Google Scholar] [CrossRef]
- Hamlyn, J.M.; Manunta, P. Endogenous cardiotonic steroids in kidney failure: A review and an hypothesis. Adv. Chronic Kidney Dis. 2015, 22, 232–244. [Google Scholar] [CrossRef] [PubMed]
- Gottlieb, S.S.; Rogowski, A.C.; Weinberg, M.; Krichten, C.M.; Hamilton, B.P.; Hamlyn, J.M. Elevated concentrations of endogenous ouabain in patients with congestive heart failure. Circulation 1992, 86, 420–425. [Google Scholar] [CrossRef] [PubMed]
- Deray, G.; Rieu, M.; Devynck, M.A.; Pernollet, M.G.; Chanson, P.; Luton, J.P.; Meyer, P. Evidence of an endogenous digitalis-like factor in the plasma of patients with acromegaly. N. Engl. J. Med. 1987, 316, 575–580. [Google Scholar] [CrossRef] [PubMed]
- Bagrov, A.Y.; Fedorova, O.V.; Dmitrieva, R.I.; Howald, W.N.; Hunter, A.P.; Kuznetsova, E.A.; Shpen, V.M. Characterization of a urinary bufodienolide Na+,K+-ATPase inhibitor in patients after acute myocardial infarction. Hypertension 1998, 31, 1097–1103. [Google Scholar] [CrossRef]
- Schoner, W.; Scheiner-Bobis, G. Endogenous and exogenous cardiac glycosides: Their roles in hypertension, salt metabolism, and cell growth. Am. J. Physiol. Cell Physiol. 2007, 293, C509–C536. [Google Scholar] [CrossRef] [PubMed]
- Hauck, C.; Frishman, W.H. Systemic hypertension: The roles of salt, vascular Na+/K+ ATPase and the endogenous glycosides, ouabain and marinobufagenin. Cardiol. Rev. 2012, 20, 130–138. [Google Scholar] [CrossRef]
- Bagrov, A.Y.; Kuznetsova, E.A.; Fedorova, O.V. Endogenous digoxin-like factor in acute myocardial infarction. J. Intern. Med. 1994, 235, 63–67. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, D.J.; Vetteth, S.; Periyasamy, S.M.; Kanj, M.; Fedorova, L.; Khouri, S.; Kahaleh, M.B.; Xie, Z.; Malhotra, D.; Kolodkin, N.I.; et al. Central role for the cardiotonic steroid marinobufagenin in the pathogenesis of experimental uremic cardiomyopathy. Hypertension 2006, 47, 488–495. [Google Scholar] [CrossRef] [PubMed]
- Gonick, H.C.; Ding, Y.; Vaziri, N.D.; Bagrov, A.Y.; Fedorova, O.V. Simultaneous measurement of marinobufagenin, ouabain, and hypertension-associated protein in various disease states. Clin. Exp. Hypertens. 1998, 20, 617–627. [Google Scholar] [CrossRef]
- Manunta, P.; Stella, P.; Rivera, R.; Ciurlino, D.; Cusi, D.; Ferrandi, M.; Hamlyn, J.M.; Bianchi, G. Left ventricular mass, stroke volume, and ouabain-like factor in essential hypertension. Hypertension 1999, 34, 450–456. [Google Scholar] [CrossRef]
- Periyasamy, S.M.; Chen, J.; Cooney, D.; Carter, P.; Omran, E.; Tian, J.; Priyadarshi, S.; Bagrov, A.; Fedorova, O.; Malhotra, D.; et al. Effects of uremic serum on isolated cardiac myocyte calcium cycling and contractile function. Kidney Int. 2001, 60, 2367–2376. [Google Scholar] [CrossRef] [PubMed]
- Drummond, C.A.; Hill, M.C.; Shi, H.; Fan, X.; Xie, J.X.; Haller, S.T.; Kennedy, D.J.; Liu, J.; Garrett, M.R.; Xie, Z.; et al. Na/K-ATPase signaling regulates collagen synthesis through microRNA-29b-3p in cardiac fibroblasts. Physiol. Genom. 2016, 48, 220–229. [Google Scholar] [CrossRef] [PubMed]
- Lichtstein, D.; Rosen, H.; Dvela, M. Cardenolides and bufadienolides as hormones: What is missing? Am. J. Physiol. Physiol. 2012, 302, F957–F958. [Google Scholar] [CrossRef][Green Version]
- Sen, A.K.; Post, R.L. Stoichiometry and localization of adenosine triphosphate-dependent sodium and potassium transport in the erythrocyte. J. Biol. Chem. 1964, 239, 345–352. [Google Scholar] [CrossRef]
- Lingrel, J.B. The physiological significance of the cardiotonic steroid/ouabain-binding site of the Na,K-ATPase. Annu. Rev. Physiol. 2010, 72, 395–412. [Google Scholar] [CrossRef]
- Bavendiek, U.; Aguirre Davila, L.; Koch, A.; Bauersachs, J. Assumption versus evidence: The case of digoxin in atrial fibrillation and heart failure. Eur. Heart J. 2017, 38, 2095–2099. [Google Scholar] [CrossRef]
- Eisen, A.; Ruff, C.T.; Braunwald, E.; Hamershock, R.A.; Lewis, B.S.; Hassager, C.; Chao, T.F.; Le Heuzey, J.Y.; Mercuri, M.; Rutman, H.; et al. Digoxin Use and Subsequent Clinical Outcomes in Patients With Atrial Fibrillation With or Without Heart Failure in the ENGAGE AF-TIMI 48 Trial. J. Am. Heart Assoc. 2017, 6, e006035. [Google Scholar] [CrossRef]
- Ziff, O.J.; Lane, D.A.; Samra, M.; Griffith, M.; Kirchhof, P.; Lip, G.Y.; Steeds, R.P.; Townend, J.; Kotecha, D. Safety and efficacy of digoxin: Systematic review and meta-analysis of observational and controlled trial data. BMJ 2015, 351, h4451. [Google Scholar] [CrossRef]
- Khundmiri, S.J. Advances in understanding the role of cardiac glycosides in control of sodium transport in renal tubules. J. Endocrinol. 2014, 222, R11–R24. [Google Scholar] [CrossRef]
- Liu, J.; Tian, J.; Haas, M.; Shapiro, J.I.; Askari, A.; Xie, Z. Ouabain Interaction with Cardiac Na+/K+-ATPase Initiates Signal Cascades Independent of Changes in Intracellular Na+ and Ca2+ Concentrations. J. Biol. Chem. 2000, 275, 27838–27844. [Google Scholar] [CrossRef]
- Tian, J.; Liu, J.; Garlid, K.D.; Shapiro, J.I.; Xie, Z. Involvement of mitogen-activated protein kinases and reactive oxygen species in the inotropic action of ouabain on cardiac myocytes. A potential role for mitochondrial K(ATP) channels. Mol. Cell. Biochem. 2003, 242, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Akimova, O.A.; Bagrov, A.Y.; Lopina, O.D.; Kamernitsky, A.V.; Tremblay, J.; Hamet, P.; Orlov, S.N. Cardiotonic steroids differentially affect intracellular Na+ and [Na+]i/[K+]i-independent signaling in C7-MDCK cells. J. Biol. Chem. 2005, 280, 832–839. [Google Scholar] [CrossRef] [PubMed]
- Scheiner-Bobis, G.; Schoner, W. A fresh facet for ouabain action. Nat. Med. 2001, 7, 1288–1289. [Google Scholar] [CrossRef] [PubMed]
- Fontana, J.M.; Burlaka, I.; Khodus, G.; Brismar, H.; Aperia, A. Calcium oscillations triggered by cardiotonic steroids. FEBS J. 2013, 280, 5450–5455. [Google Scholar] [CrossRef] [PubMed]
- Aperia, A.; Akkuratov, E.E.; Fontana, J.M.; Brismar, H. Na+-K+-ATPase, a new class of plasma membrane receptors. Am. J. Physiol. Cell Physiol. 2016, 310, C491–C495. [Google Scholar] [CrossRef]
- Xie, Z.; Askari, A. Na(+)/K(+)-ATPase as a signal transducer. Eur. J. Biochem. 2002, 269, 2434–2439. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Kometiani, P.; Liu, J.; Li, J.; Shapiro, J.I.; Askari, A. Intracellular Reactive Oxygen Species Mediate the Linkage of Na+/K+-ATPase to Hypertrophy and Its Marker Genes in Cardiac Myocytes. J. Biol. Chem. 1999, 274, 19323–19328. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Cai, T.; Yuan, Z.; Wang, H.; Liu, L.; Haas, M.; Maksimova, E.; Huang, X.Y.; Xie, Z.J. Binding of Src to Na+/K+-ATPase forms a functional signaling complex. Mol. Biol. Cell 2006, 17, 317–326. [Google Scholar] [CrossRef] [PubMed]
- Peng, M.; Huang, L.; Xie, Z.; Huang, W.-H.; Askari, A. Partial Inhibition of Na/K-ATPase by Ouabain Induces the Ca-dependent Expressions of Early-response Genes in Cardiac Myocytes. J. Biol. Chem. 1996, 271, 10372–10378. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Li, H.; Xie, Z. Ouabain-induced hypertrophy in cultured cardiac myocytes is accompanied by changes in expression of several late response genes. J. Biol. Chem. 1997, 29, 429–437. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Li, J.; Liu, J.; Yuan, Z.; Pierre, S.V.; Qu, W.; Zhao, X.; Xie, Z. Involvement of Na+/K+-ATPase in hydrogen peroxide-induced hypertrophy in cardiac myocytes. Free Radic. Biol. Med. 2006, 41, 1548–1556. [Google Scholar] [CrossRef]
- Yan, Y.; Shapiro, A.P.; Haller, S.; Katragadda, V.; Liu, L.; Tian, J.; Basrur, V.; Malhotra, D.; Xie, Z.J.; Abraham, N.G.; et al. Involvement of reactive oxygen species in a feed-forward mechanism of Na/K-ATPase-mediated signaling transduction. J. Biol. Chem. 2013, 288, 34249–34258. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Tian, J.; Chaudhry, M.; Maxwell, K.; Yan, Y.; Wang, X.; Shah, P.T.; Khawaja, A.A.; Martin, R.; Robinette, T.J.; et al. Attenuation of Na/K-ATPase Mediated Oxidant Amplification with pNaKtide Ameliorates Experimental Uremic Cardiomyopathy. Sci. Rep. 2016, 6, 34592. [Google Scholar] [CrossRef]
- Ritterhoff, J.; Young, S.; Villet, O.; Shao, D.; Neto, F.C.; Bettcher, L.F.; Hsu, Y.A.; Kolwicz, S.C., Jr.; Raftery, D.; Tian, R. Metabolic Remodeling Promotes Cardiac Hypertrophy by Directing Glucose to Aspartate Biosynthesis. Circ. Res. 2020, 126, 182–196. [Google Scholar] [CrossRef]
- Xiang, K.; Qin, Z.; Zhang, H.; Liu, X. Energy Metabolism in Exercise-Induced Physiologic Cardiac Hypertrophy. Front. Pharmacol. 2020, 11, 1133. [Google Scholar] [CrossRef] [PubMed]
- Maillet, M.; van Berlo, J.H.; Molkentin, J.D. Molecular basis of physiological heart growth: Fundamental concepts and new players. Nat. Rev. Mol. Cell Biol. 2013, 14, 38–48. [Google Scholar] [CrossRef]
- Gibb, A.A.; Hill, B.G. Metabolic Coordination of Physiological and Pathological Cardiac Remodeling. Circ. Res. 2018, 123, 107–128. [Google Scholar] [CrossRef]
- Nakamura, M.; Sadoshima, J. Mechanisms of physiological and pathological cardiac hypertrophy. Nat. Rev. Cardiol. 2018, 15, 387–407. [Google Scholar] [CrossRef]
- Quijano, C.; Trujillo, M.; Castro, L.; Trostchansky, A. Interplay between oxidant species and energy metabolism. Redox Biol. 2016, 8, 28–42. [Google Scholar] [CrossRef]
- Korla, K. Reactive oxygen species and energy machinery: An integrated dynamic model. J. Biomol. Struct. Dyn. 2016, 34, 1625–1640. [Google Scholar] [CrossRef]
- Jakubczyk, K.; Dec, K.; Kałduńska, J.; Kawczuga, D.; Kochman, J.; Janda, K. Reactive oxygen species—Sources, functions, oxidative damage. Pol. Merkur. Lek. 2020, 48, 124–127. [Google Scholar]
- Pratt, R.D.; Brickman, C.R.; Cottrill, C.L.; Shapiro, J.I.; Liu, J. The Na/K-ATPase Signaling: From Specific Ligands to General Reactive Oxygen Species. Int. J. Mol. Sci. 2018, 19, 2600. [Google Scholar] [CrossRef]
- Ichihara, S.; Senbonmatsu, T.; Price, E., Jr.; Ichiki, T.; Gaffney, F.A.; Inagami, T. Angiotensin II type 2 receptor is essential for left ventricular hypertrophy and cardiac fibrosis in chronic angiotensin II-induced hypertension. Circulation 2001, 104, 346–351. [Google Scholar] [CrossRef]
- Ni, H.; Li, W.; Zhuge, Y.; Xu, S.; Wang, Y.; Chen, Y.; Shen, G.; Wang, F. Inhibition of circHIPK3 prevents angiotensin II-induced cardiac fibrosis by sponging miR-29b-3p. Int. J. Cardiol. 2019, 292, 188–196. [Google Scholar] [CrossRef] [PubMed]
- Dostanic, I.; Schultz Jel, J.; Lorenz, J.N.; Lingrel, J.B. The alpha 1 isoform of Na,K-ATPase regulates cardiac contractility and functionally interacts and co-localizes with the Na/Ca exchanger in heart. J. Biol. Chem. 2004, 279, 54053–54061. [Google Scholar] [CrossRef] [PubMed]
- Wansapura, A.N.; Lasko, V.; Xie, Z.; Fedorova, O.V.; Bagrov, A.Y.; Lingrel, J.B.; Lorenz, J.N. Marinobufagenin enhances cardiac contractility in mice with ouabain-sensitive alpha1 Na+-K+-ATPase. Am. J. Physiol. Circ. Physiol. 2009, 296, H1833–H1839. [Google Scholar] [CrossRef]
- Wansapura, A.N.; Lasko, V.M.; Lingrel, J.B.; Lorenz, J.N. Mice expressing ouabain-sensitive α1-Na,K-ATPase have increased susceptibility to pressure overload-induced cardiac hypertrophy. Am. J. Physiol. Circ. Physiol. 2011, 300, H347–H355. [Google Scholar] [CrossRef]
- Dostanic-Larson, I.; Lorenz, J.N.; Van Huysse, J.W.; Neumann, J.C.; Moseley, A.E.; Lingrel, J.B. Physiological role of the alpha1- and alpha2-isoforms of the Na+-K+-ATPase and biological significance of their cardiac glycoside binding site. Am. J. Physiol. Integr. Comp. Physiol. 2006, 290, R524–R528. [Google Scholar] [CrossRef] [PubMed]
- Dostanic-Larson, I.; Van Huysse, J.W.; Lorenz, J.N.; Lingrel, J.B. The highly conserved cardiac glycoside binding site of Na,K-ATPase plays a role in blood pressure regulation. Proc. Natl. Acad. Sci. USA 2005, 102, 15845–15850. [Google Scholar] [CrossRef] [PubMed]
- Shioi, T.; Kang, P.M.; Douglas, P.S.; Hampe, J.; Yballe, C.M.; Lawitts, J.; Cantley, L.C.; Izumo, S. The conserved phosphoinositide 3-kinase pathway determines heart size in mice. EMBO J. 2000, 19, 2537–2548. [Google Scholar] [CrossRef]
- O’Neill, B.T.; Kim, J.; Wende, A.R.; Theobald, H.A.; Tuinei, J.; Buchanan, J.; Guo, A.; Zaha, V.G.; Davis, D.K.; Schell, J.C.; et al. A conserved role for phosphatidylinositol 3-kinase but not Akt signaling in mitochondrial adaptations that accompany physiological cardiac hypertrophy. Cell Metab. 2007, 6, 294–306. [Google Scholar] [CrossRef]
- Luo, J.; McMullen, J.R.; Sobkiw, C.L.; Zhang, L.; Dorfman, A.L.; Sherwood, M.C.; Logsdon, M.N.; Horner, J.W.; DePinho, R.A.; Izumo, S.; et al. Class IA phosphoinositide 3-kinase regulates heart size and physiological cardiac hypertrophy. Mol. Cell. Biol. 2005, 25, 9491–9502. [Google Scholar] [CrossRef]
- Sabri, A.; Hughie, H.H.; Lucchesi, P.A. Regulation of hypertrophic and apoptotic signaling pathways by reactive oxygen species in cardiac myocytes. Antioxid. Redox Signal. 2003, 5, 731–740. [Google Scholar] [CrossRef]
- Sag, C.M.; Santos, C.X.; Shah, A.M. Redox regulation of cardiac hypertrophy. J. Biol. Chem. 2014, 73, 103–111. [Google Scholar] [CrossRef]
- Anilkumar, N.; Sirker, A.; Shah, A.M. Redox sensitive signaling pathways in cardiac remodeling, hypertrophy and failure. Front. Biosci. 2009, 14, 3168–3187. [Google Scholar] [CrossRef] [PubMed]
- Schwarzer, M.; Osterholt, M.; Lunkenbein, A.; Schrepper, A.; Amorim, P.; Doenst, T. Mitochondrial reactive oxygen species production and respiratory complex activity in rats with pressure overload-induced heart failure. J. Physiol. 2014, 592, 3767–3782. [Google Scholar] [CrossRef]
- Dai, D.F.; Johnson, S.C.; Villarin, J.J.; Chin, M.T.; Nieves-Cintrón, M.; Chen, T.; Marcinek, D.J.; Dorn, G.W., 2nd; Kang, Y.J.; Prolla, T.A.; et al. Mitochondrial oxidative stress mediates angiotensin II-induced cardiac hypertrophy and Galphaq overexpression-induced heart failure. Circ. Res. 2011, 108, 837–846. [Google Scholar] [CrossRef]
- Ago, T.; Liu, T.; Zhai, P.; Chen, W.; Li, H.; Molkentin, J.D.; Vatner, S.F.; Sadoshima, J. A redox-dependent pathway for regulating class II HDACs and cardiac hypertrophy. Cell 2008, 133, 978–993. [Google Scholar] [CrossRef] [PubMed]
- Pavlovic, D. Endogenous cardiotonic steroids and cardiovascular disease, where to next? Cell Calcium 2020, 86, 102156. [Google Scholar] [CrossRef] [PubMed]
- Schoner, W.; Scheiner-Bobis, G. Role of endogenous cardiotonic steroids in sodium homeostasis. Nephrol. Dial. Transplant. 2008, 23, 2723–2729. [Google Scholar] [CrossRef] [PubMed]
- Kau, M.M.; Kan, S.F.; Wang, J.R.; Wang, P.S.; Lau, Y.T.; Wang, S.W. Acute effects of digoxin on plasma aldosterone and cortisol in monkeys. Metabolism 2009, 58, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Sophocleous, A.; Elmatzoglou, I.; Souvatzoglou, A. Circulating endogenous digitalis-like factor(s) (EDLF) in man is derived from the adrenals and its secretion is ACTH-dependent. J. Endocrinol. Investig. 2003, 26, 668–674. [Google Scholar] [CrossRef]
- Dvela-Levitt, M.; Cohen-Ben Ami, H.; Rosen, H.; Ornoy, A.; Hochner-Celnikier, D.; Granat, M.; Lichtstein, D. Reduction in Maternal Circulating Ouabain Impairs Offspring Growth and Kidney Development. Am. J. Physiol. Circ. Physiol. 2015, 26, 1103–1114. [Google Scholar] [CrossRef]
- Chen, L.; Yue, J.; Wu, H.; Yang, J.; Han, X.; Li, J.; Hu, Y. Ouabain Attenuates Cardiac Hypertrophy of Male Rat Offspring Exposed to Intrauterine Growth Restriction Following High-Salt Diet Challenge. Reprod. Sci. 2015, 22, 1587–1596. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, B.E.; Liu, Y.; Pulina, M.V.; Golovina, V.A.; Hamlyn, J.M. Normal pregnancy: Mechanisms underlying the paradox of a ouabain-resistant state with elevated endogenous ouabain, suppressed arterial sodium calcium exchange, and low blood pressure. Am. J. Physiol. Circ. Physiol. 2012, 302, H1317–H1329. [Google Scholar] [CrossRef]
- Clerico, A.; Cambi, A.; Del Chicca, M.G.; Cecchini, L.; Giaconi, S. Urinary excretion of digoxin-like immunoreactivity after physical exercise. Clin. Chem. 1988, 34, 215. [Google Scholar] [CrossRef] [PubMed]
- Yoshika, M.; Komiyama, Y.; Konishi, M.; Akizawa, T.; Kobayashi, T.; Date, M.; Kobatake, S.; Masuda, M.; Masaki, H.; Takahashi, H. Novel digitalis-like factor, marinobufotoxin, isolated from cultured Y-1 cells, and its hypertensive effect in rats. Hypertension 2007, 49, 209–214. [Google Scholar] [CrossRef]
- Ferrandi, M.; Barassi, P.; Molinari, I.; Torielli, L.; Tripodi, G.; Minotti, E.; Bianchi, G.; Ferrari, P. Ouabain antagonists as antihypertensive agents. Curr. Pharm. Des. 2005, 11, 3301–3305. [Google Scholar] [CrossRef] [PubMed]
- Maixent, J.M.; Lelièvre, L.G. Differential inactivation of inotropic and toxic digitalis receptors in ischemic dog heart. Molecular basis of the deleterious effects of digitalis. J. Biol. Chem. 1987, 262, 12458–12462. [Google Scholar] [CrossRef]
- Liu, C.C.; Fry, N.A.; Hamilton, E.J.; Chia, K.K.; Garcia, A.; Karimi Galougahi, K.; Figtree, G.A.; Clarke, R.J.; Bundgaard, H.; Rasmussen, H.H. Redox-dependent regulation of the Na⁺-K⁺ pump: New twists to an old target for treatment of heart failure. J. Biol. Chem. 2013, 61, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Mohmand, B.; Malhotra, D.K.; Shapiro, J.I. Uremic cardiomyopathy: Role of circulating digitalis like substances. Front. Biosci. 2005, 10, 2036–2044. [Google Scholar] [CrossRef]
- Taniike, M.; Yamaguchi, O.; Tsujimoto, I.; Hikoso, S.; Takeda, T.; Nakai, A.; Omiya, S.; Mizote, I.; Nakano, Y.; Higuchi, Y.; et al. Apoptosis signal-regulating kinase 1/p38 signaling pathway negatively regulates physiological hypertrophy. Circulation 2008, 117, 545–552. [Google Scholar] [CrossRef]
- Tian, Z.; Miyata, K.; Kadomatsu, T.; Horiguchi, H.; Fukushima, H.; Tohyama, S.; Ujihara, Y.; Okumura, T.; Yamaguchi, S.; Zhao, J.; et al. ANGPTL2 activity in cardiac pathologies accelerates heart failure by perturbing cardiac function and energy metabolism. Nat. Commun. 2016, 7, 13016. [Google Scholar] [CrossRef] [PubMed]
- Ruppert, C.; Deiss, K.; Herrmann, S.; Vidal, M.; Oezkur, M.; Gorski, A.; Weidemann, F.; Lohse, M.J.; Lorenz, K. Interference with ERK(Thr188) phosphorylation impairs pathological but not physiological cardiac hypertrophy. Proc. Natl. Acad. Sci. USA 2013, 110, 7440–7445. [Google Scholar] [CrossRef]
- Haller, S.T.; Kennedy, D.J.; Shidyak, A.; Budny, G.V.; Malhotra, D.; Fedorova, O.V.; Shapiro, J.I.; Bagrov, A.Y. Monoclonal antibody against marinobufagenin reverses cardiac fibrosis in rats with chronic renal failure. Am. J. Hypertens. 2012, 25, 690–696. [Google Scholar] [CrossRef]
- Orlov, S.N.; La, J.; Smolyaninova, L.V.; Dulin, N.O. Na+,K+-ATPase as a Target for Treatment of Tissue Fibrosis. Curr. Med. Chem. 2019, 26, 564–575. [Google Scholar] [CrossRef]
- Kennedy, D.J.; Shrestha, K.; Sheehey, B.; Li, X.S.; Guggilam, A.; Wu, Y.; Finucan, M.; Gabi, A.; Medert, C.M.; Westfall, K.; et al. Elevated Plasma Marinobufagenin, An Endogenous Cardiotonic Steroid, Is Associated With Right Ventricular Dysfunction and Nitrative Stress in Heart Failure. Circ. Heart Fail. 2015, 8, 1068–1076. [Google Scholar] [CrossRef]
- Strauss-Kruger, M.; Kruger, R.; Smith, W.; Gafane-Matemane, L.F.; Mokwatsi, G.; Wei, W.; Fedorova, O.V.; Schutte, A.E. The Cardiotonic Steroid Marinobufagenin Is a Predictor of Increased Left Ventricular Mass in Obesity: The African-PREDICT Study. Nutrients 2020, 12, 3185. [Google Scholar] [CrossRef] [PubMed]
- Simonini, M.; Pozzoli, S.; Bignami, E.; Casamassima, N.; Messaggio, E.; Lanzani, C.; Frati, E.; Botticelli, I.M.; Rotatori, F.; Alfieri, O.; et al. Endogenous Ouabain: An Old Cardiotonic Steroid as a New Biomarker of Heart Failure and a Predictor of Mortality after Cardiac Surgery. BioMed Res. Int. 2015, 2015, 714793. [Google Scholar] [CrossRef]
- Xu, Y.; Jiang, X.; Xu, J.; Qu, W.; Xie, Z.; Jiang, R.W.; Feng, F. A previously undescribed phenylethanoid glycoside from Callicarpa kwangtungensis Chun acts as an agonist of the Na/K-ATPase signal transduction pathway. Phytochemistry 2021, 181, 112577. [Google Scholar] [CrossRef] [PubMed]
- Manunta, P.; Ferrandi, M. Different effects of marinobufagenin and endogenous ouabain. J. Hypertens. 2004, 22, 257–259. [Google Scholar] [CrossRef] [PubMed]
- Zulian, A.; Linde, C.I.; Pulina, M.V.; Baryshnikov, S.G.; Papparella, I.; Hamlyn, J.M.; Golovina, V.A. Activation of c-SRC underlies the differential effects of ouabain and digoxin on Ca(2+) signaling in arterial smooth muscle cells. Am. J. Physiol. Cell Physiol. 2013, 304, C324–C333. [Google Scholar] [CrossRef]
- Blaustein, M.P. Endogenous ouabain: Role in the pathogenesis of hypertension. Kidney Int. 1996, 49, 1748–1753. [Google Scholar] [CrossRef]
- Blaustein, M.P.; Hamlyn, J.M. Signaling mechanisms that link salt retention to hypertension: Endogenous ouabain, the Na(+) pump, the Na(+)/Ca(2+) exchanger and TRPC proteins. Biochim. Biophys. Acta 2010, 1802, 1219–1229. [Google Scholar] [CrossRef] [PubMed]
- Tomaschitz, A.; Piecha, G.; Ritz, E.; Meinitzer, A.; Haas, J.; Pieske, B.; Wiecek, A.; Rus-Machan, J.; Toplak, H.; März, W.; et al. Marinobufagenin in essential hypertension and primary aldosteronism: A cardiotonic steroid with clinical and diagnostic implications. Clin. Exp. Hypertens. 2015, 37, 108–115. [Google Scholar] [CrossRef] [PubMed]
- Ferrandi, M.; Molinari, I.; Barassi, P.; Minotti, E.; Bianchi, G.; Ferrari, P. Organ hypertrophic signaling within caveolae membrane subdomains triggered by ouabain and antagonized by PST 2238. J. Biol. Chem. 2004, 279, 33306–33314. [Google Scholar] [CrossRef]
- Zahler, R.; Gilmore-Hebert, M.; Sun, W.; Benz, E.J. Na, K-ATPase isoform gene expression in normal and hypertrophied dog heart. Basic Res. Cardiol. 1996, 91, 256–266. [Google Scholar] [CrossRef]
- Shamraj, O.I.; Grupp, I.L.; Grupp, G.; Melvin, D.; Gradoux, N.; Kremers, W.; Lingrel, J.B.; De Pover, A. Characterisation of Na/K-ATPase, its isoforms, and the inotropic response to ouabain in isolated failing human hearts. Cardiovasc. Res. 1993, 27, 2229–2237. [Google Scholar] [CrossRef]
- Schwinger, R.H.; Bundgaard, H.; Müller-Ehmsen, J.; Kjeldsen, K. The Na, K-ATPase in the failing human heart. Cardiovasc. Res. 2003, 57, 913–920. [Google Scholar] [CrossRef]
- Tsuruya, Y.; Ikeda, U.; Yamamoto, K.; Seino, Y.; Kanbe, T.; Shimada, K. Decreased Na,K-ATPase gene expression in cardiomyopathic hamster hearts. Life Sci. 1994, 54, 71–77. [Google Scholar] [CrossRef]
- Gerbi, A.; Barbey, O.; Raccah, D.; Coste, T.; Jamme, I.; Nouvelot, A.; Ouafik, L.; Levy, S.; Vague, P.; Maixent, J.M. Alteration of Na,K-ATPase isoenzymes in diabetic cardiomyopathy: Effect of dietary supplementation with fish oil (n-3 fatty acids) in rats. Diabetologia 1997, 40, 496–505. [Google Scholar] [CrossRef]
- Duan, Q.; Madan, N.D.; Wu, J.; Kalisz, J.; Doshi, K.Y.; Haldar, S.M.; Liu, L.; Pierre, S.V. Role of phosphoinositide 3-kinase IA (PI3K-IA) activation in cardioprotection induced by ouabain preconditioning. J. Biol. Chem. 2015, 80, 114–125. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Shapiro, A.P.; Mopidevi, B.R.; Chaudhry, M.A.; Maxwell, K.; Haller, S.T.; Drummond, C.A.; Kennedy, D.J.; Tian, J.; Malhotra, D.; et al. Protein Carbonylation of an Amino Acid Residue of the Na/K-ATPase α1 Subunit Determines Na/K-ATPase Signaling and Sodium Transport in Renal Proximal Tubular Cells. J. Am. Heart Assoc. 2016, 5, e003675. [Google Scholar] [CrossRef]
- Duan, Q.; Xu, Y.; Marck, P.V.; Kalisz, J.; Morgan, E.E.; Pierre, S.V. Preconditioning and Postconditioning by Cardiac Glycosides in the Mouse Heart. J. Cardiovasc. Pharmacol. 2018, 71, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Bisserier, M.; Berthouze-Duquesnes, M.; Breckler, M.; Tortosa, F.; Fazal, L.; de Régibus, A.; Laurent, A.C.; Varin, A.; Lucas, A.; Branchereau, M.; et al. Carabin protects against cardiac hypertrophy by blocking calcineurin, Ras, and Ca2+/calmodulin-dependent protein kinase II signaling. Circulation 2015, 131, 390–400. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.; Basu, R.; Guo, D.; Chow, F.L.; Byrns, S.; Schuster, M.; Loibner, H.; Wang, X.H.; Penninger, J.M.; Kassiri, Z.; et al. Angiotensin-converting enzyme 2 suppresses pathological hypertrophy, myocardial fibrosis, and cardiac dysfunction. Circulation 2010, 122, 717–728. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Wang, J.; Jaehnig, E.J.; Shi, Z.; Zhang, B. WebGestalt 2019: Gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019, 47, W199–W205. [Google Scholar] [CrossRef] [PubMed]


| Parameter | α1r/rα2s/s (n = 9) | α1s/sα2s/s (n = 6) |
|---|---|---|
| HR (bpm) | 457 ± 23 | 475 ± 31 |
| LVID; s (mm) | 2.50 ± 0.52 | 2.60 ± 0.42 |
| LVID; d (mm) | 3.61 ± 0.54 | 3.86 ± 0.28 |
| LV Volume; s (µL) | 23.7 ± 10.6 | 25.5 ± 9.2 |
| LV Volume; d (µL) | 56.6 ± 18.7 | 64.9 ± 11.0 |
| SV (µL) | 32.9 ± 11.2 | 39.4 ± 3.8 |
| EF (%) | 59 ± 11 | 62 ± 9 |
| FS (%) | 31 ± 7 | 33 ± 7 |
| CO (mL/min) | 14.9 ± 4.7 | 18.7 ± 2.1 |
| LVAW; s (mm) | 1.34 ± 0.16 | 1.53 ± 0.12 * |
| LVAW; d (mm) | 0.95 ± 0.12 | 1.06 ± 0.11 |
| LVPW; s (mm) | 1.19 ± 0.17 | 1.24 ± 0.24 |
| LVPW; d (mm) | 0.80 ± 0.10 | 0.84 ± 0.18 |
| LV Mass (mg) | 87.1 ± 12.4 | 110.7 ± 19.9 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marck, P.V.; Pessoa, M.T.; Xu, Y.; Kutz, L.C.; Collins, D.M.; Yan, Y.; King, C.; Wang, X.; Duan, Q.; Cai, L.; et al. Cardiac Oxidative Signaling and Physiological Hypertrophy in the Na/K-ATPase α1s/sα2s/s Mouse Model of High Affinity for Cardiotonic Steroids. Int. J. Mol. Sci. 2021, 22, 3462. https://doi.org/10.3390/ijms22073462
Marck PV, Pessoa MT, Xu Y, Kutz LC, Collins DM, Yan Y, King C, Wang X, Duan Q, Cai L, et al. Cardiac Oxidative Signaling and Physiological Hypertrophy in the Na/K-ATPase α1s/sα2s/s Mouse Model of High Affinity for Cardiotonic Steroids. International Journal of Molecular Sciences. 2021; 22(7):3462. https://doi.org/10.3390/ijms22073462
Chicago/Turabian StyleMarck, Pauline V., Marco T. Pessoa, Yunhui Xu, Laura C. Kutz, Dominic M. Collins, Yanling Yan, Cierra King, Xiaoliang Wang, Qiming Duan, Liquan Cai, and et al. 2021. "Cardiac Oxidative Signaling and Physiological Hypertrophy in the Na/K-ATPase α1s/sα2s/s Mouse Model of High Affinity for Cardiotonic Steroids" International Journal of Molecular Sciences 22, no. 7: 3462. https://doi.org/10.3390/ijms22073462
APA StyleMarck, P. V., Pessoa, M. T., Xu, Y., Kutz, L. C., Collins, D. M., Yan, Y., King, C., Wang, X., Duan, Q., Cai, L., Xie, J. X., Lingrel, J. B., Xie, Z., Tian, J., & Pierre, S. V. (2021). Cardiac Oxidative Signaling and Physiological Hypertrophy in the Na/K-ATPase α1s/sα2s/s Mouse Model of High Affinity for Cardiotonic Steroids. International Journal of Molecular Sciences, 22(7), 3462. https://doi.org/10.3390/ijms22073462








