1. Introduction
Viral capsids are natural nanostructures that evolved towards the efficient delivery of viral genetic material to host cells. Their nanoscale dimensions, presence of an interior cavity, self-organization abilities and responsiveness to environmental conditions are desired features in terms of nanotechnology. Additional attributes of viral capsids, including homogeneity, susceptibility to chemical modification and, in the case of plant viruses, lack of pathogenic activity to humans, give them potential as biologically active compound carriers in the biomedical field. The reports on potential applications of viral capsids in nanotechnology and biomedical fields has been thoroughly reviewed [
1,
2,
3,
4]. The key to the utilization of viral capsids is understanding and controlling their assembly and disassembly processes.
Brome mosaic virus (BMV) is one of the most extensively studied plant viruses. Its structure and biology are well known, and numerous reports describe the potential applications of BMV as a vehicle for nanoparticle delivery. BMV is a typical representative plant virus with a genome composed of single-stranded RNA and nonenveloped spherical virions. BMV belongs to the Bromovirus genus. The BMV capsid is composed of 180 capsid protein (CP) subunits with a weight of 20 kDa. The virion has an approximately 28 nm diameter, depending on the surrounding conditions, and it shows T = 3 icosahedral symmetry [
5]. The cargo space inside the virion is limited by the internal cage with the T = 1 triangulation number. CP consists of 189 amino acids and has a globular shape with a protruding N-terminal arginine-rich motif (ARM) directed into the interior of the capsid. The ARM, due to its positive charge, interacts with negatively charged RNA and therefore is crucial for capsid assembly.
Capsid assembly is currently being extensively analyzed, and some general rules of this process have already been determined. The formation of capsid depends on the temperature, pH, ionic strength, CP concentration and features of the encapsidated core molecules [
6,
7,
8]. Due to the recent development of experimental and computational techniques in the field of physical virology, the static descriptions of viral particles have been replaced by dynamic models based on soft-mode dynamics and kinetic trapping [
9]. Still, the knowledge of the capsid assembly is not sufficient to control it or rationally design VLPs with desired features. The disassembly and reassembly of BMV virions containing their native RNA or RNA derived from another virus is possible in the course of dialysis, starting from a high-salt to a low-salt buffer at neutral pH [
10,
11,
12]. Under natural conditions, BMV capsids as well as those of similar viruses, such as cowpea chlorotic mottle virus (CCMV), begin forming via the nucleation process, driven by electrostatic interactions between CP dimers and the viral genome. The assembly of complete virions is further continued mostly due to interactions among CPs [
13]. The key to BMV virion assembly is the approximately 200-nucleotide-long 3′ end of the genomic RNA, which forms a tRNA-like structure (TLS). The importance of TLSs was initially shown by Choi et al. in an experiment in which BMV capsid assembly attempted upon RNA with truncated TLSs failed, while the addition of yeast tRNA promoted virion formation [
14]. Later, it was shown that not only RNA but also other materials, such as gold nanoparticles, quantum dots [
15], and even cubic-shaped nanoparticles [
16], may be encapsulated in the BMV capsid. The primary requirement for in vitro encapsidation of any type of cargo is the negative charge forming electrostatic interactions with the positively charged N-terminal portion of the BMV CP. Under in vitro conditions, these interactions are heavily dependent on the ionic strength of the buffer and as such can be modulated [
6,
13,
17].
The capsid assembly process is driven not only by CP-cargo interactions but also by interactions among the CPs. The importance of such interactions can clearly be seen in the formation of CP dimers, which are the basic building blocks for capsid structures. CP dimers attach to each other, creating larger protein complexes and leading to full capsid formation [
7]. The interactions among CPs are based on hydrophobic attractions and hydrophilic repulsions, in contrast to the interactions between CP and cargo [
13]. Assembly of BMV-derived empty capsids is pH-dependent and, according to molecular dynamics and dynamic light scattering (DLS) analysis, is initiated by the formation of CP trimer nuclei at pH close to 5 [
18].
Considering the utilization of plant viral capsids as biologically active compounds vehicles, attempts are made to obtain VLPs showing desired features. The easiest way to modify VLPs is to introduce mutations into the CP sequence. However, during viral replication in native host plants, most CP mutations result in the loss of viral infectivity or are reversed to the wild-type sequence [
19]. This phenomenon is related to error-prone RNA replication, RNA recombination and subsequent selective pressure, which eliminates viral variants with lower fitness compared to the wild type [
19,
20,
21,
22]. To overcome this impediment, heterologous expression systems are applied that offer CP production without selective pressure working on protein mutants.
In this paper, we describe the in vitro assembly of VLPs with the application of recombinant BMV CP (rCP) produced in a bacterial expression system. We used the rCP to study the impact of environmental factors and encapsulated cargo on the assembly process and properties of the resulting VLPs. In our assay, we applied a spectrum of methods, including protein crosslinking, electrophoresis, microscale thermophoresis (MST), cryogenic transmission electron microscopy (cryo-TEM), atomic force microscopy (AFM) and dynamic light scattering (DLS). We also produced and studied a CP mutant designed to slightly weaken the CP-CP interaction. Our results show that pH is a crucial factor influencing CP-CP interactions, but we also noticed the significant influence of ionic strength. We also observed that the physicochemical parameters of VLPs, such as diameter, polydispersity and thermal stability, are strongly influenced by the cargo. Interestingly, CP carrying mutations formed VLPs showing a wide range of possible diameters compared to the wild-type VLPs.
3. Discussion
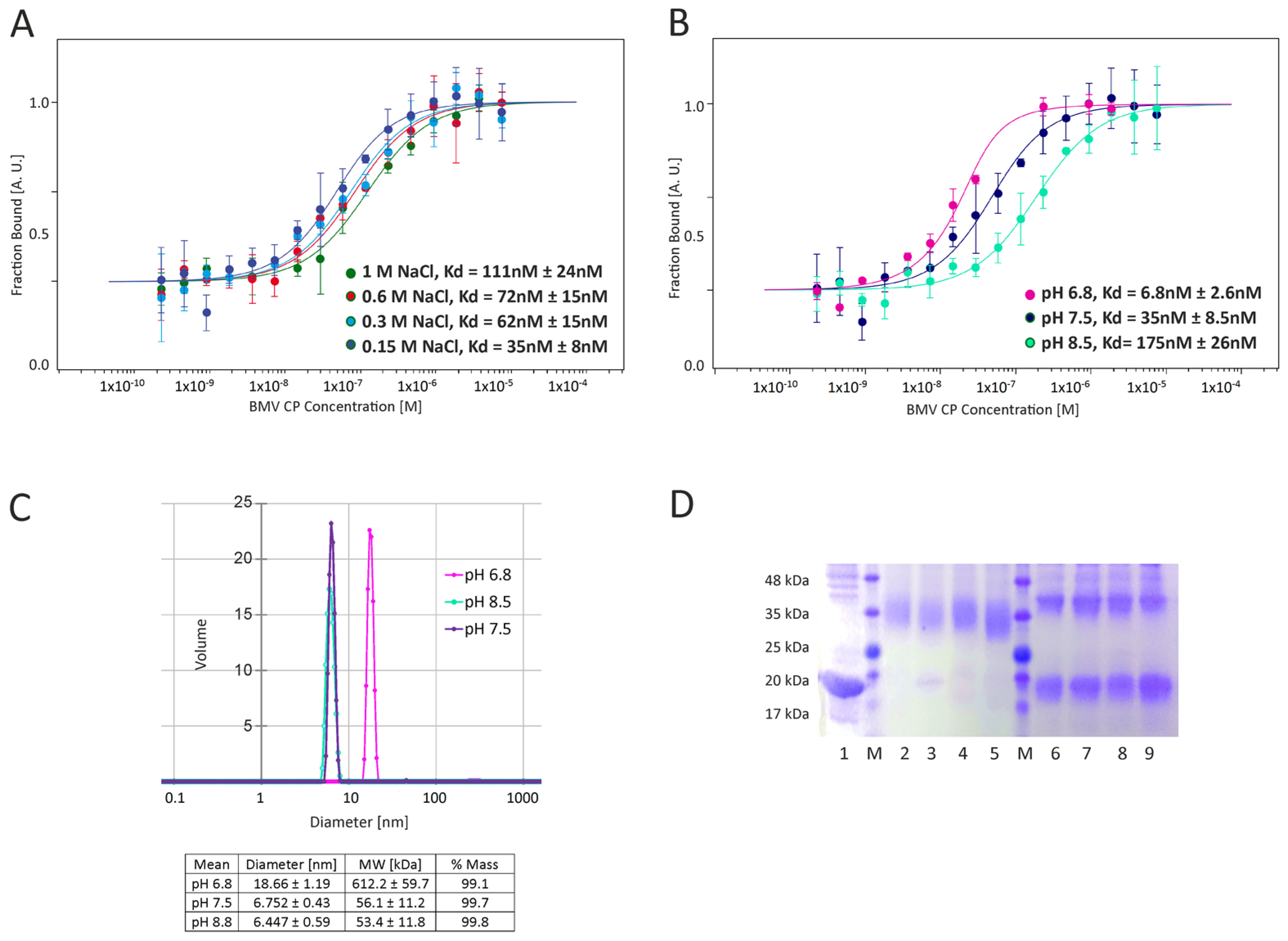
There are two main types of interactions important for BMV capsid formation: CP-CP and RNA-CP. The lack of empty capsids in BMV-infected plants indicates that in vivo, RNA-CP interactions are pivotal for the formation of viral particles [
25]. However, CP-CP interactions are sufficient to form BMV-based VLPs in vitro [
26]. Our analysis showed that strong CP-CP binding depends on pH. We noticed that at the lowest tested pH of 6.8, rCPs tended to form an oligomeric structure, most likely rCP trimers proposed to initiate the nucleation process leading to capsid formation [
18] (
Figure 1). This result is consistent with the observation of Chevreuil and coworkers that a decrease in pH leads to stronger interactions between CPs of CCMV and more compact VLP formation [
24]. Additionally, van Eldijk and coworkers showed that changing the pH can control empty CCMV capsid formation [
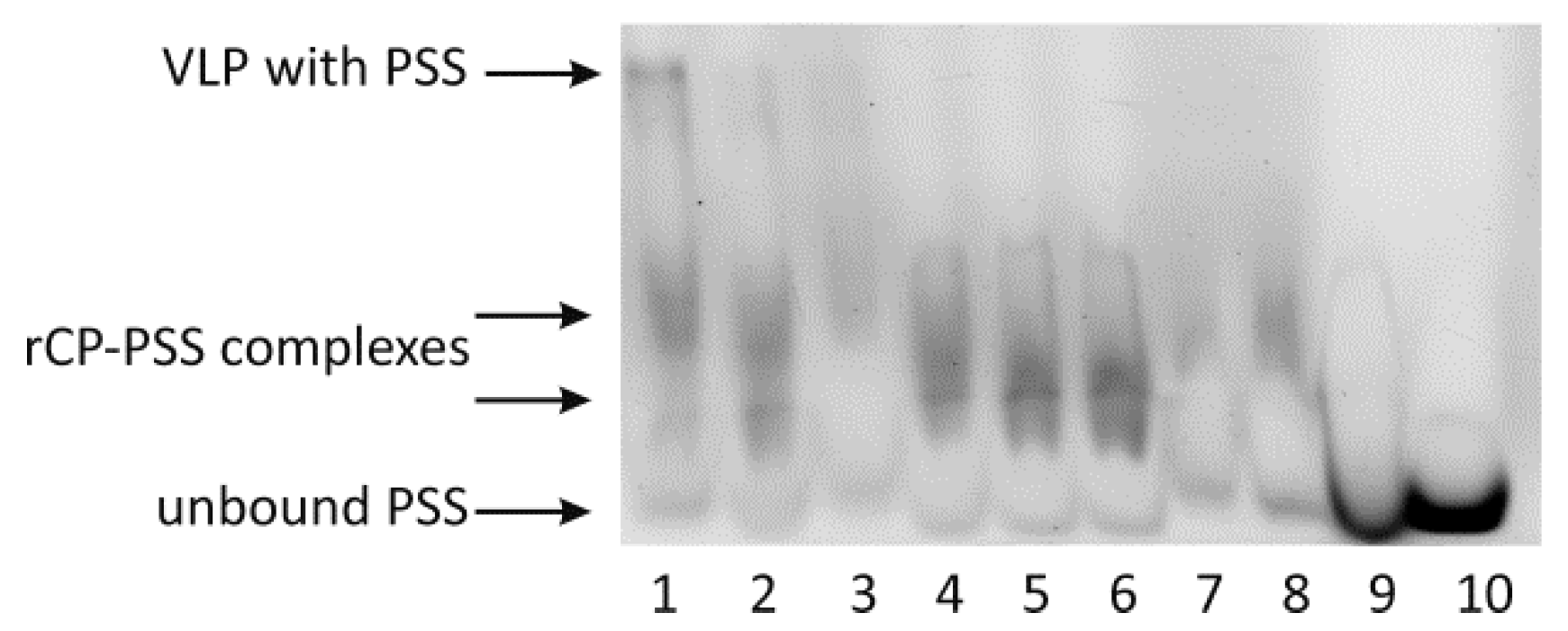
26]. Here, we showed that the ionic strength may also significantly influence CP-CP interactions, suggesting that ionic interactions might be involved in empty VLP assembly apart from hydrophobic interactions. We also observed that a rapid decrease in pH induces the transformation of CP dimers into larger CP oligomers and blocks VLP formation. Thus, we proposed that two-step dialysis gradually decreasing the ionic strength and pH is a good method to assemble both empty and cored VLPs. This protocol proves to be particularly efficient for the production of tRNA-cored VLPs. Other types of VLPs, including empty capsids, were formed less effectively (
Figure 4). Moreover, experiments involving empty capsids showed that VLPs preserve their structure even after drying (
Figure 6).
Considering that virion assembly is based upon CP-RNA binding under native conditions, Rao distinguished two types of CP-RNA interactions that impact this process [
25]. The first type depends on a specific RNA structure and involves the TLS located at the 3′ end of BMV genomic RNAs [
14]. It was shown that the encapsidation of BMV RNA lacking TLSs is not effective but can be restored after complementation with yeast tRNA or separate TLSs [
8]. We show that yeast tRNA alone is sufficient to mediate the formation of stable and homogenous VLPs. The mass ratio of yeast tRNA to rCP was similar to those shown by Cadena-Nava et al., which indicates that the length of the RNA molecule is less important [
27]. Our results also confirm that the structural features of the cargo, apart from its negative charge, are important factors influencing VLP formation and features. Accordingly, VLPs containing tRNA better resembled native BMV capsids in terms of their size than VLPs containing PSS [
28,
29].
The second type of CP-RNA interaction, crucial for capsid formation, is sequence-independent electrostatic attraction between negatively charged RNA and the positively charged CP N-terminus. The importance of those interactions was demonstrated by Cadena-Nava and coworkers, who showed that CCMV CPs were able to encapsidate longer RNA than viral genomes [
27]. The importance of a negative charge was also shown by Cheveruil and coworkers by encapsidating PSS in a VLP composed of CCMV CPs [
24]. Here, we confirmed the above observations by showing that PSS mediated the formation of BMV rCP-based VLPs. In addition, our data suggest that due to the electrostatic interaction between rCP and cargo, VLPs are more compact. Thus, tVLPs and PVLP were smaller than eVLPs. As expected, the dimensions of VLPs also depended on the cargo size; accordingly, PVLPs were smaller than tVLPs and BMV capsids. Although PSS particles are longer than tRNA and do not form complex spatial structures, due to the hydrophobic core, they adopt compact structures in water environments. Chevreuil and coworkers obtained similar results studying VLPs based on CCMV CP [
24]. The authors also explain this effect by the hydrophobic properties of PSS, which is poorly soluble in water and exists in a collapsed form [
24]. Moreover, they noted that linear, flexible PSS chains require less free energy to be encapsulated than particles with stiff conformations, such as RNA [
24,
30].
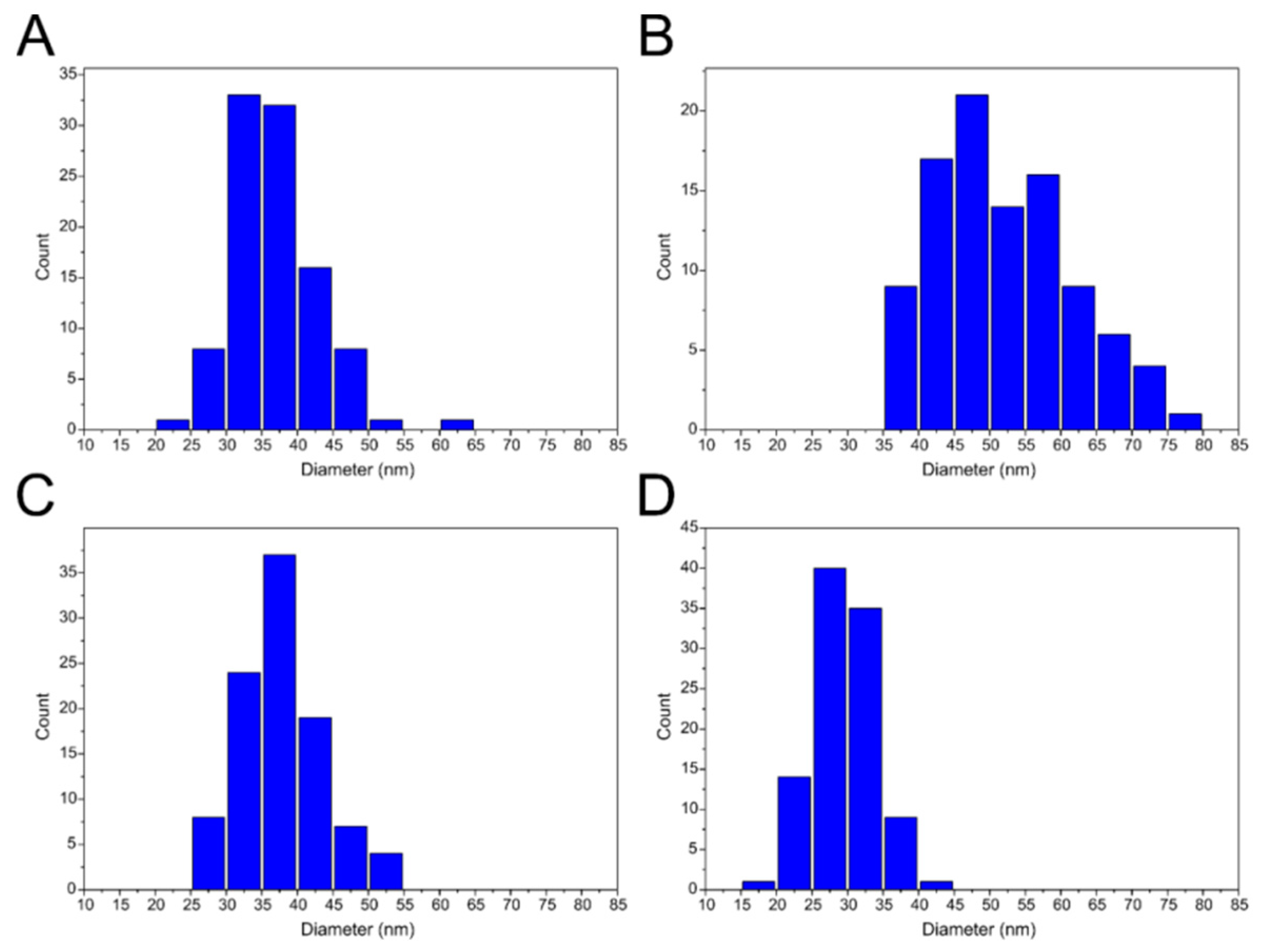
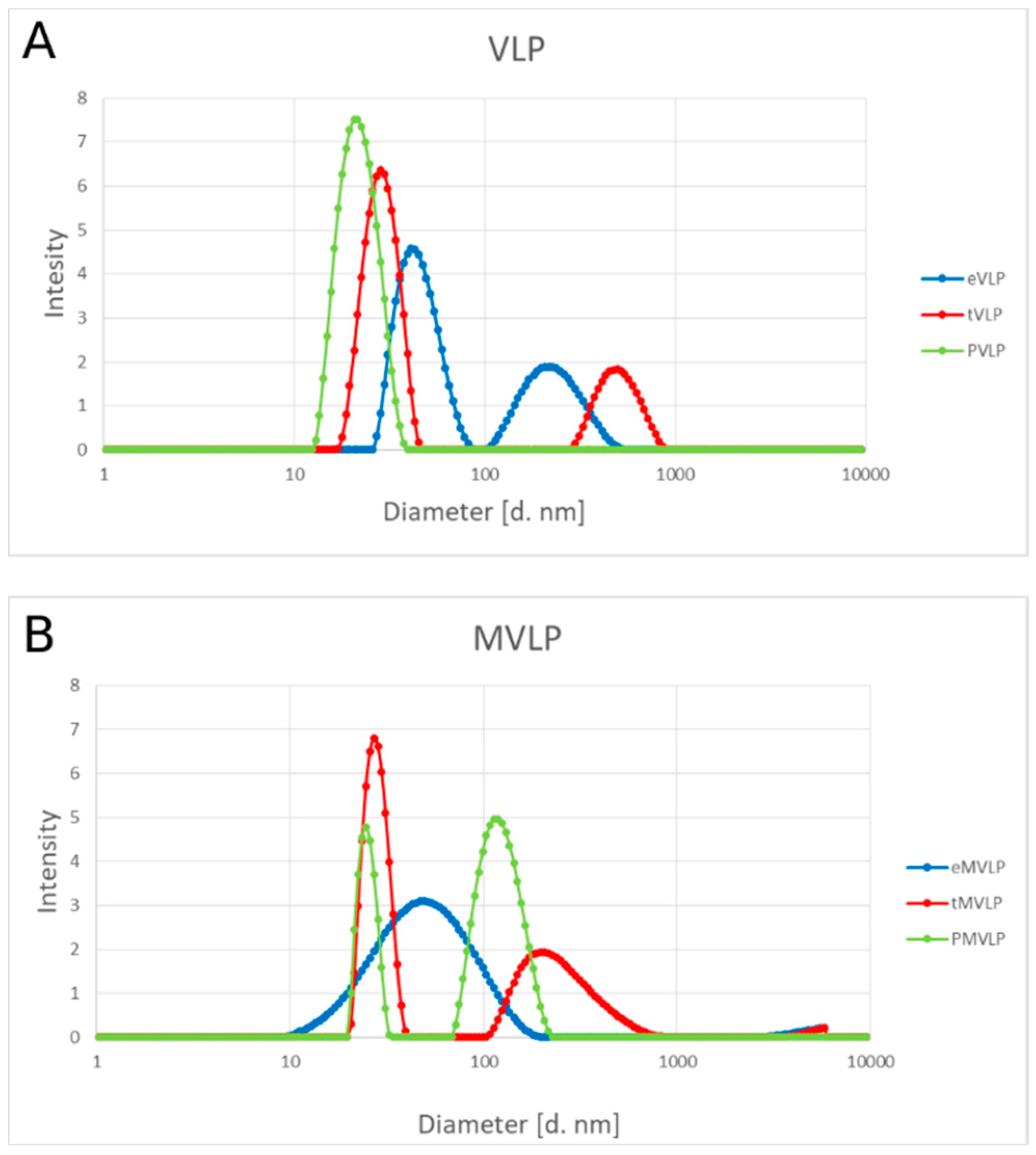
In contrast to tVLPs and PVLPs, empty VLPs that rely only on weak CP-CP interactions are more flexible and less stable than those relying on strong CP-RNA or CP-PSS interactions. Accordingly, eVLPs, both wild-type and mutated, have a broader range of diameters than the corresponding loaded VLPs (
Figure 5 and
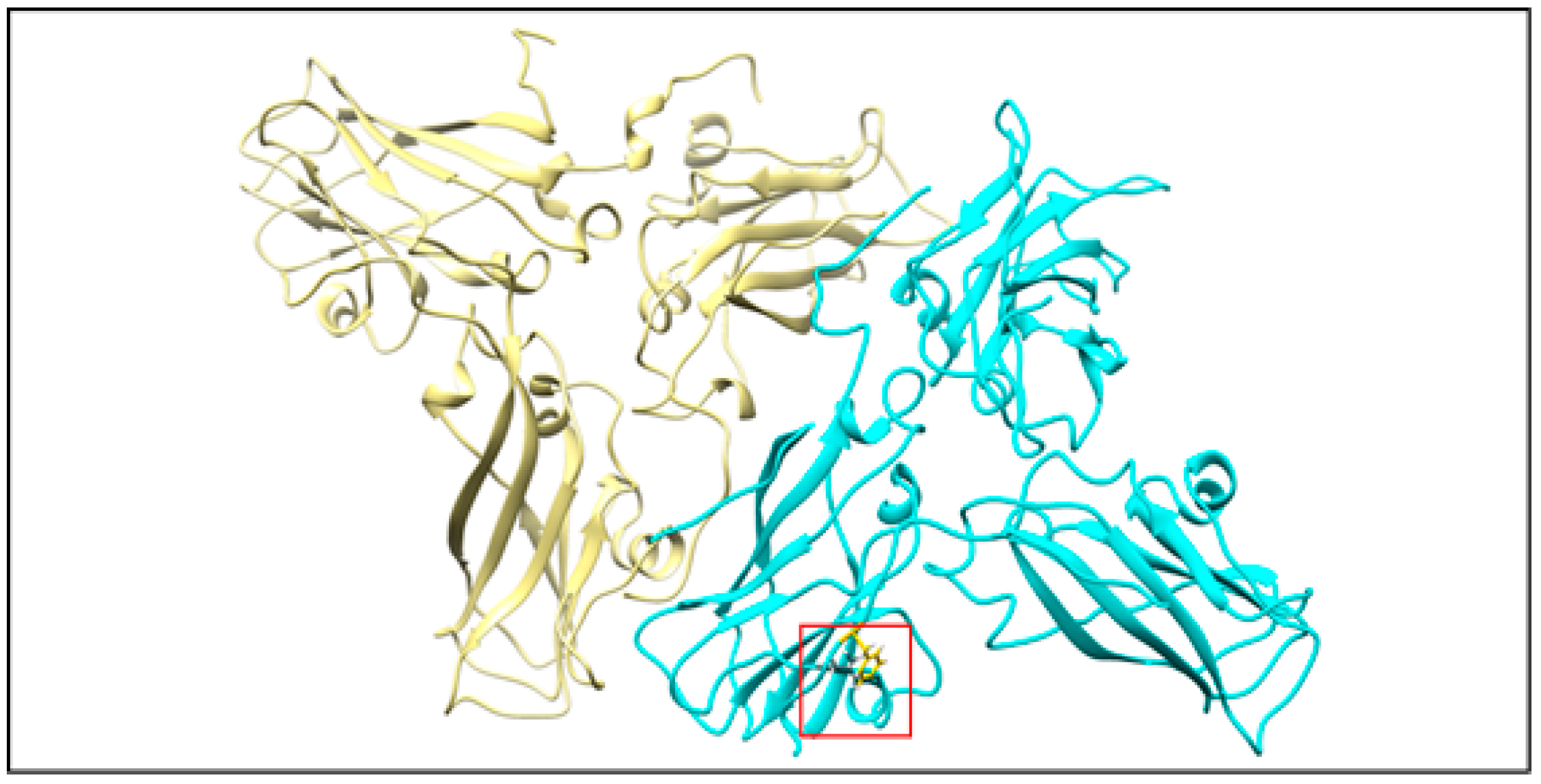
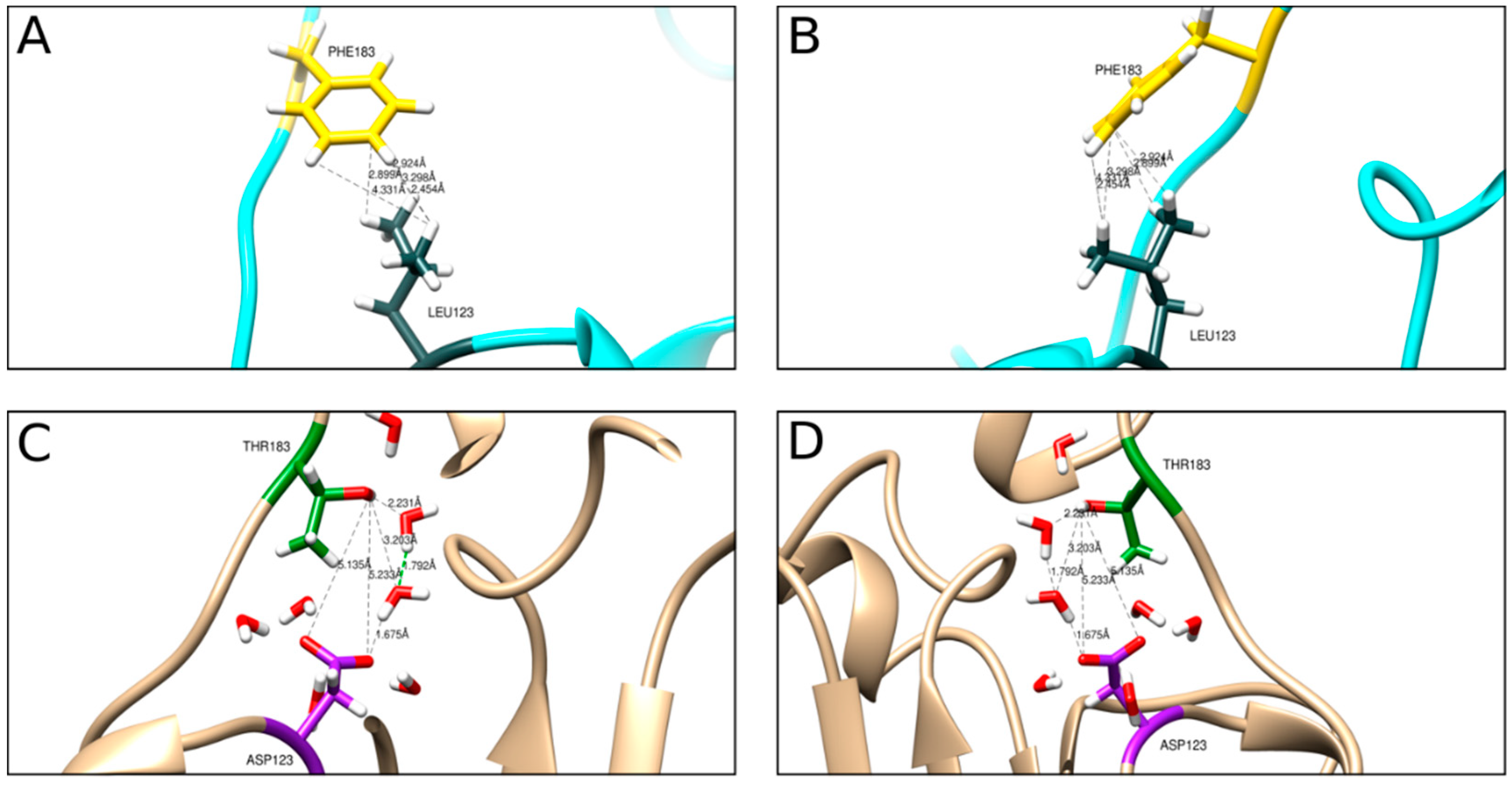
Figure 7). However, our designed eMVLP exhibits considerably higher assembly potential. The introduced mutations change the hydrophobic interaction between amino acid residues of two CPs into hydrophilic ones, which results in VLPs showing a wider range of diameters (
Figure 10). Higher amounts of formed eVLPs in comparison to eMVLPs also indicate that hydrophobic interactions between two CPs (L123 and F183) are significant for BMV capsid assembly. The addition of cargo strongly influences MVLP formation (
Figure 8B). Mutated VLPs containing tRNA were similar in size to wild-type tVLPs. This suggests that CP interactions with the tRNA structure strongly compensate for the weaker CP-CP interactions among the mutated CPs and are crucial for the formation of VLPs resembling native capsids. This observation once again confirms that the spatial conformation of tRNA strongly enhances VLP assembly, which was also presented by Rao et al. [
25]. On the other hand, MVLPc-containing PSSs are smaller than wild-type PSSs and are more homogenous. PSS, as mentioned above, can form a collapsed structure that, together with weaker CP-CP interactions and higher flexibility of the mutated VLP shell, might result in better adjustment of rCPs to its cargo and bring it closer to the VLP’s core.
The results showing the thermal stability of the produced VLP are consistent with the above observations. We found that TLS and the negative charge of the cargo stabilize VLPs. PSS and tRNA stabilized wild-type capsid VLPs up to 40 °C. However, above this temperature, PVLPs were unstable, unlike tVLPs. As concluded earlier, TLSs and electrostatic interactions with rCPs significantly stabilize VLPs better than negative charge alone (
Figure 8A). Our results indicate that tRNA presence is not mandatory for VLP formation but is crucial for particle stabilization. Mutated VLPs had slightly different thermal stability than wild-type VLPs. eMVLPs showed a wider range of sizes at room temperature than eVLPs. Interestingly, the aggregation of eMVLPs started to appear at 51 °C, while wild-type VLPs began to aggregate at 45 °C. This could be the effect of the stronger hydrophilic interactions resulting from the substitutions of two nonpolar amino acids with polar amino acids in the mutated CP. However, explaining this observation demands further study. For tMVLPs, 49 °C seems to be the temperature at which large aggregates start to prevail. Thus, tMVLPs seem to be less stable than tVLPs, which also indicates the significance of lateral CP-CP interactions on capsid stability. Interestingly, better adjustment of the cargo and CP in MVLPs was also confirmed by their higher thermal stability (aggregation proceeds at 42–51 °C) than that of VLPs (aggregation proceeds at 40–44 °C).
In summary, we have shown that recombinant BMV CP produced in a bacterial expression system is capable of self-assembly mediated by both CP-CP and CP-cargo interactions. By encapsidating tRNA and PSS, we highlighted the influence of cargo features on the structure and stability of BMV-based VLPs. We noted the profound influence of TLSs on the features of VLPs, while the negative charge of the cargo and CP-CP interactions seems to play a secondary but also important role. Moreover, we demonstrated that using genetic engineering, one can obtain VLPs with altered properties in comparison to VLPs formed by wild-type CP. Thus, the usage of a bacterial expression system for BMV CP production provides potential for the rational design of VLPs and research on their properties.
4. Materials and Methods
4.1. Protein Production and Purification
DNA coding for brome mosaic virus CP was cloned into the pMCSG48 expression plasmid and expressed in RosettaTM 2(DE3)pLysS competent E. coli cells (Novagen, Merck KGaA, Darmstadt, Germany) according to the manufacturer’s instructions. Briefly, bacterial cells were transformed with plasmids encoding wt or mutated BMV CP by a heat shock method. Then, each transformant was used to inoculate 250 mL of LB medium containing 100 mg/L ampicillin and 34 mg/L chloramphenicol. Bacteria were grown at 37 °C with shaking. When the OD600 of the culture reached 0.5–0.8, IPTG was added to a final concentration of 0.5 mM. After induction, bacteria were cultured for 16 h at 20 °C. The cells were harvested and frozen on dry ice for storage at −20 °C. Approximately 5 g of cells was suspended in 40 mL of lysis buffer I [25 mM Tris pH 8, 0.5 M NaCl, 0.2 mg/mL lysozyme (BioShop, Ontario, Canada), 250 U benzonase nuclease (Novagen,)] and sonicated. Cell debris was removed by centrifugation, and the supernatant was mixed with the same amount of buffer II [25 mM Tris pH 8, 0.5 M NaCl, 20 mM imidazole (BioShop)]. To purify BMV CP, affinity chromatography was used. After protein binding with Ni-NTA resin, the column was washed with buffer II. Next, the protein was eluted from the column using an imidazole gradient: 200, 300 and 500 mM in buffer II. The eluted protein was dialyzed using buffer III (25 mM Tris pH 8, 0.5 M NaCl). To remove the N-terminal tag, 1 mg of TEV protease was added per 10 mg of eluted protein. After TEV digestion, the sample was once again applied to a Ni-NTA charged column to remove the tag and any undigested protein. The first flowthrough was collected, and the purified proteins were concentrated by ultrafiltration (Amicon Ultra-10,000 MWCO, Millipore, Burlington, MA, USA). The concentration of the protein was determined by measuring the absorbance at 280 nm (NanoPhotometer N60, Implen, Westlake Village, CA, USA). The purity of the protein samples was assessed by SDS-PAGE, and the monodisperse state of the protein solutions was confirmed by dynamic light scattering (DLS) measurements using a Zetasizer µV (Malvern Panalytical, Malvern, GB) with the application of 6 µL of sample in a quartz cuvette with a 1 cm path length (Hellma QS 105.231). The correct molecular masses of all recombinant proteins were confirmed with Ultraflextreme MALDI Tof/Tof MS (Bruker Daltonics, Billerica, MA USA).
Two mutations, L123D and F183T, were introduced into the DNA sequence coding for BMV CP through PCR with specific primers. The mutated BMV CP sequence was then cloned into the pMCSG48 expression plasmid and amplified in DH5α competent E. coli cells (Thermo Fisher Scientific, Waltham, MA, USA). The plasmids were isolated and verified for L123D and F183T presence via DNA sequencing. Mutated protein expression, isolation and purification were performed as described above for the wild-type BMV CP. Isolated and purified BMV rCP was dialyzed against protein storage buffer (PSB) [1 M NaCl (BioShop), 1 mM EDTA (Merck), 20 mM Tris (BioShop), pH 7.2].
4.2. VLP Assembly
rCP (stored in the PSB) solution was subjected to a two-step dialysis process with the application of SnakeSkin Dialysis Tubing with a 3500 MWCO (Thermo Fisher Scientific). First, dialysis was performed against VLP assemble buffer (1) [50 mM NaCl (BioShop), 10 mM KCl (BioShop), 5 mM MgCl2·6H2O (Merck), 50 mM Tris (BioShop), pH 7.2] for 16 h at 4 °C. Next, dialysis was performed against VLP assemble buffer (2) [25 mM NaCl (BioShop), 10 mM KCl (BioShop), 25 mM NaOAc (Merck), 5 mM MgCl2·6H2O (Merck), 50 mM Tris (BioShop), pH 7.8] for 16 h at 4 °C. Afterwards, the samples were centrifuged (10 min, 10,000 rcf, 4 °C), and the supernatant was collected for further examination.
Assembly reactions of the VLP with tRNA or PSS were performed in a mass ratio gradient from 1:1 to 1:6 for tRNA:rCP and 1:1 to 1:8 for PSS:rCP. Assembly reactions proceeded as a two-step dialysis, as described above. Afterwards, the samples were centrifuged (10 min, 10,000 rcf, 4 °C), and the supernatant was transferred for further examination. Agarose gel electrophoresis was conducted to visualize the results of the VLP-core assembly gradient. A 1% agarose gel was used and stained with Midori Green (ABO). 10 µL of tVLP sample with 2 µL of loading dye (EURX, Gdańsk, Poland) and 25 µL of PVLP sample with 5 µL of loading dye was loaded on the gel. Samples were electrophoresed for 1.5 h at 80 V in 0.5× TBE buffer (BioShop). Gel was depicted on the ChemicDoc XRS+ System (BioRad, Hercules, CA, USA).
4.3. Cryo-TEM Measurements
Specimens for cryo-TEM imaging were prepared using CryoPunge 3 System (Gatan, Pleasanton, CA, USA). At room temperature and 95% relative humidity in the chamber, 4 µL droplet of each sample was put on a lacey carbon coated copper grid (Lacey C only, Ted Pella Inc., Redding, CA, USA) which were previously hydrophilized for 1 min using ELMO glow discharge system (Cordouan Technologies, Bordeaux, France). Subsequently to blotting the excess liquid, specimens were instantaneously plunged into a liquid ethan and shock frozen to ca. –183.15 °C. Vitrified samples were then transferred to Gatan 626 cryo-holder (Gatan, Pleasanton, CA, USA) that maintained temperature below −178.15 °C during the imaging. Imaging was carried out using Jeol JEM1440 transmission electron microscopy (Jeol Ltd., Tokyo, Japan) at accelerating voltage of 120 kV.
4.4. AFM Measurements
The VLP solution was diluted to a low concentration of 5 μg/mL, applied to a mica sample disc and left to dry. The dried sample was loaded onto a Multimode 8 instrument (Bruker, Billerica, MA, USA), and AFM tapping mode with an SNL-10 probe (Bruker) was used to record the shape of the prepared eVLPs. The results of the measurement were analyzed with NanoScope Analysis 1.5 software.
4.5. DLS Measurements
DLS measurements were recorded using a Zetasizer uV instrument (Malvern Instruments). The measurements were performed at a wavelength λ = 488 nm and at a 90° incidence angle of the light beam; a 2 µL quartz cuvette was used. The concentration of rCP was 0.1 mg/mL. Twelve measurements were performed for each VLP at 25 °C for standard DLS measurements and at temperatures ranging from 30–52 °C to analyze the thermal stability of the VLPs.
4.6. MST Experiments
Purified rCP was transferred to MST assay buffer for ionic strength studies (50 mM Tris, pH 7.5: 5 mM MgCl2 and 150, 300, 600, or 1000 mM NaCl) or pH studies (50 mM Tris, pH 8.5, 7.5 or 6.8:5 mM MgCl2 and 150 mM NaCl) using Zeba™ Spin desalting columns (ThermoFisher Scientific). CP was fluorescently labeled following the manufacturer’s instructions (Nanotemper Technology, Munich, Germany), i.e., using Monolith NTTM Protein Labeling Kit RED-NHS 647. The concentration of labeled proteins was adjusted to 50 nM. A dilution series (starting from 15 μM) of up to 16 unlabeled protein concentrations was prepared in a final volume of 10 μL. Ten microliters of labeled protein was added to each dilution and mixed with a pipette. All samples were centrifuged for 5 min at 10,000 g prior to MST measurement. Capillaries were filled and loaded into a Monolith NT.115 instrument, and the thermophoresis experiment was performed. The results were analyzed with MO.Affinity Analysis software (Nanotemper).
4.7. Protein Cross-Linking
CP crosslinking was performed using 5 mM disuccinimidyl suberate (DSS, Thermo Scientific) in DMSO according to the manufacturer’s instructions using rCP in MST buffer described above at pH 7.5 and 6.8. The reaction was stopped after 5, 10, 15 and 30 min by adding 1 M TRIS-HCl, pH 7.5. The crosslinking results were visualized by SDS-PAGE.
4.8. CP Mutant Design
The hexamer model of the BMV coat protein and of the mutant variant were constructed using the UCSF Chimera program [
31] on the basis of the 1JS9 structure from the PDB repository. Proper protonation and parameterization of protein models was performed using the pdb2gmx module of the GROMACS 4.6.7 package [
32], employing the Amber99SB-ILDN [
33] force field. CP hexamers were immersed in a truncated dodecahedral simulation box containing TIP3P model water molecules. Solvated systems were subsequently neutralized, and an excess of Na
+ and Cl
− ions was added to obtain a final concentration of 0.1 M NaCl.
The geometry of the systems was optimized using the energy minimization technique with the steepest descent algorithm. Next, optimized systems were relaxed during 200 ps NVT ensemble simulation runs and subsequently 200 ps NPT runs. Such relaxed systems were used as starting structures for final, 50 ns production NPT runs. During all runs, the time step was 2 fs. During equilibration runs, the output frequency was set to 100 time steps (200 fs), whereas in production simulations, it was set to 1000 time steps (2 ps). Long-range electrostatic interactions were calculated using the particle mesh Ewald method, with a cutoff value of 1 nm. Velocity rescaling was used to scale the temperature and Parinello-Rahman barostat to maintain constant pressure in the simulated systems.