Insights of Phenolic Pathway in Fruits: Transcriptional and Metabolic Profiling in Apricot (Prunus armeniaca)
Abstract
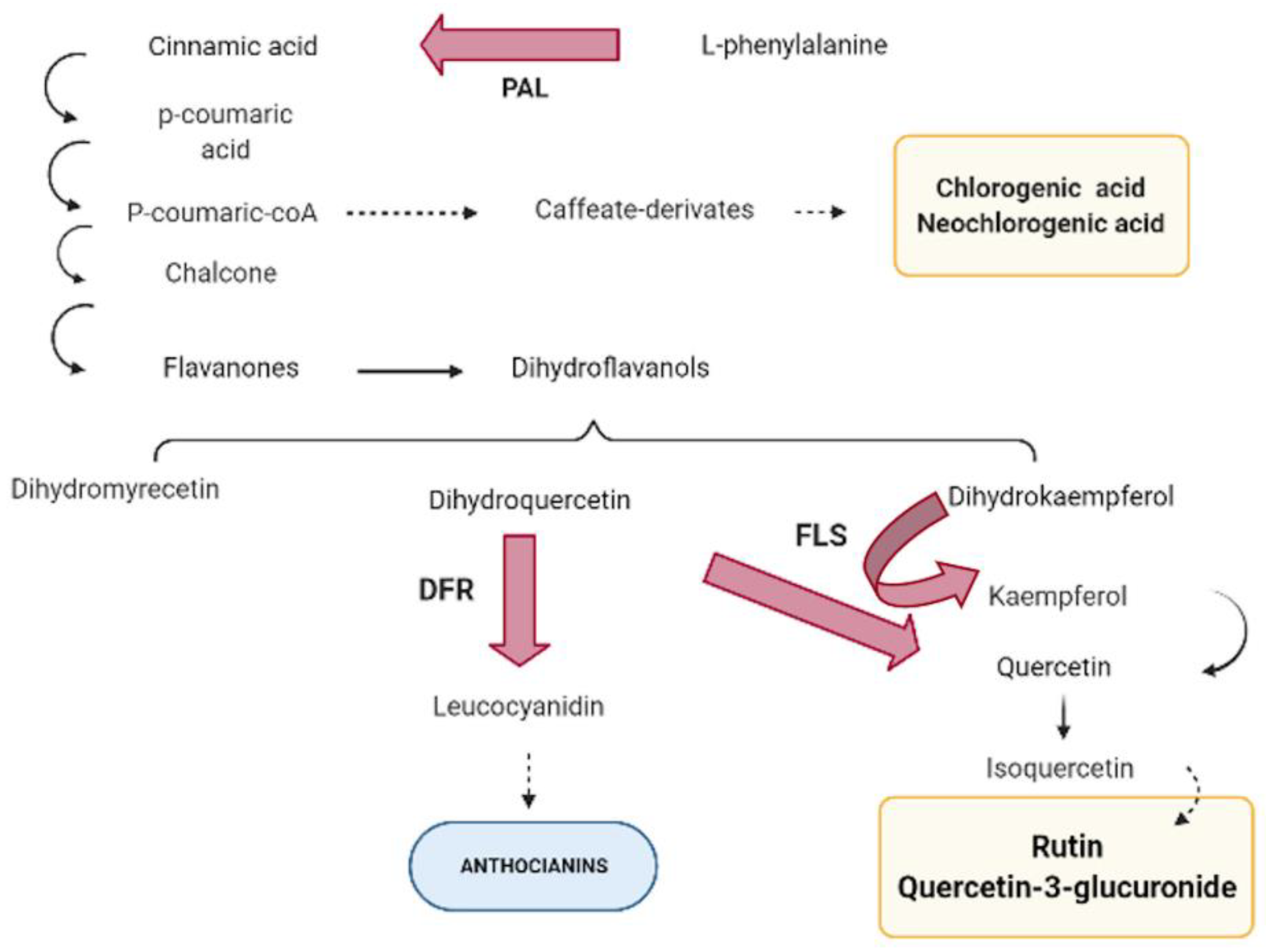
1. Introduction
2. Results
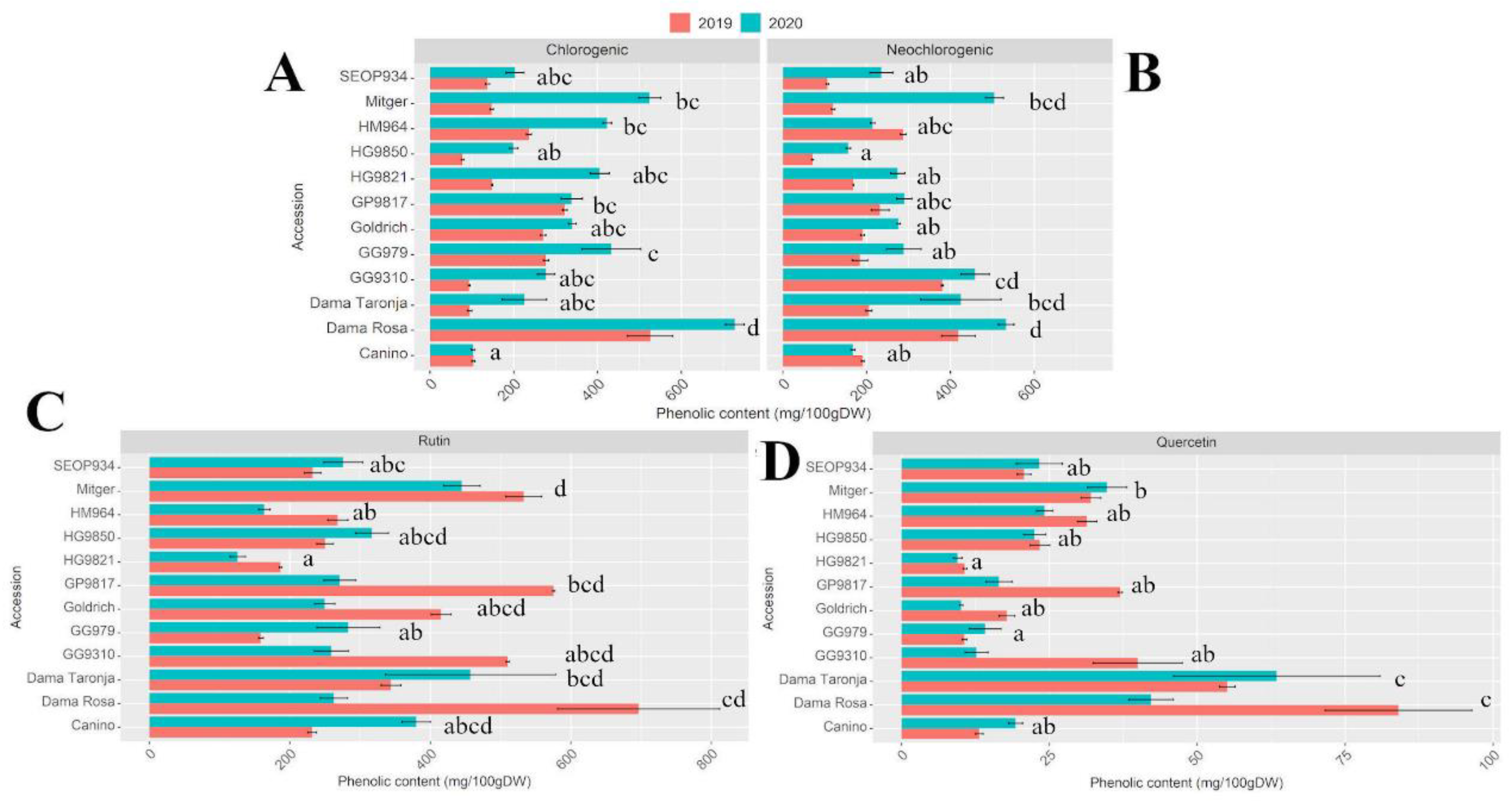
2.1. Apricot Polyphenol Content
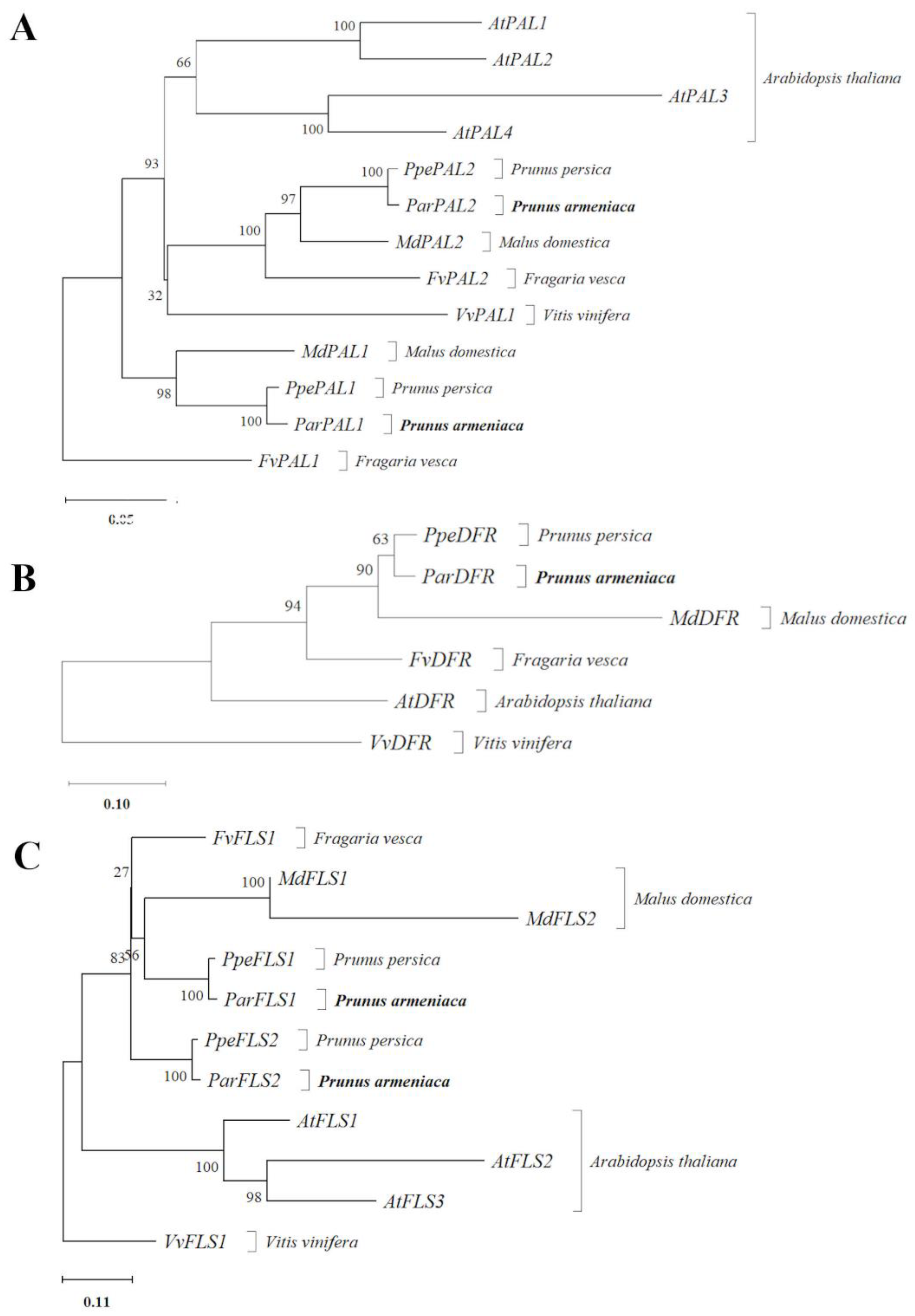
2.2. Putative Orthologous and Phylogenetic Analysis
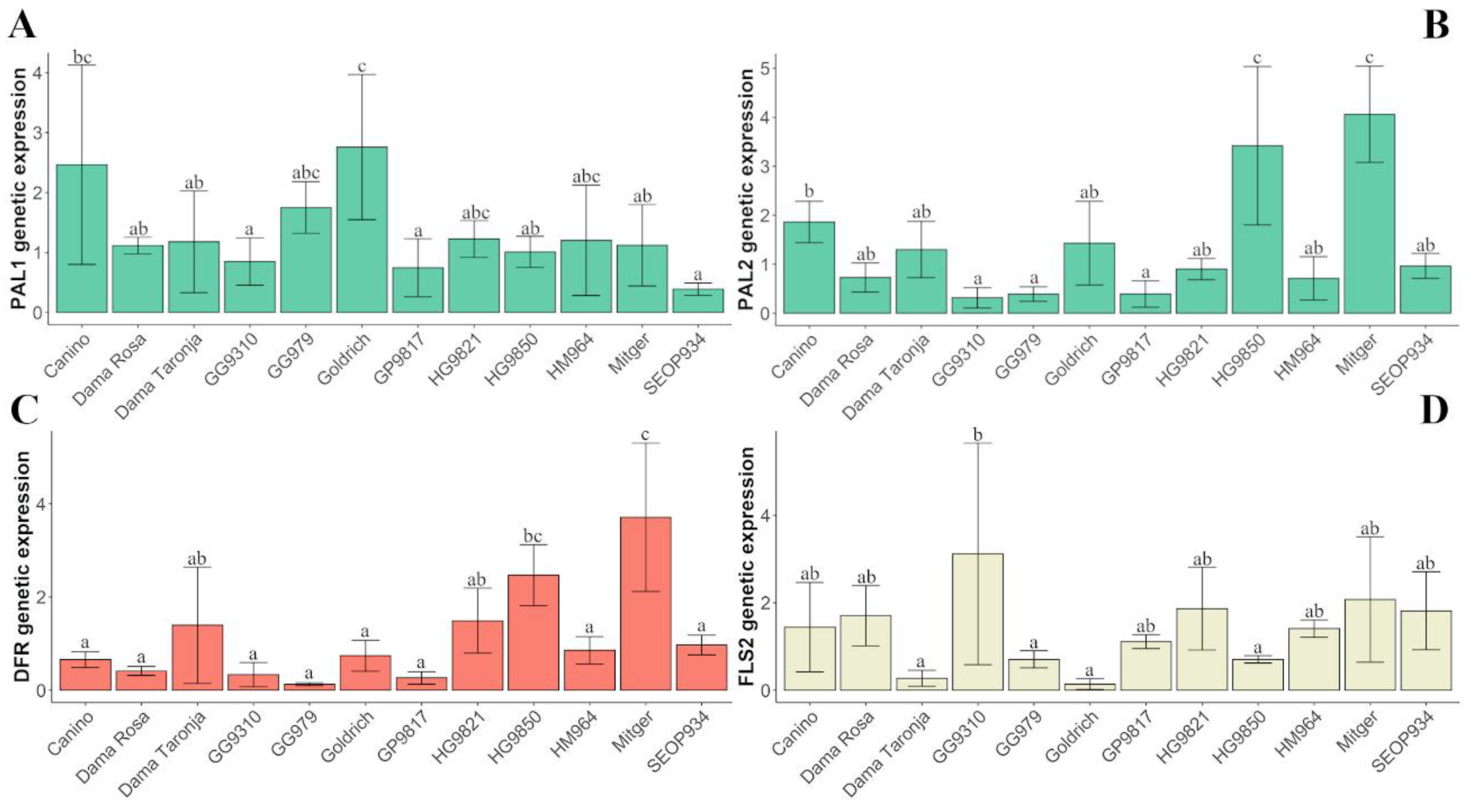
2.3. Gene Expression
2.4. Contribution of ‘Goldrich’ to Phenolic Compounds Content and Genetic Expression
2.5. Relationships between Gene Expression and Phenolic Compound Accumulation
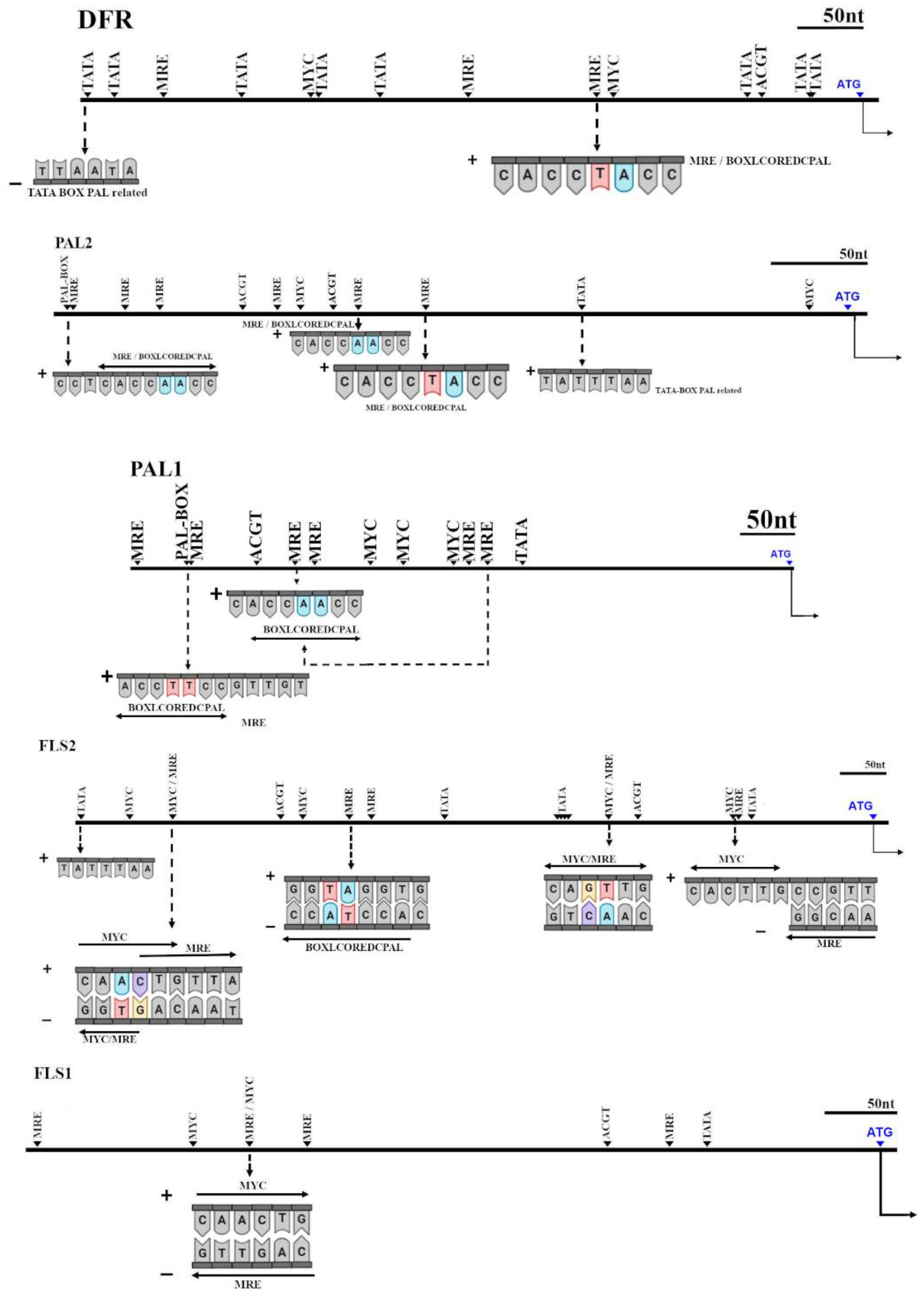
2.6. Cis-Acting Elements Analysis
3. Discussion
3.1. Polyphenol Content
3.2. Genes and Its Inference in Polyphenols Pathway
4. Materials and Methods
4.1. Plant Material
4.2. HPLC Analysis
4.3. Obtention of Gene Sequences and Cis-Acting Elements Motif Identification
4.4. Gene Expression
4.5. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bailey, C.H.; Hough, L.F. Advances in Fruit Breeding; Purdue University Press: West Lafayette, IN, USA, 1975; ISBN 0-911198-36-9. [Google Scholar]
- Roussos, P.A.; Sefferou, V.; Denaxa, N.-K.; Tsantili, E.; Stathis, V. Apricot (Prunus armeniaca L.) fruit quality attributes and phytochemicals under different crop load. Sci. Hortic. 2011, 129, 472–478. [Google Scholar] [CrossRef]
- Ruiz, D.; Egea, J. Phenotypic diversity and relationships of fruit quality traits in apricot (Prunus armeniaca L.) germplasm. Euphytica 2007, 163, 143–158. [Google Scholar] [CrossRef]
- Ferrer, J.-L.; Austin, M.; Stewart, C.; Noel, J. Structure and function of enzymes involved in the biosynthesis of phenylpropanoids. Plant Physiol. Biochem. 2008, 46, 356–370. [Google Scholar] [CrossRef] [PubMed]
- Harborne, J.B. Plant Secondary Metabolism. In Plant Ecology; Blackwell Publishing Ltd.: Oxford, UK, 2009; pp. 132–155. ISBN 9780632036394. [Google Scholar]
- Bizjak, J.; Mikulic-Petkovsek, M.; Stampar, F.; Veberic, R. Changes in Primary Metabolites and Polyphenols in the Peel of “Braeburn” Apples (Malus domestica Borkh.) during Advanced Maturation. J. Agric. Food Chem. 2013, 61, 10283–10292. [Google Scholar] [CrossRef] [PubMed]
- Campbell, O.E.; Padilla-Zakour, O.I. Phenolic and carotenoid composition of canned peaches (Prunus persica) and apricots (Prunus armeniaca) as affected by variety and peeling. Food Res. Int. 2013, 54, 448–455. [Google Scholar] [CrossRef]
- Gómez-Martínez, H.; Bermejo, A.; Zuriaga, E.; Badenes, M. Polyphenol content in apricot fruits. Sci. Hortic. 2021, 277, 109828. [Google Scholar] [CrossRef]
- Fu, L.; Xu, B.-T.; Xu, X.-R.; Qin, X.-S.; Gan, R.-Y.; Li, H.-B. Antioxidant Capacities and Total Phenolic Contents of 56 Wild Fruits from South China. Molecules 2010, 15, 8602–8617. [Google Scholar] [CrossRef]
- Gan, R.-Y.; Deng, Z.-Q.; Yan, A.-X.; Shah, N.P.; Lui, W.-Y.; Chan, C.-L.; Corke, H. Pigmented edible bean coats as natural sources of polyphenols with antioxidant and antibacterial effects. LWT 2016, 73, 168–177. [Google Scholar] [CrossRef]
- Mokrani, A.; Krisa, S.; Cluzet, S.; Da Costa, G.; Temsamani, H.; Renouf, E.; Mérillon, J.-M.; Madani, K.; Mesnil, M.; Monvoisin, A.; et al. Phenolic contents and bioactive potential of peach fruit extracts. Food Chem. 2016, 202, 212–220. [Google Scholar] [CrossRef] [PubMed]
- Erdogan-Orhan, I.; Kartal, M. Insights into research on phytochemistry and biological activities of Prunus armeniaca L. (apricot). Food Res. Int. 2011, 44, 1238–1243. [Google Scholar] [CrossRef]
- Fraser, C.M.; Chapple, C. The Phenylpropanoid Pathway in Arabidopsis. Arab. Book 2011, 9, e0152. [Google Scholar] [CrossRef]
- Raes, J.; Rohde, A.; Christensen, J.H.; Van De Peer, Y.; Boerjan, W. Genome-Wide Characterization of the Lignification Toolbox in Arabidopsis. Plant Physiol. 2003, 133, 1051–1071. [Google Scholar] [CrossRef]
- Fukasawa-Akada, T.; Kung, S.-D.; Watson, J.C. Phenylalanine ammonia-lyase gene structure, expression, and evolution in Nicotiana. Plant Mol. Biol. 1996, 30, 711–722. [Google Scholar] [CrossRef] [PubMed]
- Reichert, A.I.; He, X.-Z.; Dixon, R.A. Phenylalanine ammonia-lyase (PAL) from tobacco (Nicotiana tabacum): Characterization of the four tobacco PAL genes and active heterotetrameric enzymes. Biochem. J. 2009, 424, 233–242. [Google Scholar] [CrossRef]
- Hamberger, B.; Ellis, M.; Friedmann, M.; Souza, C.D.A.; Barbazuk, B.; Douglas, C.J. Genome-wide analyses of phenylpropanoid-related genes in Populus trichocarpa, Arabidopsis thaliana, and Oryza sativa: The Populus lignin toolbox and conservation and diversification of angiosperm gene familiesThis article is one of a selection of papers published in the Special Issue on Poplar Research in Canada. Can. J. Bot. 2007, 85, 1182–1201. [Google Scholar] [CrossRef]
- Irisarri, P.; Zhebentyayeva, T.; Errea, P.; Pina, A. Differential expression of phenylalanine ammonia lyase (PAL) genes implies distinct roles in development of graft incompatibility symptoms in Prunus. Sci. Hortic. 2016, 204, 16–24. [Google Scholar] [CrossRef]
- Pelletier, M.K.; Burbulis, I.E.; Winkel-Shirley, B. Disruption of specific flavonoid genes enhances the accumulation of flavonoid enzymes and end-products in Arabidopsis seedlings. Plant Mol. Biol. 1999, 40, 45–54. [Google Scholar] [CrossRef]
- Owens, D.K.; Alerding, A.B.; Crosby, K.C.; Bandara, A.B.; Westwood, J.H.; Winkel, B.S. Functional Analysis of a Predicted Flavonol Synthase Gene Family in Arabidopsis. Plant Physiol. 2008, 147, 1046–1061. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, B.M.; Geisler, M.; Bigler, L.; Ringli, C. Flavonols Accumulate Asymmetrically and Affect Auxin Transport in Arabidopsis. Plant Physiol. 2011, 156, 585–595. [Google Scholar] [CrossRef]
- Martens, S.; Knott, J.; Seitz, C.A.; Janvari, L.; Yu, S.-N.; Forkmann, G. Impact of biochemical pre-studies on specific metabolic engineering strategies of flavonoid biosynthesis in plant tissues. Biochem. Eng. J. 2003, 14, 227–235. [Google Scholar] [CrossRef]
- Shimada, N.; Sasaki, R.; Sato, S.; Kaneko, T.; Tabata, S.; Aoki, T.; Ayabe, S.-I. A comprehensive analysis of six dihydroflavonol 4-reductases encoded by a gene cluster of the Lotus japonicus genome. J. Exp. Bot. 2005, 56, 2573–2585. [Google Scholar] [CrossRef]
- Lopiero, A.; Puglisi, I.; Petrone, G. Gene characterization, analysis of expression and in vitro synthesis of dihydroflavonol 4-reductase from [Citrus sinensis (L.) Osbeck]. Phytochemistry 2006, 67, 684–695. [Google Scholar] [CrossRef] [PubMed]
- Xie, D.-Y.; Jackson, L.A.; Cooper, J.D.; Ferreira, D.; Paiva, N.L. Molecular and Biochemical Analysis of Two cDNA Clones Encoding Dihydroflavonol-4-Reductase from Medicago truncatula. Plant Physiol. 2004, 134, 979–994. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.; Kumar, S.; Yadav, S.K.; Ahuja, P.S. Characterization of dihydroflavonol 4-reductase cDNA in tea [Camellia sinensis (L.) O. Kuntze]. Plant Biotechnol. Rep. 2008, 3, 95–101. [Google Scholar] [CrossRef]
- Huang, Y.; Gou, J.; Jia, Z.; Yang, L.; Sun, Y.; Xiao, X.; Song, F.; Luo, K. Molecular Cloning and Characterization of Two Genes Encoding Dihydroflavonol-4-Reductase from Populus trichocarpa. PLoS ONE 2012, 7, e30364. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, U.; Sagasser, M.; Mehrtens, F.; Stracke, R.; Weisshaar, B. Differential combinatorial interactions of cis-acting elements recognized by R2R3-MYB, BZIP, and BHLH factors control light-responsive and tissue-specific activation of phenylpropanoid biosynthesis genes. Plant Mol. Biol. 2005, 57, 155–171. [Google Scholar] [CrossRef]
- Jin, X.; Huang, H.; Wang, L.; Sun, Y.; Dai, S. Transcriptomics and Metabolite Analysis Reveals the Molecular Mechanism of Anthocyanin Biosynthesis Branch Pathway in Different Senecio cruentus Cultivars. Front. Plant Sci. 2016, 7, 1307. [Google Scholar] [CrossRef]
- Eluo, P.; Ening, G.; Ewang, Z.; Eshen, Y.; Ejin, H.; Eli, P.; Ehuang, S.; Ezhao, J.; Ebao, M. Disequilibrium of Flavonol Synthase and Dihydroflavonol-4-Reductase Expression Associated Tightly to White vs. Red Color Flower Formation in Plants. Front. Plant Sci. 2016, 6, 1257. [Google Scholar] [CrossRef]
- Dossett, M.; Lee, J.; Finn, C.E. Characterization of a novel anthocyanin profile in wild black raspberry mutants: An opportunity for studying the genetic control of pigment and color. J. Funct. Foods 2011, 3, 207–214. [Google Scholar] [CrossRef]
- Kayesh, E.; Shangguan, L.; Korir, N.K.; Sun, X.; Bilkish, N.; Zhang, Y.; Han, J.; Song, C.; Cheng, Z.-M.; Fang, J. Fruit skin color and the role of anthocyanin. Acta Physiol. Plant. 2013, 35, 2879–2890. [Google Scholar] [CrossRef]
- Wang, K.; Jin, P.; Han, L.; Shang, H.; Tang, S.; Rui, H.; Duan, Y.; Kong, F.; Kai, X.; Zheng, Y. Methyl jasmonate induces resistance against Penicillium citrinum in Chinese bayberry by priming of defense responses. Postharvest Biol. Technol. 2014, 98, 90–97. [Google Scholar] [CrossRef]
- Badenes, M.L.; Martínez-Calvo, J.; Gómez, H.; Zuriaga, E. ‘Dama Taronja’ and ‘Dama Rosa’ Apricot Cultivars that are Resistant to Sharka (Plum pox virus). HortScience 2018, 53, 1228–1229. [Google Scholar] [CrossRef]
- Bureau, S.; Renard, C.M.; Reich, M.; Ginies, C.; Audergon, J.-M. Change in anthocyanin concentrations in red apricot fruits during ripening. LWT 2009, 42, 372–377. [Google Scholar] [CrossRef]
- Roulin, A.; Auer, P.L.; Libault, M.; Schlueter, J.; Farmer, A.; May, G.; Stacey, G.; Doerge, R.W.; Jackson, S.A. The fate of duplicated genes in a polyploid plant genome. Plant J. 2012, 73, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Huang, C.-H.; Hu, Y.; Wen, J.; Li, S.; Yi, T.; Chen, H.; Xiang, J.; Ma, H. Evolution of Rosaceae Fruit Types Based on Nuclear Phylogeny in the Context of Geological Times and Genome Duplication. Mol. Biol. Evol. 2016, 34, 262–281. [Google Scholar] [CrossRef]
- Bomblies, K.; Weigel, D. Arabidopsis—A model genus for speciation. Curr. Opin. Genet. Dev. 2007, 17, 500–504. [Google Scholar] [CrossRef] [PubMed]
- Bomblies, K.; Madlung, A. Polyploidy in the Arabidopsis genus. Chromosom. Res. 2014, 22, 117–134. [Google Scholar] [CrossRef]
- Del Pozo, J.C.; Ramirez-Parra, E. Whole genome duplications in plants: An overview from Arabidopsis. J. Exp. Bot. 2015, 66, 6991–7003. [Google Scholar] [CrossRef] [PubMed]
- Edger, P.P.; Pires, J.C. Gene and genome duplications: The impact of dosage-sensitivity on the fate of nuclear genes. Chromosom. Res. 2009, 17, 699–717. [Google Scholar] [CrossRef]
- Lian, S.; Zhou, Y.; Liu, Z.; Gong, A.; Cheng, L. The differential expression patterns of paralogs in response to stresses indicate expression and sequence divergences. BMC Plant Biol. 2020, 20, 277. [Google Scholar] [CrossRef]
- Cano, A.; Medina, A.; Bermejo, A. Bioactive compounds in different citrus varieties. Discrimination among cultivars. J. Food Compos. Anal. 2008, 21, 377–381. [Google Scholar] [CrossRef]
- Cano, A.; Bermejo, A. Influence of rootstock and cultivar on bioactive compounds in citrus peels. J. Sci. Food Agric. 2011, 91, 1702–1711. [Google Scholar] [CrossRef] [PubMed]
- Genome Dabase of Rosaceae. Available online: https://www.rosaceae.org/ (accessed on 21 July 2020).
- Higo, K.; Ugawa, Y.; Iwamoto, M.; Korenaga, T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999, 27, 297–300. [Google Scholar] [CrossRef]
- Expasy. Available online: https://web.expasy.org/translate/ (accessed on 19 August 2020).
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. Package ‘Dplyr’. 2020. Available online: https://dplyr.tidyverse.org (accessed on 14 September 2020).
- Wickham, H. Readxl: Read Excel Files. 2016; pp. 1–9. Available online: https://readxl.tidyverse.org (accessed on 14 September 2020).
- Wickham, H. Package ‘Plyr’. 2020. Available online: http://had.co.nz/plyr (accessed on 14 September 2020).
- Wickham, H.; Seidel, D. Package ‘Scales’. 2020. Available online: https://scales.r-lib.org (accessed on 14 September 2020).
- Wickham, H. Ggplot2; Springer: New York, NY, USA, 2009; ISBN 978-0-387-98140-6. [Google Scholar]


| Genotype | 2019 | 2020 | Two-Years Average |
|---|---|---|---|
| Canino | 539.63 ± 12.81 ab | 669.52 ± 30.03 a | 604.58 ± 74.08 a |
| Dama Rosa * | 1725.21 ± 222.12 g | 1565.50 ± 64.26 e | 1645.36 ± 170.41 d |
| Dama Taronja * | 699.71 ± 27.00 bcd | 1171.19 ± 286.97 cd | 935.45 ± 316.10 abc |
| GG9310 * | 1024.98 ± 9.58 ef | 1008.08 ± 80.72 abc | 1016.53 ± 52.23 bc |
| GG979 * | 630.10 ± 11.57 abc | 1018.73 ± 159.22 bc | 824.41 ± 235.59 abc |
| Goldrich | 894.02 ± 25.15 de | 876.18 ± 22.06 abc | 885.10 ± 23.31 abc |
| GP9817 * | 1167.16 ± 17.91 f | 916.08 ± 67.15 abc | 1041.62 ± 144.38 bc |
| HG9821 | 514.38 ± 3.73 ab | 814.96 ± 51.46 ab | 664.67 ± 167.84 ab |
| HG9850 | 422.75 ± 18.96 a | 695.63 ± 39.43 ab | 559.19 ± 152.00 a |
| HM964 * | 822.83 ± 28.91 cde | 825.43 ± 23.56 ab | 824.13 ± 23.63 abc |
| Mitger | 832.44 ± 28.91 cde | 1509.14 ± 76.83 de | 1170.79 ± 374.27 c |
| SEOP934 | 497.04 ± 20.18 ab | 738.04 ± 75.04 ab | 617.54 ± 140.85 a |
| Year | Goldrich | Year × Goldrich | Residual | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SSt | SSr | p-v | SSt | SSr | p-v | SSt | SSr | p-v | SSt | SSr | Total | R2 | |
| Neochlogenic | 108,653 | 0.095 | 0.0039 | 120,718 | 0.1051 | 0.0024 | 10.0892 | 0.000 | 0.9771 | 826,863 | 0.720 | 1.15 × 106 | 0.280 |
| Chlorogenic | 288,022 | 0.149 | 0.0004 | 134,852 | 0.0700 | 0.0124 | 12,565.3 | 0.007 | 0.4359 | 1.39 × 106 | 0.722 | 1.93 × 106 | 0.278 |
| Rutin | 1140.39 | 0.001 | 0.8023 | 90,359.6 | 0.0596 | 0.0286 | 92,447.4 | 0.061 | 0.0269 | 1.23 × 106 | 0.811 | 1.51 × 106 | 0.189 |
| Quercetin-3-glucurunide | 26.687 | 0.001 | 0.7803 | 523.664 | 0.0208 | 0.219 | 605.821 | 0.024 | 0.1865 | 23,134.6 | 0.921 | 25,129.6 | 0.079 |
| Total content | 684,536 | 0.083 | 0.0089 | 1.08 × 106 | 0.1312 | 0.0012 | 191,481 | 0.023 | 0.1591 | 6.42 × 106 | 0.782 | 8.21 × 106 | 0.218 |
| Constant | Goldrich | ||||||||||||
| Mean | Lower Lim | Error | Mean | Lower Lim | Error | Gr | Sig | ||||||
| Neochlogenic | 228.557 | 193.471 | 35.086 | 127.94 | 46.913 | 81.027 | 56.0% | ** | |||||
| Chlorogenic | 236.938 | 191.437 | 45.501 | 135.222 | 30.1403 | 105.082 | 57.1% | * | |||||
| Rutin | 296.716 | 253.959 | 42.757 | 110.689 | 11.9464 | 98.743 | 37.3% | * | |||||
| Quercetin-3-glucurunide | 25.4043 | 19.5356 | 5.869 | 8.4265 | −5.12673 | 13.553 | 33.2% | NS | |||||
| Total content | 787.615 | 689.815 | 97.800 | 382.278 | 156.418 | 225.860 | 48.5% | ** | |||||
| Year | Goldrich | Year × Goldrich | Residual | Total | R2 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SSt | SSr | p-v | SSt | SSr | p-v | SSt | SSr | p-v | SSt | SSr | |||
| ParDFR | 2.2294 | 0.023 | 0.1686 | 17.738 | 0.1795 | 0.0002 | 2.4002 | 0.024 | 0.1534 | 78.2977 | 0.792 | 98.8062 | 0.208 |
| ParFLS1 | 17.9388 | 0.073 | 0.002 | 8.0677 | 0.0327 | 0.0362 | 11.5896 | 0.047 | 0.0124 | 219.4 | 0.888 | 246.939 | 0.112 |
| ParFLS2 | 0.0474 | 0.000 | 0.8526 | 6.7223 | 0.0636 | 0.0297 | 2.8529 | 0.027 | 0.1527 | 92.7271 | 0.878 | 105.665 | 0.122 |
| ParPAL1 | 0.3709 | 0.006 | 0.5249 | 4.2923 | 0.0655 | 0.0332 | 0.3443 | 0.005 | 0.5401 | 60.8265 | 0.928 | 65.5523 | 0.072 |
| ParPAL2 | 1.0437 | 0.009 | 0.4138 | 16.03 | 0.1310 | 0.0020 | 0.1313 | 0.001 | 0.7714 | 104.974 | 0.859 | 122.217 | 0.141 |
| Constant | Goldrich | ||||||||||||
| Mean | Lower Lim | Error | Mean | Lower Lim | Error | Gr | Sig | ||||||
| ParDFR | 1.57183 | 1.2304 | 0.341 | −1.5509 | −2.3393 | 0.788 | −98.7% | ** | |||||
| ParFLS1 | 1.54347 | 1.2259 | 0.318 | −0.7697 | −1.4891 | 0.719 | −49.9% | * | |||||
| ParFLS2 | 1.64119 | 1.2696 | 0.372 | −0.9547 | −1.8128 | 0.858 | −58.2% | * | |||||
| ParParPAL1 | 1.08334 | 0.7799 | 0.303 | 0.7652 | 0.0628 | 0.702 | 70.6% | * | |||||
| ParPAL2 | 1.80279 | 1.4075 | 0.395 | −1.4731 | −2.3861 | 0.913 | −81.7% | ** | |||||
| Parameter | ParPAL1 | ParPAL2 | ParDFR | ParFLS1 | ParFLS2 | Neochlorogenic | Chlorogenic | Rutin |
|---|---|---|---|---|---|---|---|---|
| ParPAL1 | ||||||||
| ParPAL2 | 0.1507 | |||||||
| ParDFR | −0.0163 | 0.8098 ** | ||||||
| ParFLS1 | −0.1899 | 0.3408 ** | 0.3139 ** | |||||
| ParFLS2 | −0.2726 * | 0.0273 | 0.1629 | 0.1261 | ||||
| Neochlorogenic | −0.1951 | −0.1369 | −0.0258 | −0.1043 | 0.2919 * | |||
| Chlorogenic | −0.1317 | −0.1216 | −0.017 | 0.0051 | −0.011 | 0.6835 ** | ||
| Rutin | 0.0283 | 0.1635 | 0.0568 | −0.2062 | 0.1943 | 0.2929 * | 0.0734 | |
| Quercetin-3-glucuronide | −0.1638 | 0.0083 | 0.0477 | −0.144 | 0.1216 | 0.4452 ** | 0.2233 | 0.7407 ** |
| Genotype | Pedigree | Origin |
|---|---|---|
| Canino | Unknown | Spain |
| Dama Rosa | Goldrich × Ginesta | IVIA |
| Dama Taronja | Goldrich × Katy | IVIA |
| GG9310 | Goldrich × Ginesta | IVIA |
| GG979 | Goldrich × Ginesta | IVIA |
| Goldrich | Sunglo × Perfection | USA |
| GP9817 | Goldrich × Palau | IVIA |
| HG9821 | Harcot × Ginesta | IVIA |
| HG9850 | Harcot × Ginesta | IVIA |
| HM964 | Harcot × Mitger | IVIA |
| Mitger | Unknown | Spain |
| SEOP934 | SEO × Palau | IVIA |
| Gen | Forward | Reverse |
|---|---|---|
| ParPAL1 | CGACTGGGTTATGGATAGCATGA | CAATGTGTGGGTAGATTCTGTGC |
| ParPAL2 | TAAAGAGGTGGATAGTGCAAGGG | GAGAACACCTTGTCGCATTCTTC |
| ParFLS1F | TGGAGGGGATGACATGGTTTATC | CCGTTGCTCATAATCTCCATCTG |
| ParFLS2F | ACAGGAGGAAAAGGAGGCTTATG | GGCCAGAACCGGTAATTAATGAC |
| ParDFR | GTTCGAAGGCTGGTGTTTACATC | GAGAAATGGGCCAATCACAAGAG |
| ACTIN | CTTCTTACTGAGGCACCCCTGAAT | AGCATAGAGGGAGAGAACTGCTTG |
| SAND | TCGTGGGTACCAGGAAAACGACAT | CCTGCTAGCTTGTGTTCATCTCCA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gómez-Martínez, H.; Gil-Muñoz, F.; Bermejo, A.; Zuriaga, E.; Badenes, M.L. Insights of Phenolic Pathway in Fruits: Transcriptional and Metabolic Profiling in Apricot (Prunus armeniaca). Int. J. Mol. Sci. 2021, 22, 3411. https://doi.org/10.3390/ijms22073411
Gómez-Martínez H, Gil-Muñoz F, Bermejo A, Zuriaga E, Badenes ML. Insights of Phenolic Pathway in Fruits: Transcriptional and Metabolic Profiling in Apricot (Prunus armeniaca). International Journal of Molecular Sciences. 2021; 22(7):3411. https://doi.org/10.3390/ijms22073411
Chicago/Turabian StyleGómez-Martínez, Helena, Francisco Gil-Muñoz, Almudena Bermejo, Elena Zuriaga, and Maria L. Badenes. 2021. "Insights of Phenolic Pathway in Fruits: Transcriptional and Metabolic Profiling in Apricot (Prunus armeniaca)" International Journal of Molecular Sciences 22, no. 7: 3411. https://doi.org/10.3390/ijms22073411
APA StyleGómez-Martínez, H., Gil-Muñoz, F., Bermejo, A., Zuriaga, E., & Badenes, M. L. (2021). Insights of Phenolic Pathway in Fruits: Transcriptional and Metabolic Profiling in Apricot (Prunus armeniaca). International Journal of Molecular Sciences, 22(7), 3411. https://doi.org/10.3390/ijms22073411







