On the Origins of Homology Directed Repair in Mammalian Cells
Abstract
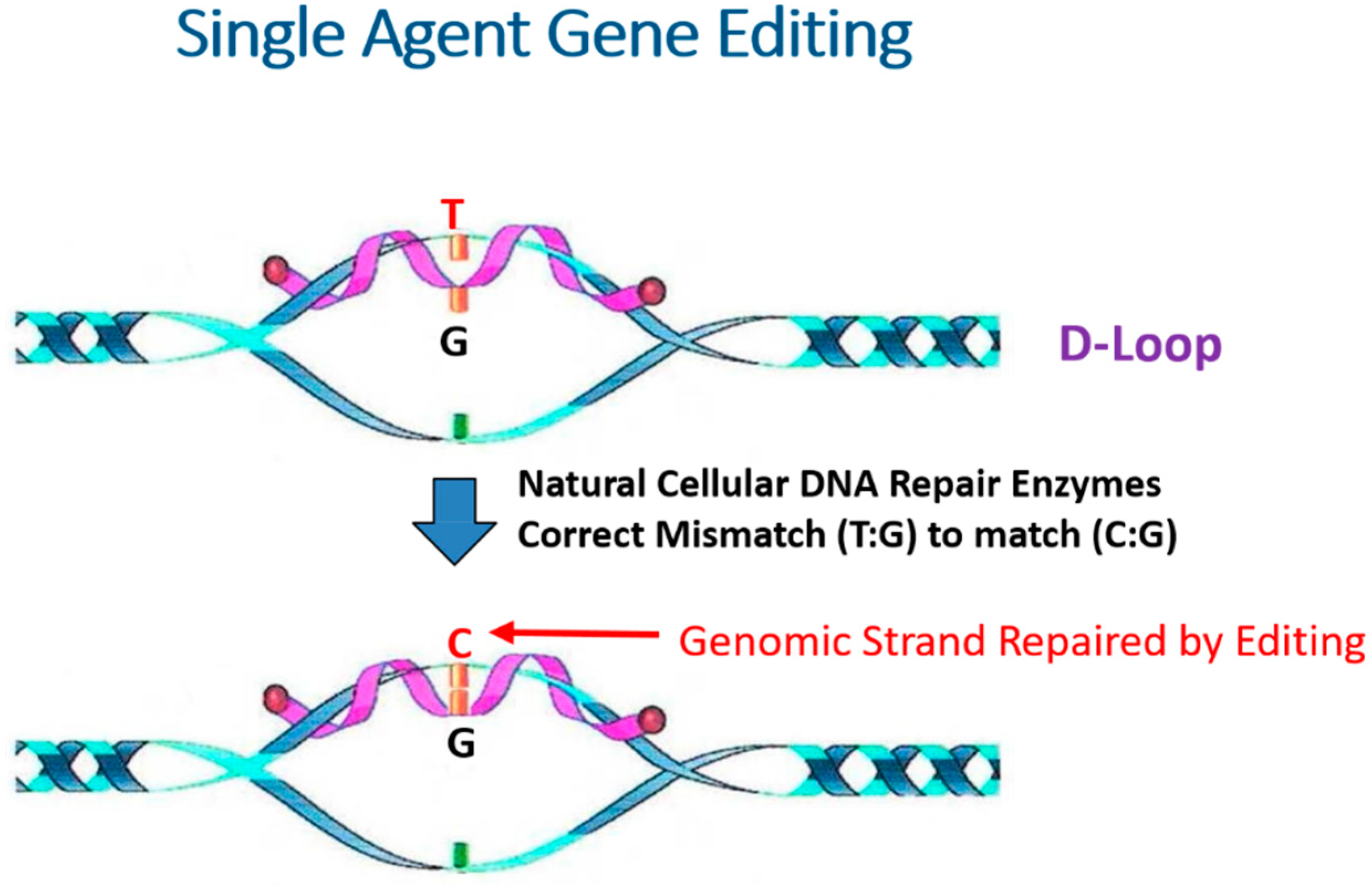
1. Introduction
2. Improving Efficiency by Modifying the Donor DNA
3. Enhancing the Frequency of Single Agent Gene Editing by Changing the Cellular Environment
4. Enhancing the Frequency of Single Agent Gene Editing Using Double-Strand DNA Breakage
5. The Cellular Cost of Genome Engineering
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Engstrom, J.U.; Kmiec, E.B. DNA Replication, cell cycle progression and the targeted gene repair reaction. Cell Cycle 2008, 7, 1402–1414. [Google Scholar] [CrossRef]
- Parekh-Olmedo, H.; Ferrara, L.; Brachman, E.; Kmiec, E.B. Gene therapy progress and prospects: Targeted gene repair. Gene Ther. 2005, 12, 639–646. [Google Scholar] [CrossRef]
- Cole-Strauss, A.; Gamper, H.; Holloman, W.K.; Muñoz, M.; Cheng, N.; Kmiec, E.B. Targeted gene repair directed by the chimeric RNA/DNA oligonucleotide in a mammalian cell-free extract. Nucleic Acids Res. 1999, 27, 1323–1330. [Google Scholar] [CrossRef] [PubMed]
- Mandecki, W. Oligonucleotide-directed double-strand break repair in plasmids of Escherichia coli: A method for site-specific mutagenesis. Proc. Natl. Acad. Sci. USA 1986, 83, 7177–7181. [Google Scholar] [CrossRef]
- Moerschell, R.P.; Tsunasawa, S.; Sherman, F. Transformation of yeast with synthetic oligonucleotides. Proc. Natl. Acad. Sci. USA 1988, 85, 524–528. [Google Scholar] [CrossRef]
- Yamamoto, T.; Moerschell, R.P.; Wakem, L.P.; Komar-Panicucci, S.; Sherman, F. Strand-specificity in the transformation of yeast with synthetic oligonucleotides. Genetics 1992, 131, 811–819. [Google Scholar] [CrossRef]
- Campbell, C.R.; Keown, W.; Lowe, L.; Kirschling, D.; Kucherlapati, R. Homologous recombination involving small single-stranded oligonucleotides in human cells. New Biol. 1989, 1, 223–227. [Google Scholar]
- Symington, L.S. End resection at double-strand breaks: Mechanism and regulation. Cold Spring Harb. Perspect. Biol. 2014, 6. [Google Scholar] [CrossRef] [PubMed]
- Kotani, H.; Kmiec, E.B. A role for RNA synthesis in homologous pairing events. Mol. Cell. Biol. 1994, 14, 6097–6106. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Culver, K.W.; Hsieh, W.T.; Huyen, Y.; Chen, V.; Liu, J.; Khripine, Y.; Khorlin, A. Correction of chromosomal point mutations in human cells with bifunctional oligonucleotides. Nat. Biotechnol. 1999, 17, 989–993. [Google Scholar] [CrossRef] [PubMed]
- Rogers, F.A.; Vasquez, K.M.; Egholm, M.; Glazer, P.M. Site-directed recombination via bifunctional PNA-DNA conjugates. Proc. Natl. Acad. Sci. USA 2002, 99, 16695–16700. [Google Scholar] [CrossRef] [PubMed]
- Chan, P.P.; Lin, M.; Fawad Faruqi, A.; Powell, J.; Seidman, M.M.; Glazer, P.M. Targeted correction of an episomal gene in mammalian cells by a short DNA fragment tethered to a triplex-forming oligonucleotide. J. Biol. Chem. 1999, 274, 11541–11548. [Google Scholar] [CrossRef]
- McNeer, N.A.; Anandalingam, K.; Fields, R.J.; Caputo, C.; Kopic, S.; Gupta, A.; Quijano, E.; Polikoff, L.; Kong, Y.; Bahal, R.; et al. Nanoparticles that deliver triplex-forming peptide nucleic acid molecules correct F508del CFTR in airway epithelium. Nat. Commun. 2015, 6, 6952. [Google Scholar] [CrossRef] [PubMed]
- Gamper, H.B.; Parekh, H.; Rice, M.C.; Bruner, M.; Youkey, H.; Kmiec, E.B. The DNA strand of chimeric RNA/DNA oligonucleotides can direct gene repair/conversion activity in mammalian and plant cell-free extracts. Nucleic Acids Res. 2000, 28, 4332–4339. [Google Scholar] [CrossRef]
- Gamper, H. A plausible mechanism for gene correction by chimeric oligonucleotides. Biochemistry 2000, 39, 5808–5816. [Google Scholar] [CrossRef]
- Drury, M.D.; Kmiec, E.B. Double displacement loops (double d-loops) are templates for oligonucleotide-directed mutagenesis and gene repair. Oligonucleotides 2004, 14, 274–286. [Google Scholar] [CrossRef] [PubMed]
- Aarts, M.; Te Riele, H. Progress and prospects: Oligonucleotide-directed gene modification in mouse embryonic stem cells: A route to therapeutic application. Gene Ther. 2011, 18, 213–219. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Olsen, P.A.; Randol, M.; Krauss, S. Implications of cell cycle progression on functional sequence correction by short single-stranded DNA oligonucleotides. Gene Ther. 2005, 12, 546–551. [Google Scholar] [CrossRef] [PubMed]
- EE Brachman, E.K.; Brachman, E.E.; Kmiec, E.B. Gene repair in mammalian cells is stimulated by the elongation of S phase and transient stalling of replication forks. DNA Repair 2005, 4, 445–457. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef]
- Bialk, P.; Rivera-Torres, N.; Strouse, B.; Kmiec, E.B. Regulation of Gene Editing Activity Directed by Single-Stranded Oligonucleotides and CRISPR/Cas9 Systems. PLoS ONE 2015, 10, e0129308. [Google Scholar] [CrossRef]
- Schumann, K.; Lin, S.; Boyer, E.; Simeonov, D.R.; Subramaniam, M.; Gate, R.E.; Haliburton, G.E.; Ye, C.J.; Bluestone, J.A.; Doudna, J.A.; et al. Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proc. Natl. Acad. Sci. USA 2015, 112, 10437–10442. [Google Scholar] [CrossRef]
- Brandsma, I.; Gent, D.C. Pathway choice in DNA double strand break repair: Observations of a balancing act. Genome Integr. 2012, 3, 9. [Google Scholar] [CrossRef]
- Rodgers, K.; McVey, M. Error-prone repair of DNA double-strand breaks. J. Cell. Physiol. 2016, 231, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Helleday, T.; Lo, J.; van Gent, D.C.; Engelward, B.P. DNA double-strand break repair: From mechanistic understanding to cancer treatment. DNA Repair 2007, 6, 923–935. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.H.Y.; Pannunzio, N.R.; Adachi, N.; Lieber, M.R. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 2017, 18, 495–506. [Google Scholar] [CrossRef]
- Wang, H.; Xu, X. Microhomology-mediated end joining: New players join the team. Cell Biosci. 2017, 7, 6. [Google Scholar] [CrossRef]
- Ferrara, L.; Engstrom, J.U.; Schwartz, T.; Parekh-Olmedo, H.; Kmiec, E.B. Recovery of cell cycle delay following targeted gene repair by oligonucleotides. DNA Repair 2007, 6, 1529–1535. [Google Scholar] [CrossRef]
- Wu, X.S.; Xin, L.; Yin, W.X.; Shang, X.Y.; Lu, L.; Watt, R.M.; Cheah, K.S.E.; Huang, J.D.; Liu, D.P.; Liang, C.C. Increased efficiency of oligonucleotide-mediated gene repair through slowing replication fork progression. Proc. Natl. Acad. Sci. USA 2005, 102, 2508–2513. [Google Scholar] [CrossRef] [PubMed]
- Engstrom, J.U.; Suzuki, T.; Kmiec, E.B. Regulation of targeted gene repair by intrinsic cellular processes. 2BioEssays 2009, 31, 159–168. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sansbury, B.M.; Kmiec, E.B. On the Origins of Homology Directed Repair in Mammalian Cells. Int. J. Mol. Sci. 2021, 22, 3348. https://doi.org/10.3390/ijms22073348
Sansbury BM, Kmiec EB. On the Origins of Homology Directed Repair in Mammalian Cells. International Journal of Molecular Sciences. 2021; 22(7):3348. https://doi.org/10.3390/ijms22073348
Chicago/Turabian StyleSansbury, Brett M., and Eric B. Kmiec. 2021. "On the Origins of Homology Directed Repair in Mammalian Cells" International Journal of Molecular Sciences 22, no. 7: 3348. https://doi.org/10.3390/ijms22073348
APA StyleSansbury, B. M., & Kmiec, E. B. (2021). On the Origins of Homology Directed Repair in Mammalian Cells. International Journal of Molecular Sciences, 22(7), 3348. https://doi.org/10.3390/ijms22073348





