Extracellular Vesicles and Their Role in the Spatial and Temporal Expansion of Tumor–Immune Interactions
Abstract
1. Introduction
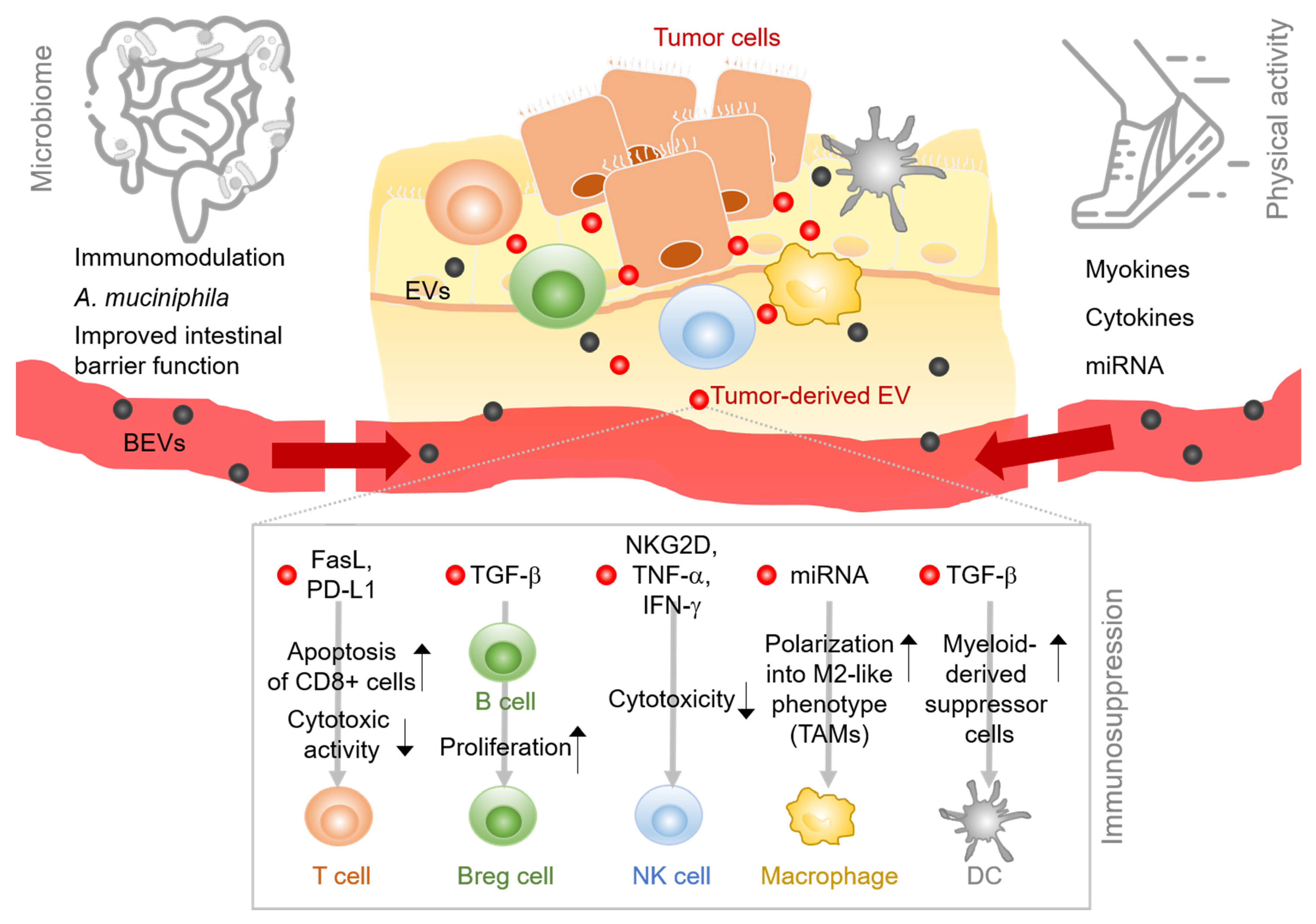
2. EVs Expand the Tumor–Immune Interactions
2.1. EVs in the Tumor-Immune Environment Cross-Talk
2.1.1. T Cells
2.1.2. B Cells and Plasma Cells
2.1.3. NK Cells
2.1.4. Professional APCs
2.2. EVs in the Microbiota—Gut Barrier Tumor Cross-Talk
2.3. EVs in Physical Exercise-Tumor Cross-Talk
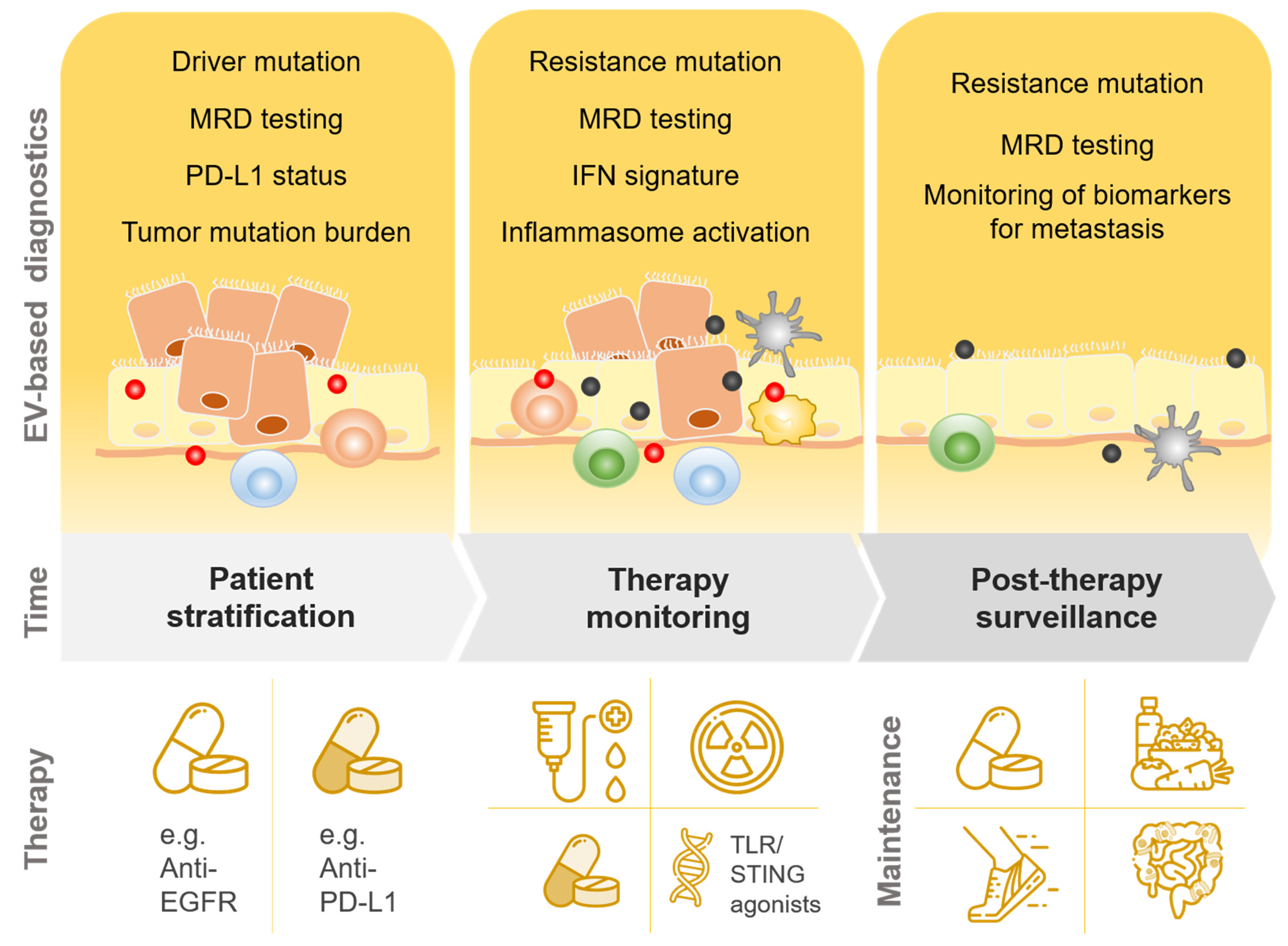
3. EVs in Tumor Development—Role of Immune Responses and Implications for Diagnostics
3.1. EVs in the Promotion of Cell Proliferation and Immune Escape
3.2. EVs in Intervention-Induced Innate Immune Responses
3.3. Implications for Diagnostics
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Théry, C.; Ostrowski, M.; Segura, E. Membrane Vesicles as Conveyors of Immune Responses. Nat. Rev. Immunol. 2009, 9, 581–593. [Google Scholar] [CrossRef]
- Raposo, G.; Stahl, P.D. Extracellular Vesicles: A New Communication Paradigm? Nat. Rev. Mol. Cell Biol. 2019, 20, 509–510. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Meehan, K.; Vella, L.J. The Contribution of Tumour-Derived Exosomes to the Hallmarks of Cancer. Crit. Rev. Clin. Lab. Sci. 2016, 53, 121–131. [Google Scholar] [CrossRef]
- Whiteside, T.L. Exosomes and Tumor-Mediated Immune Suppression. J. Clin. Investig. 2016, 126, 1216–1223. [Google Scholar] [CrossRef]
- Robbins, P.D.; Morelli, A.E. Regulation of Immune Responses by Extracellular Vesicles. Nat. Rev. Immunol. 2014, 14, 195–208. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal Information for Studies of Extracellular Vesicles 2018 (MISEV2018): A Position Statement of the International Society for Extracellular Vesicles and Update of the MISEV2014 Guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed]
- Catalano, V.; Turdo, A.; Di Franco, S.; Dieli, F.; Todaro, M.; Stassi, G. Tumor and Its Microenvironment: A Synergistic Interplay. Semin. Cancer Biol. 2013, 23, 522–532. [Google Scholar] [CrossRef] [PubMed]
- Quail, D.F.; Joyce, J.A. Microenvironmental Regulation of Tumor Progression and Metastasis. Nat. Med. 2013, 19, 1423–1437. [Google Scholar] [CrossRef] [PubMed]
- Bissell, M.J.; Hines, W.C. Why Don’t We Get More Cancer? A Proposed Role of the Microenvironment in Restraining Cancer Progression. Nat. Med. 2011, 17, 320–329. [Google Scholar] [CrossRef]
- McAllister, S.S.; Weinberg, R.A. The Tumour-Induced Systemic Environment as a Critical Regulator of Cancer Progression and Metastasis. Nat. Cell Biol. 2014, 16, 717–727. [Google Scholar] [CrossRef]
- Laplane, L.; Duluc, D.; Larmonier, N.; Pradeu, T.; Bikfalvi, A. The Multiple Layers of the Tumor Environment. Trends Cancer 2018, 4, 802–809. [Google Scholar] [CrossRef] [PubMed]
- Laplane, L.; Duluc, D.; Bikfalvi, A.; Larmonier, N.; Pradeu, T. Beyond the Tumour Microenvironment. Int. J. Cancer 2019, 145, 2611–2618. [Google Scholar] [CrossRef]
- Zitvogel, L.; Galluzzi, L.; Viaud, S.; Vétizou, M.; Daillère, R.; Merad, M.; Kroemer, G. Cancer and the Gut Microbiota: An Unexpected Link. Sci. Transl. Med. 2015, 7, 271. [Google Scholar] [CrossRef]
- Fridman, W.H.; Pagès, F.; Sautès-Fridman, C.; Galon, J. The Immune Contexture in Human Tumours: Impact on Clinical Outcome. Nat. Rev. Cancer 2012, 12, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Mittal, D.; Gubin, M.M.; Schreiber, R.D.; Smyth, M.J. New Insights into Cancer Immunoediting and Its Three Component Phases—Elimination, Equilibrium and Escape. Curr. Opin. Immunol. 2014, 27, 16–25. [Google Scholar] [CrossRef]
- Palucka, A.K.; Coussens, L.M. The Basis of Oncoimmunology. Cell 2016, 164, 1233–1247. [Google Scholar] [CrossRef] [PubMed]
- Bosque, A.; Dietz, L.; Gallego-Lleyda, A.; Sanclemente, M.; Iturralde, M.; Naval, J.; Alava, M.A.; Martínez-Lostao, L.; Thierse, H.-J.; Anel, A. Comparative Proteomics of Exosomes Secreted by Tumoral Jurkat T Cells and Normal Human T Cell Blasts Unravels a Potential Tumorigenic Role for Valosin-Containing Protein. Oncotarget 2016, 7, 29287–29305. [Google Scholar] [CrossRef] [PubMed]
- Azoulay-Alfaguter, I.; Mor, A. Proteomic Analysis of Human T Cell-Derived Exosomes Reveals Differential RAS/MAPK Signaling. Eur. J. Immunol. 2018, 48, 1915–1917. [Google Scholar] [CrossRef]
- Torralba, D.; Baixauli, F.; Villarroya-Beltri, C.; Fernández-Delgado, I.; Latorre-Pellicer, A.; Acín-Pérez, R.; Martín-Cófreces, N.B.; Jaso-Tamame, Á.L.; Iborra, S.; Jorge, I.; et al. Priming of Dendritic Cells by DNA-Containing Extracellular Vesicles from Activated T Cells through Antigen-Driven Contacts. Nat. Commun. 2018, 9, 2658. [Google Scholar] [CrossRef]
- Villarroya-Beltri, C.; Gutiérrez-Vázquez, C.; Sánchez-Cabo, F.; Pérez-Hernández, D.; Vázquez, J.; Martin-Cofreces, N.; Martinez-Herrera, D.J.; Pascual-Montano, A.; Mittelbrunn, M.; Sánchez-Madrid, F. Sumoylated HnRNPA2B1 Controls the Sorting of MiRNAs into Exosomes through Binding to Specific Motifs. Nat. Commun. 2013, 4, 2980. [Google Scholar] [CrossRef] [PubMed]
- Seo, N.; Shirakura, Y.; Tahara, Y.; Momose, F.; Harada, N.; Ikeda, H.; Akiyoshi, K.; Shiku, H. Activated CD8+ T Cell Extracellular Vesicles Prevent Tumour Progression by Targeting of Lesional Mesenchymal Cells. Nat. Commun. 2018, 9, 435. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Jiang, J.; Xia, W.; Huang, J. Tumor-Related Exosomes Contribute to Tumor-Promoting Microenvironment: An Immunological Perspective. J. Immunol. Res. 2017, 2017, 1073947. [Google Scholar] [CrossRef]
- Hellwinkel, J.E.; Redzic, J.S.; Harland, T.A.; Gunaydin, D.; Anchordoquy, T.J.; Graner, M.W. Glioma-Derived Extracellular Vesicles Selectively Suppress Immune Responses. Neuro-Oncol. 2016, 18, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Diergaarde, B.; Ferrone, S.; Kirkwood, J.M.; Whiteside, T.L. Melanoma Cell-Derived Exosomes in Plasma of Melanoma Patients Suppress Functions of Immune Effector Cells. Sci. Rep. 2020, 10, 92. [Google Scholar] [CrossRef]
- Chen, G.; Huang, A.C.; Zhang, W.; Zhang, G.; Wu, M.; Xu, W.; Yu, Z.; Yang, J.; Wang, B.; Sun, H.; et al. Exosomal PD-L1 Contributes to Immunosuppression and Is Associated with Anti-PD-1 Response. Nature 2018, 560, 382–386. [Google Scholar] [CrossRef] [PubMed]
- Sharonov, G.V.; Serebrovskaya, E.O.; Yuzhakova, D.V.; Britanova, O.V.; Chudakov, D.M. B Cells, Plasma Cells and Antibody Repertoires in the Tumour Microenvironment. Nat. Rev. Immunol. 2020, 20, 294–307. [Google Scholar] [CrossRef]
- Wubbolts, R.; Leckie, R.S.; Veenhuizen, P.T.M.; Schwarzmann, G.; Möbius, W.; Hoernschemeyer, J.; Slot, J.-W.; Geuze, H.J.; Stoorvogel, W. Proteomic and Biochemical Analyses of Human B Cell-Derived Exosomes. Potential Implications for Their Function and Multivesicular Body Formation. J. Biol. Chem. 2003, 278, 10963–10972. [Google Scholar] [CrossRef]
- Admyre, C.; Bohle, B.; Johansson, S.M.; Focke-Tejkl, M.; Valenta, R.; Scheynius, A.; Gabrielsson, S. B Cell-Derived Exosomes Can Present Allergen Peptides and Activate Allergen-Specific T Cells to Proliferate and Produce TH2-like Cytokines. J. Allergy Clin. Immunol. 2007, 120, 1418–1424. [Google Scholar] [CrossRef] [PubMed]
- Saunderson, S.C.; Dunn, A.C.; Crocker, P.R.; McLellan, A.D. CD169 Mediates the Capture of Exosomes in Spleen and Lymph Node. Blood 2014, 123, 208–216. [Google Scholar] [CrossRef]
- Saunderson, S.C.; McLellan, A.D. Role of Lymphocyte Subsets in the Immune Response to Primary B Cell-Derived Exosomes. J. Immunol. 2017, 199, 2225–2235. [Google Scholar] [CrossRef]
- Yang, C.; Chalasani, G.; Ng, Y.-H.; Robbins, P.D. Exosomes Released from Mycoplasma Infected Tumor Cells Activate Inhibitory B Cells. PLoS ONE 2012, 7, e36138. [Google Scholar] [CrossRef]
- Li, Y.; An, J.; Huang, S.; He, J.; Zhang, J. Esophageal Cancer-Derived Microvesicles Induce Regulatory B Cells. Cell Biochem. Funct. 2015, 33, 308–313. [Google Scholar] [CrossRef] [PubMed]
- Pucci, F.; Garris, C.; Lai, C.P.; Newton, A.; Pfirschke, C.; Engblom, C.; Alvarez, D.; Sprachman, M.; Evavold, C.; Magnuson, A.; et al. SCS Macrophages Suppress Melanoma by Restricting Tumor-Derived Vesicle-B Cell Interactions. Science 2016, 352, 242–246. [Google Scholar] [CrossRef]
- Lugini, L.; Cecchetti, S.; Huber, V.; Luciani, F.; Macchia, G.; Spadaro, F.; Paris, L.; Abalsamo, L.; Colone, M.; Molinari, A.; et al. Immune Surveillance Properties of Human NK Cell-Derived Exosomes. J. Immunol. 2012, 189, 2833–2842. [Google Scholar] [CrossRef] [PubMed]
- Jong, A.Y.; Wu, C.-H.; Li, J.; Sun, J.; Fabbri, M.; Wayne, A.S.; Seeger, R.C. Large-Scale Isolation and Cytotoxicity of Extracellular Vesicles Derived from Activated Human Natural Killer Cells. J. Extracell. Vesicles 2017, 6, 1294368. [Google Scholar] [CrossRef] [PubMed]
- Neviani, P.; Wise, P.M.; Murtadha, M.; Liu, C.W.; Wu, C.-H.; Jong, A.Y.; Seeger, R.C.; Fabbri, M. Natural Killer-Derived Exosomal MiR-186 Inhibits Neuroblastoma Growth and Immune Escape Mechanisms. Cancer Res. 2019, 79, 1151–1164. [Google Scholar] [CrossRef] [PubMed]
- Federici, C.; Shahaj, E.; Cecchetti, S.; Camerini, S.; Casella, M.; Iessi, E.; Camisaschi, C.; Paolino, G.; Calvieri, S.; Ferro, S.; et al. Natural-Killer-Derived Extracellular Vesicles: Immune Sensors and Interactors. Front. Immunol. 2020, 11, 262. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-H.; Li, J.; Li, L.; Sun, J.; Fabbri, M.; Wayne, A.S.; Seeger, R.C.; Jong, A.Y. Extracellular Vesicles Derived from Natural Killer Cells Use Multiple Cytotoxic Proteins and Killing Mechanisms to Target Cancer Cells. J. Extracell. Vesicles 2019, 8, 1588538. [Google Scholar] [CrossRef]
- Zhao, J.; Schlößer, H.A.; Wang, Z.; Qin, J.; Li, J.; Popp, F.; Popp, M.C.; Alakus, H.; Chon, S.-H.; Hansen, H.P.; et al. Tumor-Derived Extracellular Vesicles Inhibit Natural Killer Cell Function in Pancreatic Cancer. Cancers 2019, 11, 874. [Google Scholar] [CrossRef] [PubMed]
- Hedlund, M.; Nagaeva, O.; Kargl, D.; Baranov, V.; Mincheva-Nilsson, L. Thermal- and Oxidative Stress Causes Enhanced Release of NKG2D Ligand-Bearing Immunosuppressive Exosomes in Leukemia/Lymphoma T and B Cells. PLoS ONE 2011, 6, e16899. [Google Scholar] [CrossRef]
- Clayton, A.; Mitchell, J.P.; Court, J.; Linnane, S.; Mason, M.D.; Tabi, Z. Human Tumor-Derived Exosomes down-Modulate NKG2D Expression. J. Immunol. 2008, 180, 7249–7258. [Google Scholar] [CrossRef]
- Szczepanski, M.J.; Szajnik, M.; Welsh, A.; Whiteside, T.L.; Boyiadzis, M. Blast-Derived Microvesicles in Sera from Patients with Acute Myeloid Leukemia Suppress Natural Killer Cell Function via Membrane-Associated Transforming Growth Factor-Beta1. Haematologica 2011, 96, 1302–1309. [Google Scholar] [CrossRef] [PubMed]
- Bhatnagar, S.; Shinagawa, K.; Castellino, F.J.; Schorey, J.S. Exosomes Released from Macrophages Infected with Intracellular Pathogens Stimulate a Proinflammatory Response in Vitro and in Vivo. Blood 2007, 110, 3234–3244. [Google Scholar] [CrossRef] [PubMed]
- Ferrante, C.J.; Pinhal-Enfield, G.; Elson, G.; Cronstein, B.N.; Hasko, G.; Outram, S.; Leibovich, S.J. The Adenosine-Dependent Angiogenic Switch of Macrophages to an M2-like Phenotype Is Independent of Interleukin-4 Receptor Alpha (IL-4Rα) Signaling. Inflammation 2013, 36, 921–931. [Google Scholar] [CrossRef]
- Zhou, J.; Li, X.; Wu, X.; Zhang, T.; Zhu, Q.; Wang, X.; Wang, H.; Wang, K.; Lin, Y.; Wang, X. Exosomes Released from Tumor-Associated Macrophages Transfer MiRNAs That Induce a Treg/Th17 Cell Imbalance in Epithelial Ovarian Cancer. Cancer Immunol. Res. 2018, 6, 1578–1592. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.; Ma, T.; Huang, B.; Lin, L.; Zhou, Y.; Yan, J.; Zou, Y.; Chen, S. Macrophage-Derived Exosomal MicroRNA-501-3p Promotes Progression of Pancreatic Ductal Adenocarcinoma through the TGFBR3-Mediated TGF-β Signaling Pathway. J. Exp. Clin. Cancer Res. 2019, 38, 310. [Google Scholar] [CrossRef]
- Cianciaruso, C.; Beltraminelli, T.; Duval, F.; Nassiri, S.; Hamelin, R.; Mozes, A.; Gallart-Ayala, H.; Ceada Torres, G.; Torchia, B.; Ries, C.H.; et al. Molecular Profiling and Functional Analysis of Macrophage-Derived Tumor Extracellular Vesicles. Cell Rep. 2019, 27, 3062–3080. [Google Scholar] [CrossRef]
- Cooks, T.; Pateras, I.S.; Jenkins, L.M.; Patel, K.M.; Robles, A.I.; Morris, J.; Forshew, T.; Appella, E.; Gorgoulis, V.G.; Harris, C.C. Mutant P53 Cancers Reprogram Macrophages to Tumor Supporting Macrophages via Exosomal MiR-1246. Nat. Commun. 2018, 9, 771. [Google Scholar] [CrossRef] [PubMed]
- Kanlikilicer, P.; Bayraktar, R.; Denizli, M.; Rashed, M.H.; Ivan, C.; Aslan, B.; Mitra, R.; Karagoz, K.; Bayraktar, E.; Zhang, X.; et al. Exosomal MiRNA Confers Chemo Resistance via Targeting Cav1/p-Gp/M2-Type Macrophage Axis in Ovarian Cancer. EBioMedicine 2018, 38, 100–112. [Google Scholar] [CrossRef]
- Harris, A.L. Hypoxia—A Key Regulatory Factor in Tumour Growth. Nat. Rev. Cancer 2002, 2, 38–47. [Google Scholar] [CrossRef]
- Chen, X.; Zhou, J.; Li, X.; Wang, X.; Lin, Y.; Wang, X. Exosomes Derived from Hypoxic Epithelial Ovarian Cancer Cells Deliver MicroRNAs to Macrophages and Elicit a Tumor-Promoted Phenotype. Cancer Lett. 2018, 435, 80–91. [Google Scholar] [CrossRef] [PubMed]
- van der Vos, K.E.; Abels, E.R.; Zhang, X.; Lai, C.; Carrizosa, E.; Oakley, D.; Prabhakar, S.; Mardini, O.; Crommentuijn, M.H.W.; Skog, J.; et al. Directly Visualized Glioblastoma-Derived Extracellular Vesicles Transfer RNA to Microglia/Macrophages in the Brain. Neuro-Oncol. 2016, 18, 58–69. [Google Scholar] [CrossRef] [PubMed]
- Casadei, L.; Calore, F.; Creighton, C.J.; Guescini, M.; Batte, K.; Iwenofu, O.H.; Zewdu, A.; Braggio, D.A.; Bill, K.L.; Fadda, P.; et al. Exosome-Derived MiR-25-3p and MiR-92a-3p Stimulate Liposarcoma Progression. Cancer Res. 2017, 77, 3846–3856. [Google Scholar] [CrossRef]
- Sekirov, I.; Russell, S.L.; Antunes, L.C.M.; Finlay, B.B. Gut Microbiota in Health and Disease. Physiol. Rev. 2010, 90, 859–904. [Google Scholar] [CrossRef] [PubMed]
- Kaparakis-Liaskos, M.; Ferrero, R.L. Immune Modulation by Bacterial Outer Membrane Vesicles. Nat. Rev. Immunol. 2015, 15, 375–387. [Google Scholar] [CrossRef] [PubMed]
- Toyofuku, M.; Nomura, N.; Eberl, L. Types and Origins of Bacterial Membrane Vesicles. Nat. Rev. Microbiol. 2019, 17, 13–24. [Google Scholar] [CrossRef]
- Chronopoulos, A.; Kalluri, R. Emerging Role of Bacterial Extracellular Vesicles in Cancer. Oncogene 2020, 39, 6951–6960. [Google Scholar] [CrossRef] [PubMed]
- Brown, L.; Wolf, J.M.; Prados-Rosales, R.; Casadevall, A. Through the Wall: Extracellular Vesicles in Gram-Positive Bacteria, Mycobacteria and Fungi. Nat. Rev. Microbiol. 2015, 13, 620–630. [Google Scholar] [CrossRef]
- Tulkens, J.; Vergauwen, G.; Van Deun, J.; Geeurickx, E.; Dhondt, B.; Lippens, L.; De Scheerder, M.-A.; Miinalainen, I.; Rappu, P.; De Geest, B.G.; et al. Increased Levels of Systemic LPS-Positive Bacterial Extracellular Vesicles in Patients with Intestinal Barrier Dysfunction. Gut 2020, 69, 191–193. [Google Scholar] [CrossRef]
- Jones, E.J.; Booth, C.; Fonseca, S.; Parker, A.; Cross, K.; Miquel-Clopés, A.; Hautefort, I.; Mayer, U.; Wileman, T.; Stentz, R.; et al. The Uptake, Trafficking, and Biodistribution of Bacteroides Thetaiotaomicron Generated Outer Membrane Vesicles. Front. Microbiol. 2020, 11, 57. [Google Scholar] [CrossRef]
- Bhatt, A.P.; Redinbo, M.R.; Bultman, S.J. The Role of the Microbiome in Cancer Development and Therapy. CA Cancer J. Clin. 2017, 67, 326–344. [Google Scholar] [CrossRef]
- Gopalakrishnan, V.; Helmink, B.A.; Spencer, C.N.; Reuben, A.; Wargo, J.A. The Influence of the Gut Microbiome on Cancer, Immunity, and Cancer Immunotherapy. Cancer Cell 2018, 33, 570–580. [Google Scholar] [CrossRef]
- Xavier, J.B.; Young, V.B.; Skufca, J.; Ginty, F.; Testerman, T.; Pearson, A.T.; Macklin, P.; Mitchell, A.; Shmulevich, I.; Xie, L.; et al. The Cancer Microbiome: Distinguishing Direct and Indirect Effects Requires a Systemic View. Trends Cancer 2020, 6, 192–204. [Google Scholar] [CrossRef]
- Zitvogel, L.; Ayyoub, M.; Routy, B.; Kroemer, G. Microbiome and Anticancer Immunosurveillance. Cell 2016, 165, 276–287. [Google Scholar] [CrossRef]
- Shen, Y.; Torchia, M.L.G.; Lawson, G.W.; Karp, C.L.; Ashwell, J.D.; Mazmanian, S.K. Outer Membrane Vesicles of a Human Commensal Mediate Immune Regulation and Disease Protection. Cell Host Microbe 2012, 12, 509–520. [Google Scholar] [CrossRef]
- Alaniz, R.C.; Deatherage, B.L.; Lara, J.C.; Cookson, B.T. Membrane Vesicles Are Immunogenic Facsimiles of Salmonella Typhimurium That Potently Activate Dendritic Cells, Prime B and T Cell Responses, and Stimulate Protective Immunity in Vivo. J. Immunol. 2007, 179, 7692–7701. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Wang, X.; Fan, G.-C. Versatile Effects of Bacterium-Released Membrane Vesicles on Mammalian Cells and Infectious/Inflammatory Diseases. Acta Pharmacol. Sin. 2018, 39, 514–533. [Google Scholar] [CrossRef] [PubMed]
- Geerlings, S.Y.; Kostopoulos, I.; de Vos, W.M.; Belzer, C. Akkermansia Muciniphila in the Human Gastrointestinal Tract: When, Where, and How? Microorganisms 2018, 6, 75. [Google Scholar] [CrossRef]
- Kang, C.-S.; Ban, M.; Choi, E.-J.; Moon, H.-G.; Jeon, J.-S.; Kim, D.-K.; Park, S.-K.; Jeon, S.G.; Roh, T.-Y.; Myung, S.-J.; et al. Extracellular Vesicles Derived from Gut Microbiota, Especially Akkermansia Muciniphila, Protect the Progression of Dextran Sulfate Sodium-Induced Colitis. PLoS ONE 2013, 8, e76520. [Google Scholar] [CrossRef] [PubMed]
- Chelakkot, C.; Choi, Y.; Kim, D.-K.; Park, H.T.; Ghim, J.; Kwon, Y.; Jeon, J.; Kim, M.-S.; Jee, Y.-K.; Gho, Y.S.; et al. Akkermansia Muciniphila-Derived Extracellular Vesicles Influence Gut Permeability through the Regulation of Tight Junctions. Exp. Mol. Med. 2018, 50, e450. [Google Scholar] [CrossRef]
- Ashrafian, F.; Behrouzi, A.; Shahriary, A.; Ahmadi Badi, S.; Davari, M.; Khatami, S.; Rahimi Jamnani, F.; Fateh, A.; Vaziri, F.; Siadat, S.D. Comparative Study of Effect of Akkermansia Muciniphila and Its Extracellular Vesicles on Toll-like Receptors and Tight Junction. Gastroenterol. Hepatol. Bed Bench 2019, 12, 163–168. [Google Scholar]
- Ashrafian, F.; Shahriary, A.; Behrouzi, A.; Moradi, H.R.; Keshavarz Azizi Raftar, S.; Lari, A.; Hadifar, S.; Yaghoubfar, R.; Ahmadi Badi, S.; Khatami, S.; et al. Akkermansia Muciniphila-Derived Extracellular Vesicles as a Mucosal Delivery Vector for Amelioration of Obesity in Mice. Front. Microbiol. 2019, 10, 2155. [Google Scholar] [CrossRef]
- Yaghoubfar, R.; Behrouzi, A.; Ashrafian, F.; Shahryari, A.; Moradi, H.R.; Choopani, S.; Hadifar, S.; Vaziri, F.; Nojoumi, S.A.; Fateh, A.; et al. Modulation of Serotonin Signaling/Metabolism by Akkermansia Muciniphila and Its Extracellular Vesicles through the Gut-Brain Axis in Mice. Sci. Rep. 2020, 10, 22119. [Google Scholar] [CrossRef]
- Routy, B.; Le Chatelier, E.; Derosa, L.; Duong, C.P.M.; Alou, M.T.; Daillère, R.; Fluckiger, A.; Messaoudene, M.; Rauber, C.; Roberti, M.P.; et al. Gut Microbiome Influences Efficacy of PD-1-Based Immunotherapy against Epithelial Tumors. Science 2018, 359, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Moore, S.C.; Lee, I.-M.; Weiderpass, E.; Campbell, P.T.; Sampson, J.N.; Kitahara, C.M.; Keadle, S.K.; Arem, H.; Berrington de Gonzalez, A.; Hartge, P.; et al. Association of Leisure-Time Physical Activity with Risk of 26 Types of Cancer in 1.44 Million Adults. JAMA Intern. Med. 2016, 176, 816–825. [Google Scholar] [CrossRef] [PubMed]
- Guercio, B.J.; Zhang, S.; Ou, F.-S.; Venook, A.P.; Niedzwiecki, D.; Lenz, H.-J.; Innocenti, F.; O’Neil, B.H.; Shaw, J.E.; Polite, B.N.; et al. Associations of Physical Activity with Survival and Progression in Metastatic Colorectal Cancer: Results from Cancer and Leukemia Group B (Alliance)/SWOG 80405. J. Clin. Oncol. 2019, 37, 2620–2631. [Google Scholar] [CrossRef] [PubMed]
- Hong, B.S.; Lee, K.P. A Systematic Review of the Biological Mechanisms Linking Physical Activity and Breast Cancer. Phys. Act. Nutr. 2020, 24, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Frühbeis, C.; Helmig, S.; Tug, S.; Simon, P.; Krämer-Albers, E.-M. Physical Exercise Induces Rapid Release of Small Extracellular Vesicles into the Circulation. J. Extracell. Vesicles 2015, 4, 28239. [Google Scholar] [CrossRef] [PubMed]
- Whitham, M.; Parker, B.L.; Friedrichsen, M.; Hingst, J.R.; Hjorth, M.; Hughes, W.E.; Egan, C.L.; Cron, L.; Watt, K.I.; Kuchel, R.P.; et al. Extracellular Vesicles Provide a Means for Tissue Crosstalk during Exercise. Cell Metab. 2018, 27, 237–251. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, G.P.; Porto, W.F.; Palu, C.C.; Pereira, L.M.; Petriz, B.; Almeida, J.A.; Viana, J.; Filho, N.N.A.; Franco, O.L.; Pereira, R.W. Effects of Acute Aerobic Exercise on Rats Serum Extracellular Vesicles Diameter, Concentration and Small RNAs Content. Front. Physiol. 2018, 9, 532. [Google Scholar] [CrossRef] [PubMed]
- Guescini, M.; Canonico, B.; Lucertini, F.; Maggio, S.; Annibalini, G.; Barbieri, E.; Luchetti, F.; Papa, S.; Stocchi, V. Muscle Releases Alpha-Sarcoglycan Positive Extracellular Vesicles Carrying MiRNAs in the Bloodstream. PLoS ONE 2015, 10, e0125094. [Google Scholar] [CrossRef]
- Hou, Z.; Qin, X.; Hu, Y.; Zhang, X.; Li, G.; Wu, J.; Li, J.; Sha, J.; Chen, J.; Xia, J.; et al. Longterm Exercise-Derived Exosomal MiR-342-5p: A Novel Exerkine for Cardioprotection. Circ. Res. 2019, 124, 1386–1400. [Google Scholar] [CrossRef]
- Ma, C.; Wang, J.; Liu, H.; Chen, Y.; Ma, X.; Chen, S.; Chen, Y.; Bihl, J.I.; Yang, Y.I. Moderate Exercise Enhances Endothelial Progenitor Cell Exosomes Release and Function. Med. Sci. Sports Exerc. 2018, 50, 2024–2032. [Google Scholar] [CrossRef] [PubMed]
- Brahmer, A.; Neuberger, E.; Esch-Heisser, L.; Haller, N.; Jorgensen, M.M.; Baek, R.; Möbius, W.; Simon, P.; Krämer-Albers, E.-M. Platelets, Endothelial Cells and Leukocytes Contribute to the Exercise-Triggered Release of Extracellular Vesicles into the Circulation. J. Extracell. Vesicles 2019, 8, 1615820. [Google Scholar] [CrossRef]
- Brahmer, A.; Neuberger, E.W.I.; Simon, P.; Krämer-Albers, E.-M. Considerations for the Analysis of Small Extracellular Vesicles in Physical Exercise. Front. Physiol. 2020, 11, 576150. [Google Scholar] [CrossRef]
- Pedersen, L.; Idorn, M.; Olofsson, G.H.; Lauenborg, B.; Nookaew, I.; Hansen, R.H.; Johannesen, H.H.; Becker, J.C.; Pedersen, K.S.; Dethlefsen, C.; et al. Voluntary Running Suppresses Tumor Growth through Epinephrine- and IL-6-Dependent NK Cell Mobilization and Redistribution. Cell Metab. 2016, 23, 554–562. [Google Scholar] [CrossRef]
- Gannon, N.P.; Vaughan, R.A.; Garcia-Smith, R.; Bisoffi, M.; Trujillo, K.A. Effects of the Exercise-Inducible Myokine Irisin on Malignant and Non-Malignant Breast Epithelial Cell Behavior in Vitro. Int. J. Cancer 2015, 136, E197–202. [Google Scholar] [CrossRef] [PubMed]
- Hojman, P.; Dethlefsen, C.; Brandt, C.; Hansen, J.; Pedersen, L.; Pedersen, B.K. Exercise-Induced Muscle-Derived Cytokines Inhibit Mammary Cancer Cell Growth. Am. J. Physiol. Endocrinol. Metab. 2011, 301, E504–510. [Google Scholar] [CrossRef]
- Aoi, W.; Naito, Y.; Takagi, T.; Tanimura, Y.; Takanami, Y.; Kawai, Y.; Sakuma, K.; Hang, L.P.; Mizushima, K.; Hirai, Y.; et al. A Novel Myokine, Secreted Protein Acidic and Rich in Cysteine (SPARC), Suppresses Colon Tumorigenesis via Regular Exercise. Gut 2013, 62, 882–889. [Google Scholar] [CrossRef] [PubMed]
- Pulliero, A.; You, M.; Chaluvally-Raghavan, P.; Marengo, B.; Domenicotti, C.; Banelli, B.; Degan, P.; Molfetta, L.; Gianiorio, F.; Izzotti, A. Anticancer Effect of Physical Activity Is Mediated by Modulation of Extracellular MicroRNA in Blood. Oncotarget 2020, 11, 2106–2119. [Google Scholar] [CrossRef]
- Alizadeh, S.; Isanejad, A.; Sadighi, S.; Khalighfard, S.; Alizadeh, A.M. Effect of a High-Intensity Interval Training on Serum MicroRNA Levels in Women with Breast Cancer Undergoing Hormone Therapy. A Single-Blind Randomized Trial. Ann. Phys. Rehabil. Med. 2019, 62, 329–335. [Google Scholar] [CrossRef]
- Polakovičová, M.; Musil, P.; Laczo, E.; Hamar, D.; Kyselovič, J. Circulating MicroRNAs as Potential Biomarkers of Exercise Response. Int. J. Mol. Sci. 2016, 17, 1553. [Google Scholar] [CrossRef] [PubMed]
- Silva, G.J.J.; Bye, A.; El Azzouzi, H.; Wisløff, U. MicroRNAs as Important Regulators of Exercise Adaptation. Prog. Cardiovasc. Dis. 2017, 60, 130–151. [Google Scholar] [CrossRef] [PubMed]
- Georgantas, R.W.; Streicher, K.; Greenberg, S.A.; Greenlees, L.M.; Zhu, W.; Brohawn, P.Z.; Higgs, B.W.; Czapiga, M.; Morehouse, C.A.; Amato, A.; et al. Inhibition of Myogenic MicroRNAs 1, 133, and 206 by Inflammatory Cytokines Links Inflammation and Muscle Degeneration in Adult Inflammatory Myopathies: TNFα, Micrornas, and Muscle Pathology in Myositis. Arthritis Rheumatol. 2014, 66, 1022–1033. [Google Scholar] [CrossRef] [PubMed]
- Isanejad, A.; Alizadeh, A.M.; Amani Shalamzari, S.; Khodayari, H.; Khodayari, S.; Khori, V.; Khojastehnjad, N. MicroRNA-206, Let-7a and MicroRNA-21 Pathways Involved in the Anti-Angiogenesis Effects of the Interval Exercise Training and Hormone Therapy in Breast Cancer. Life Sci. 2016, 151, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Lazar, I.; Clement, E.; Dauvillier, S.; Milhas, D.; Ducoux-Petit, M.; LeGonidec, S.; Moro, C.; Soldan, V.; Dalle, S.; Balor, S.; et al. Adipocyte Exosomes Promote Melanoma Aggressiveness through Fatty Acid Oxidation: A Novel Mechanism Linking Obesity and Cancer. Cancer Res. 2016, 76, 4051–4057. [Google Scholar] [CrossRef]
- Clement, E.; Lazar, I.; Attané, C.; Carrié, L.; Dauvillier, S.; Ducoux-Petit, M.; Esteve, D.; Menneteau, T.; Moutahir, M.; Le Gonidec, S.; et al. Adipocyte Extracellular Vesicles Carry Enzymes and Fatty Acids That Stimulate Mitochondrial Metabolism and Remodeling in Tumor Cells. EMBO J. 2020, 39, e102525. [Google Scholar] [CrossRef]
- Lytle, N.K.; Barber, A.G.; Reya, T. Stem Cell Fate in Cancer Growth, Progression and Therapy Resistance. Nat. Rev. Cancer 2018, 18, 669–680. [Google Scholar] [CrossRef]
- Fittall, M.W.; Van Loo, P. Translating Insights into Tumor Evolution to Clinical Practice: Promises and Challenges. Genome Med. 2019, 11, 20. [Google Scholar] [CrossRef]
- McGranahan, N.; Swanton, C. Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future. Cell 2017, 168, 613–628. [Google Scholar] [CrossRef] [PubMed]
- Martincorena, I.; Campbell, P.J. Somatic Mutation in Cancer and Normal Cells. Science 2015, 349, 1483–1489. [Google Scholar] [CrossRef]
- Trinchieri, G. Cancer and Inflammation: An Old Intuition with Rapidly Evolving New Concepts. Annu. Rev. Immunol. 2012, 30, 677–706. [Google Scholar] [CrossRef] [PubMed]
- Landskron, G.; De la Fuente, M.; Thuwajit, P.; Thuwajit, C.; Hermoso, M.A. Chronic Inflammation and Cytokines in the Tumor Microenvironment. J. Immunol. Res. 2014, 2014, 149185. [Google Scholar] [CrossRef]
- Liu, J.; Wu, S.; Zheng, X.; Zheng, P.; Fu, Y.; Wu, C.; Lu, B.; Ju, J.; Jiang, J. Immune Suppressed Tumor Microenvironment by Exosomes Derived from Gastric Cancer Cells via Modulating Immune Functions. Sci. Rep. 2020, 10, 14749. [Google Scholar] [CrossRef]
- Karin, M. Nuclear Factor-KappaB in Cancer Development and Progression. Nature 2006, 441, 431–436. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Kortylewski, M.; Pardoll, D. Crosstalk between Cancer and Immune Cells: Role of STAT3 in the Tumour Microenvironment. Nat. Rev. Immunol. 2007, 7, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.P.; Hofseth, L.J.; Harris, C.C. Radical Causes of Cancer. Nat. Rev. Cancer 2003, 3, 276–285. [Google Scholar] [CrossRef]
- Söderberg, A.; Barral, A.M.; Söderström, M.; Sander, B.; Rosén, A. Redox-Signaling Transmitted in Trans to Neighboring Cells by Melanoma-Derived TNF-Containing Exosomes. Free Radic. Biol. Med. 2007, 43, 90–99. [Google Scholar] [CrossRef]
- Roccaro, A.M.; Sacco, A.; Maiso, P.; Azab, A.K.; Tai, Y.-T.; Reagan, M.; Azab, F.; Flores, L.M.; Campigotto, F.; Weller, E.; et al. BM Mesenchymal Stromal Cell-Derived Exosomes Facilitate Multiple Myeloma Progression. J. Clin. Investig. 2013, 123, 1542–1555. [Google Scholar] [CrossRef]
- Kucharzewska, P.; Christianson, H.C.; Welch, J.E.; Svensson, K.J.; Fredlund, E.; Ringnér, M.; Mörgelin, M.; Bourseau-Guilmain, E.; Bengzon, J.; Belting, M. Exosomes Reflect the Hypoxic Status of Glioma Cells and Mediate Hypoxia-Dependent Activation of Vascular Cells during Tumor Development. Proc. Natl. Acad. Sci. USA 2013, 110, 7312–7317. [Google Scholar] [CrossRef]
- Wang, Y.; Yi, J.; Chen, X.; Zhang, Y.; Xu, M.; Yang, Z. The Regulation of Cancer Cell Migration by Lung Cancer Cell-Derived Exosomes through TGF-β and IL-10. Oncol. Lett. 2016, 11, 1527–1530. [Google Scholar] [CrossRef] [PubMed]
- Schubert, A.; Boutros, M. Extracellular Vesicles and Oncogenic Signaling. Mol. Oncol. 2021, 15, 3–26. [Google Scholar] [CrossRef] [PubMed]
- Rong, L.; Li, R.; Li, S.; Luo, R. Immunosuppression of Breast Cancer Cells Mediated by Transforming Growth Factor-β in Exosomes from Cancer Cells. Oncol. Lett. 2016, 11, 500–504. [Google Scholar] [CrossRef] [PubMed]
- Umakoshi, M.; Takahashi, S.; Itoh, G.; Kuriyama, S.; Sasaki, Y.; Yanagihara, K.; Yashiro, M.; Maeda, D.; Goto, A.; Tanaka, M. Macrophage-Mediated Transfer of Cancer-Derived Components to Stromal Cells Contributes to Establishment of a pro-Tumor Microenvironment. Oncogene 2019, 38, 2162–2176. [Google Scholar] [CrossRef] [PubMed]
- Cha, J.-H.; Chan, L.-C.; Li, C.-W.; Hsu, J.L.; Hung, M.-C. Mechanisms Controlling PD-L1 Expression in Cancer. Mol. Cell 2019, 76, 359–370. [Google Scholar] [CrossRef]
- Garcia-Diaz, A.; Shin, D.S.; Moreno, B.H.; Saco, J.; Escuin-Ordinas, H.; Rodriguez, G.A.; Zaretsky, J.M.; Sun, L.; Hugo, W.; Wang, X.; et al. Interferon Receptor Signaling Pathways Regulating PD-L1 and PD-L2 Expression. Cell Rep. 2017, 19, 1189–1201. [Google Scholar] [CrossRef]
- Keir, M.E.; Liang, S.C.; Guleria, I.; Latchman, Y.E.; Qipo, A.; Albacker, L.A.; Koulmanda, M.; Freeman, G.J.; Sayegh, M.H.; Sharpe, A.H. Tissue Expression of PD-L1 Mediates Peripheral T Cell Tolerance. J. Exp. Med. 2006, 203, 883–895. [Google Scholar] [CrossRef]
- Dong, H.; Strome, S.E.; Salomao, D.R.; Tamura, H.; Hirano, F.; Flies, D.B.; Roche, P.C.; Lu, J.; Zhu, G.; Tamada, K.; et al. Tumor-Associated B7-H1 Promotes T-Cell Apoptosis: A Potential Mechanism of Immune Evasion. Nat. Med. 2002, 8, 793–800. [Google Scholar] [CrossRef] [PubMed]
- Schildberg, F.A.; Klein, S.R.; Freeman, G.J.; Sharpe, A.H. Coinhibitory Pathways in the B7-CD28 Ligand-Receptor Family. Immunity 2016, 44, 955–972. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Han, X. Anti-PD-1/PD-L1 Therapy of Human Cancer: Past, Present, and Future. J. Clin. Investig. 2015, 125, 3384–3391. [Google Scholar] [CrossRef]
- Powles, T.; Eder, J.P.; Fine, G.D.; Braiteh, F.S.; Loriot, Y.; Cruz, C.; Bellmunt, J.; Burris, H.A.; Petrylak, D.P.; Teng, S.; et al. MPDL3280A (Anti-PD-L1) Treatment Leads to Clinical Activity in Metastatic Bladder Cancer. Nature 2014, 515, 558–562. [Google Scholar] [CrossRef]
- Robert, C.; Long, G.V.; Brady, B.; Dutriaux, C.; Maio, M.; Mortier, L.; Hassel, J.C.; Rutkowski, P.; McNeil, C.; Kalinka-Warzocha, E.; et al. Nivolumab in Previously Untreated Melanoma without BRAF Mutation. N. Engl. J. Med. 2015, 372, 320–330. [Google Scholar] [CrossRef] [PubMed]
- Ricklefs, F.L.; Alayo, Q.; Krenzlin, H.; Mahmoud, A.B.; Speranza, M.C.; Nakashima, H.; Hayes, J.L.; Lee, K.; Balaj, L.; Passaro, C.; et al. Immune Evasion Mediated by PD-L1 on Glioblastoma-Derived Extracellular Vesicles. Sci. Adv. 2018, 4, eaar2766. [Google Scholar] [CrossRef]
- Fan, Y.; Che, X.; Qu, J.; Hou, K.; Wen, T.; Li, Z.; Li, C.; Wang, S.; Xu, L.; Liu, Y.; et al. Exosomal PD-L1 Retains Immunosuppressive Activity and Is Associated with Gastric Cancer Prognosis. Ann. Surg. Oncol. 2019, 26, 3745–3755. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Kim, H.; Choi, Y.J.; Kim, S.Y.; Lee, J.-E.; Sung, K.J.; Sung, Y.H.; Pack, C.-G.; Jung, M.-K.; Han, B.; et al. Exosomal PD-L1 Promotes Tumor Growth through Immune Escape in Non-Small Cell Lung Cancer. Exp. Mol. Med. 2019, 51, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, C.-W.; Chan, L.-C.; Wei, Y.; Hsu, J.-M.; Xia, W.; Cha, J.-H.; Hou, J.; Hsu, J.L.; Sun, L.; et al. Exosomal PD-L1 Harbors Active Defense Function to Suppress T Cell Killing of Breast Cancer Cells and Promote Tumor Growth. Cell Res. 2018, 28, 862–864. [Google Scholar] [CrossRef]
- Allavena, P.; Sica, A.; Garlanda, C.; Mantovani, A. The Yin-Yang of Tumor-Associated Macrophages in Neoplastic Progression and Immune Surveillance. Immunol. Rev. 2008, 222, 155–161. [Google Scholar] [CrossRef]
- Saccani, A.; Schioppa, T.; Porta, C.; Biswas, S.K.; Nebuloni, M.; Vago, L.; Bottazzi, B.; Colombo, M.P.; Mantovani, A.; Sica, A. P50 Nuclear Factor-KappaB Overexpression in Tumor-Associated Macrophages Inhibits M1 Inflammatory Responses and Antitumor Resistance. Cancer Res. 2006, 66, 11432–11440. [Google Scholar] [CrossRef]
- Hagemann, T.; Lawrence, T.; McNeish, I.; Charles, K.A.; Kulbe, H.; Thompson, R.G.; Robinson, S.C.; Balkwill, F.R. “Re-Educating” Tumor-Associated Macrophages by Targeting NF-KappaB. J. Exp. Med. 2008, 205, 1261–1268. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Liu, C.; Bi, Z.-Y.; Zhou, Q.; Zhang, H.; Li, L.-L.; Zhang, J.; Zhu, W.; Song, Y.-Y.-Y.; Zhang, F.; et al. Comprehensive Landscape of Extracellular Vesicle-Derived RNAs in Cancer Initiation, Progression, Metastasis and Cancer Immunology. Mol. Cancer 2020, 19, 102. [Google Scholar] [CrossRef] [PubMed]
- Gajewski, T.F. The Next Hurdle in Cancer Immunotherapy: Overcoming the Non-T-Cell-Inflamed Tumor Microenvironment. Semin. Oncol. 2015, 42, 663–671. [Google Scholar] [CrossRef] [PubMed]
- Spranger, S.; Bao, R.; Gajewski, T.F. Melanoma-Intrinsic β-Catenin Signalling Prevents Anti-Tumour Immunity. Nature 2015, 523, 231–235. [Google Scholar] [CrossRef]
- Spranger, S.; Spaapen, R.M.; Zha, Y.; Williams, J.; Meng, Y.; Ha, T.T.; Gajewski, T.F. Up-Regulation of PD-L1, IDO, and T(Regs) in the Melanoma Tumor Microenvironment Is Driven by CD8(+) T Cells. Sci. Transl. Med. 2013, 5, 200ra116. [Google Scholar] [CrossRef] [PubMed]
- Zemek, R.M.; Chin, W.L.; Nowak, A.K.; Millward, M.J.; Lake, R.A.; Lesterhuis, W.J. Sensitizing the Tumor Microenvironment to Immune Checkpoint Therapy. Front. Immunol. 2020, 11, 223. [Google Scholar] [CrossRef]
- Woo, S.-R.; Fuertes, M.B.; Corrales, L.; Spranger, S.; Furdyna, M.J.; Leung, M.Y.K.; Duggan, R.; Wang, Y.; Barber, G.N.; Fitzgerald, K.A.; et al. STING-Dependent Cytosolic DNA Sensing Mediates Innate Immune Recognition of Immunogenic Tumors. Immunity 2014, 41, 830–842. [Google Scholar] [CrossRef]
- Kitai, Y.; Kawasaki, T.; Sueyoshi, T.; Kobiyama, K.; Ishii, K.J.; Zou, J.; Akira, S.; Matsuda, T.; Kawai, T. DNA-Containing Exosomes Derived from Cancer Cells Treated with Topotecan Activate a STING-Dependent Pathway and Reinforce Antitumor Immunity. J. Immunol. 2017, 198, 1649–1659. [Google Scholar] [CrossRef]
- Diamond, J.M.; Vanpouille-Box, C.; Spada, S.; Rudqvist, N.-P.; Chapman, J.R.; Ueberheide, B.M.; Pilones, K.A.; Sarfraz, Y.; Formenti, S.C.; Demaria, S. Exosomes Shuttle TREX1-Sensitive IFN-Stimulatory DsDNA from Irradiated Cancer Cells to DCs. Cancer Immunol. Res. 2018, 6, 910–920. [Google Scholar] [CrossRef] [PubMed]
- Dutta, S.; Warshall, C.; Bandyopadhyay, C.; Dutta, D.; Chandran, B. Interactions between Exosomes from Breast Cancer Cells and Primary Mammary Epithelial Cells Leads to Generation of Reactive Oxygen Species Which Induce DNA Damage Response, Stabilization of P53 and Autophagy in Epithelial Cells. PLoS ONE 2014, 9, e97580. [Google Scholar] [CrossRef]
- Hornung, V.; Ablasser, A.; Charrel-Dennis, M.; Bauernfeind, F.; Horvath, G.; Caffrey, D.R.; Latz, E.; Fitzgerald, K.A. AIM2 Recognizes Cytosolic DsDNA and Forms a Caspase-1-Activating Inflammasome with ASC. Nature 2009, 458, 514–518. [Google Scholar] [CrossRef]
- Lian, Q.; Xu, J.; Yan, S.; Huang, M.; Ding, H.; Sun, X.; Bi, A.; Ding, J.; Sun, B.; Geng, M. Chemotherapy-Induced Intestinal Inflammatory Responses Are Mediated by Exosome Secretion of Double-Strand DNA via AIM2 Inflammasome Activation. Cell Res. 2017, 27, 784–800. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, S.; Pfeuffer, S.; Arnold, P.; Treitz, C.; Aden, K.; Ebsen, H.; Falk-Paulsen, M.; Gisch, N.; Fazio, A.; Kuiper, J.; et al. Prdx4 Limits Caspase-1 Activation and Restricts Inflammasome-mediated Signaling by Extracellular Vesicles. EMBO J. 2019, 38. [Google Scholar] [CrossRef]
- Papademetrio, D.L.; Lompardía, S.L.; Simunovich, T.; Costantino, S.; Mihalez, C.Y.; Cavaliere, V.; Álvarez, É. Inhibition of Survival Pathways MAPK and NF-KB Triggers Apoptosis in Pancreatic Ductal Adenocarcinoma Cells via Suppression of Autophagy. Target. Oncol. 2016, 11, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Verzella, D.; Pescatore, A.; Capece, D.; Vecchiotti, D.; Ursini, M.V.; Franzoso, G.; Alesse, E.; Zazzeroni, F. Life, Death, and Autophagy in Cancer: NF-ΚB Turns up Everywhere. Cell Death Dis. 2020, 11, 210. [Google Scholar] [CrossRef] [PubMed]
- Kaplanov, I.; Carmi, Y.; Kornetsky, R.; Shemesh, A.; Shurin, G.V.; Shurin, M.R.; Dinarello, C.A.; Voronov, E.; Apte, R.N. Blocking IL-1β Reverses the Immunosuppression in Mouse Breast Cancer and Synergizes with Anti-PD-1 for Tumor Abrogation. Proc. Natl. Acad. Sci. USA 2019, 116, 1361–1369. [Google Scholar] [CrossRef] [PubMed]
- Theivanthiran, B.; Evans, K.S.; DeVito, N.C.; Plebanek, M.; Sturdivant, M.; Wachsmuth, L.P.; Salama, A.K.; Kang, Y.; Hsu, D.; Balko, J.M.; et al. A Tumor-Intrinsic PD-L1/NLRP3 Inflammasome Signaling Pathway Drives Resistance to Anti-PD-1 Immunotherapy. J. Clin. Investig. 2020, 130, 2570–2586. [Google Scholar] [CrossRef]
- von Bubnoff, N. Liquid Biopsy: Approaches to Dynamic Genotyping in Cancer. Oncol. Res. Treat. 2017, 40, 409–416. [Google Scholar] [CrossRef]
- Bardelli, A.; Pantel, K. Liquid Biopsies, What We Do Not Know (Yet). Cancer Cell 2017, 31, 172–179. [Google Scholar] [CrossRef]
- Schatz, S.; Falk, M.; Jóri, B.; Ramdani, H.O.; Schmidt, S.; Willing, E.-M.; Menon, R.; Groen, H.J.M.; Diehl, L.; Kröger, M.; et al. Integration of Tumor Mutation Burden and PD-L1 Testing in Routine Laboratory Diagnostics in Non-Small Cell Lung Cancer. Cancers 2020, 12, 1685. [Google Scholar] [CrossRef]
- Marrugo-Ramírez, J.; Mir, M.; Samitier, J. Blood-Based Cancer Biomarkers in Liquid Biopsy: A Promising Non-Invasive Alternative to Tissue Biopsy. Int. J. Mol. Sci. 2018, 19, 2877. [Google Scholar] [CrossRef]
- Kahlert, C.; Melo, S.A.; Protopopov, A.; Tang, J.; Seth, S.; Koch, M.; Zhang, J.; Weitz, J.; Chin, L.; Futreal, A.; et al. Identification of Double-Stranded Genomic DNA Spanning All Chromosomes with Mutated KRAS and P53 DNA in the Serum Exosomes of Patients with Pancreatic Cancer. J. Biol. Chem. 2014, 289, 3869–3875. [Google Scholar] [CrossRef]
- Thakur, B.K.; Zhang, H.; Becker, A.; Matei, I.; Huang, Y.; Costa-Silva, B.; Zheng, Y.; Hoshino, A.; Brazier, H.; Xiang, J.; et al. Double-Stranded DNA in Exosomes: A Novel Biomarker in Cancer Detection. Cell Res. 2014, 24, 766–769. [Google Scholar] [CrossRef] [PubMed]
- Klump, J.; Phillipp, U.; Follo, M.; Eremin, A.; Lehmann, H.; Nestel, S.; von Bubnoff, N.; Nazarenko, I. Extracellular Vesicles or Free Circulating DNA: Where to Search for BRAF and CKIT Mutations? Nanomed. Nanotechnol. Biol. Med. 2018, 14, 875–882. [Google Scholar] [CrossRef] [PubMed]
- Krug, A.K.; Enderle, D.; Karlovich, C.; Priewasser, T.; Bentink, S.; Spiel, A.; Brinkmann, K.; Emenegger, J.; Grimm, D.G.; Castellanos-Rizaldos, E.; et al. Improved EGFR Mutation Detection Using Combined Exosomal RNA and Circulating Tumor DNA in NSCLC Patient Plasma. Ann. Oncol. 2018, 29, 700–706. [Google Scholar] [CrossRef] [PubMed]
- Theodoraki, M.-N.; Yerneni, S.S.; Hoffmann, T.K.; Gooding, W.E.; Whiteside, T.L. Clinical Significance of PD-L1 + Exosomes in Plasma of Head and Neck Cancer Patients. Clin. Cancer Res. 2018, 24, 896–905. [Google Scholar] [CrossRef]
- Lux, A.; Kahlert, C.; Grützmann, R.; Pilarsky, C. C-Met and PD-L1 on Circulating Exosomes as Diagnostic and Prognostic Markers for Pancreatic Cancer. Int. J. Mol. Sci. 2019, 20, 3305. [Google Scholar] [CrossRef]
- Huang, M.; Yang, J.; Wang, T.; Song, J.; Xia, J.; Wu, L.; Wang, W.; Wu, Q.; Zhu, Z.; Song, Y.; et al. Homogeneous, Low-volume, Efficient, and Sensitive Quantitation of Circulating Exosomal PD-L1 for Cancer Diagnosis and Immunotherapy Response Prediction. Angew. Chem. Int. Ed. 2020, 59, 4800–4805. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lipinski, S.; Tiemann, K. Extracellular Vesicles and Their Role in the Spatial and Temporal Expansion of Tumor–Immune Interactions. Int. J. Mol. Sci. 2021, 22, 3374. https://doi.org/10.3390/ijms22073374
Lipinski S, Tiemann K. Extracellular Vesicles and Their Role in the Spatial and Temporal Expansion of Tumor–Immune Interactions. International Journal of Molecular Sciences. 2021; 22(7):3374. https://doi.org/10.3390/ijms22073374
Chicago/Turabian StyleLipinski, Simone, and Katharina Tiemann. 2021. "Extracellular Vesicles and Their Role in the Spatial and Temporal Expansion of Tumor–Immune Interactions" International Journal of Molecular Sciences 22, no. 7: 3374. https://doi.org/10.3390/ijms22073374
APA StyleLipinski, S., & Tiemann, K. (2021). Extracellular Vesicles and Their Role in the Spatial and Temporal Expansion of Tumor–Immune Interactions. International Journal of Molecular Sciences, 22(7), 3374. https://doi.org/10.3390/ijms22073374




