Effects of Low Dose Space Radiation Exposures on the Splenic Metabolome
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
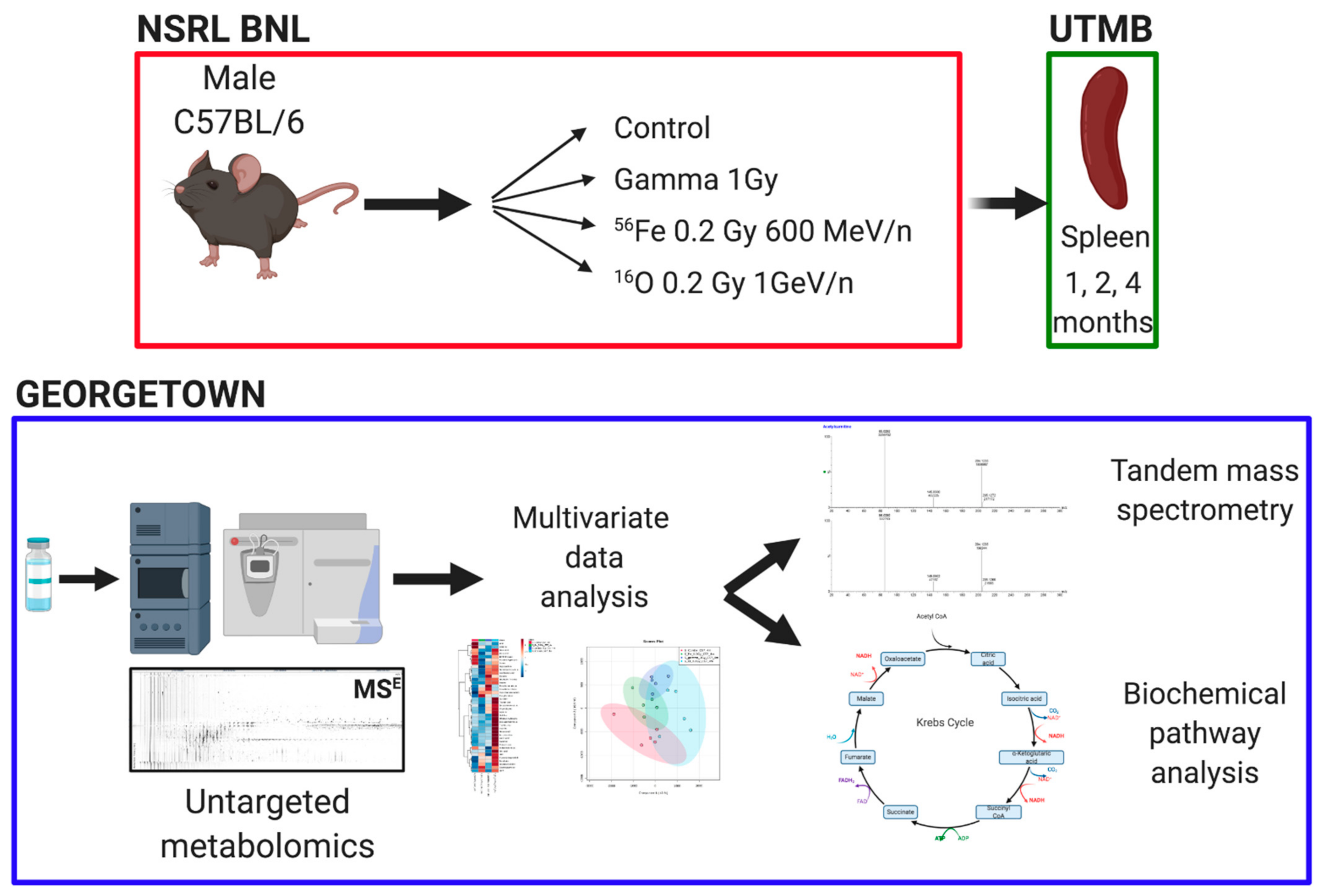
2.2. Animals and Irradiations
2.3. Untargeted Metabolomics and Data Analysis
2.4. Regression Analysis
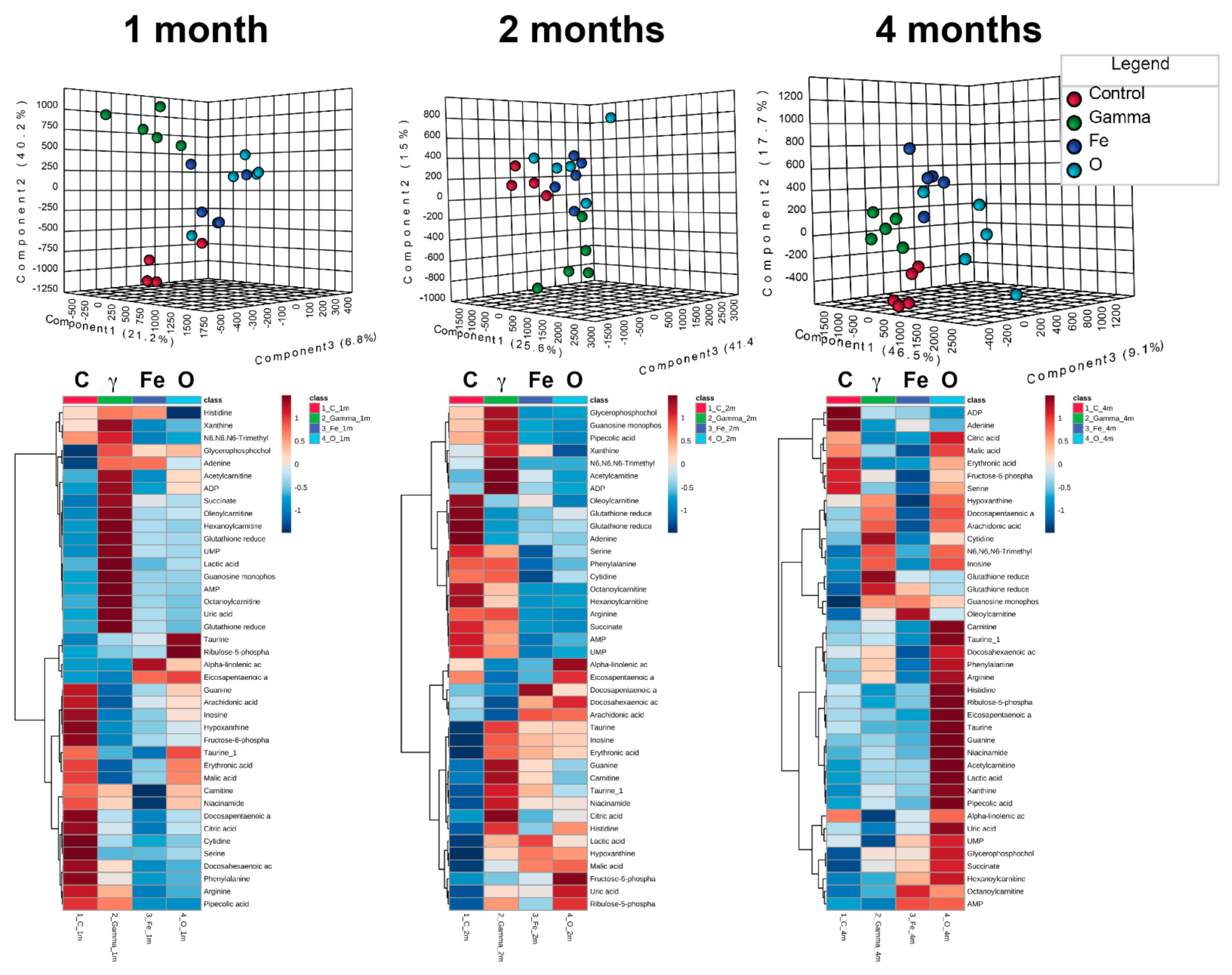
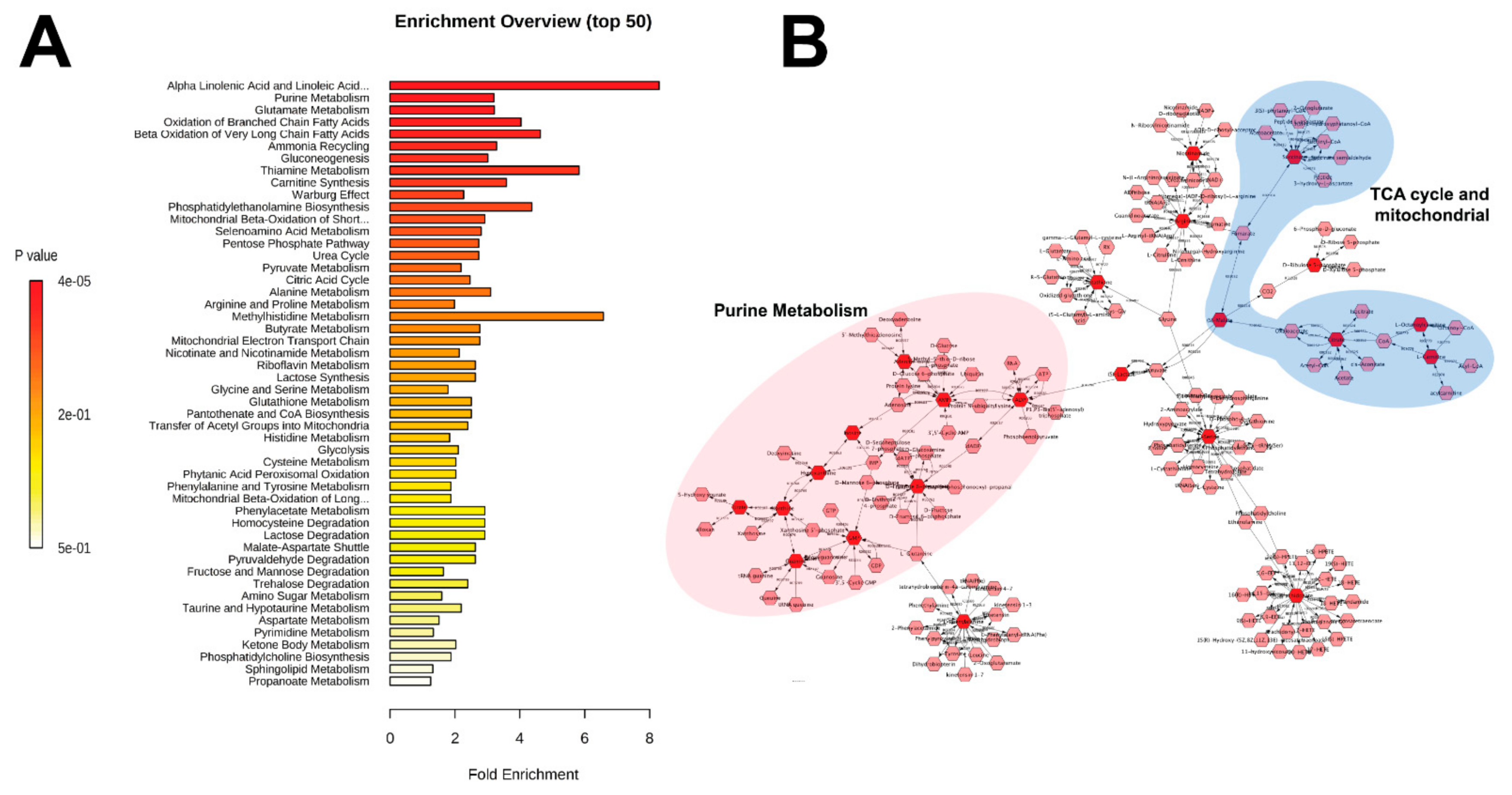
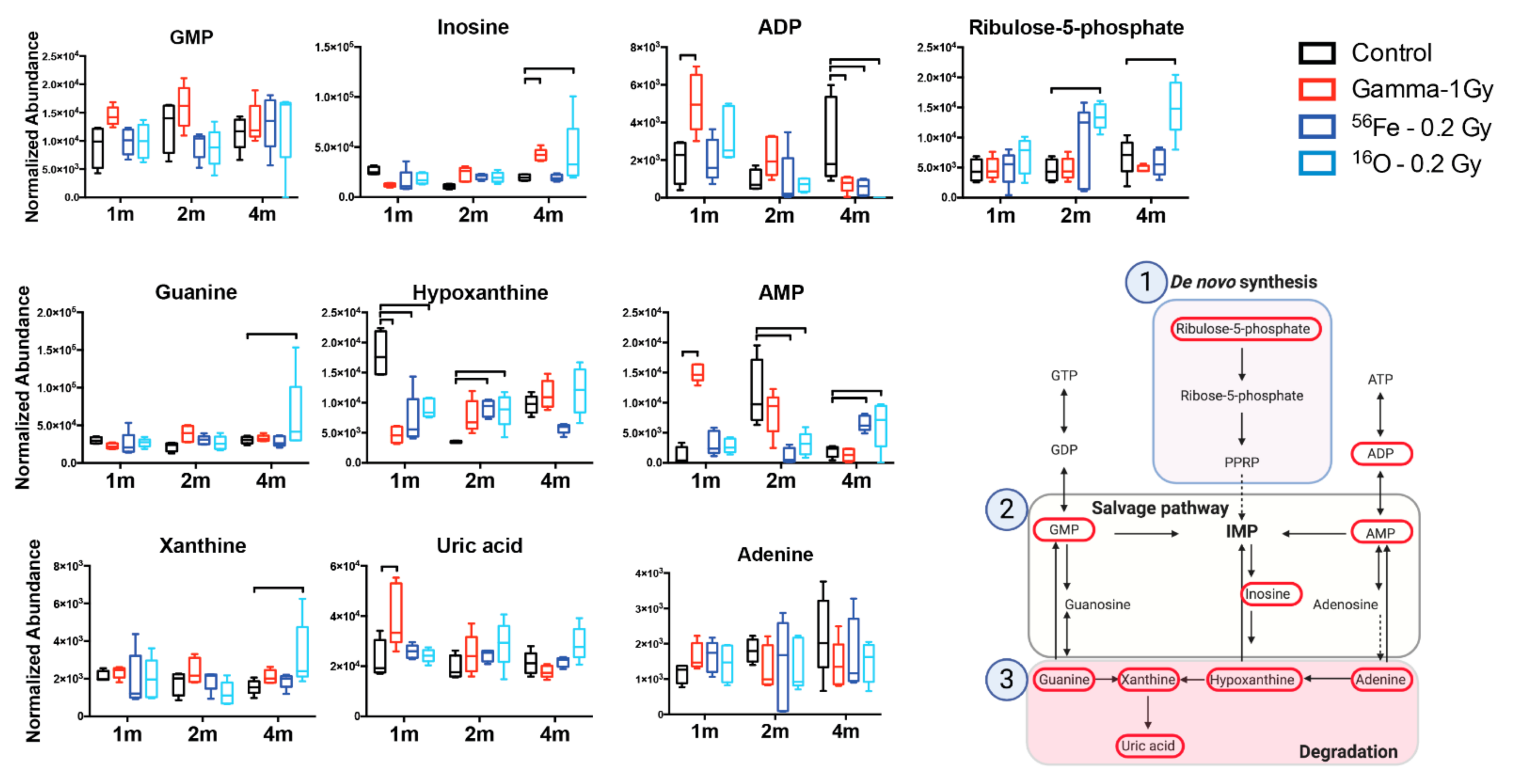
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Demontis, G.C.; Germani, M.M.; Caiani, E.G.; Barravecchia, I.; Passino, C.; Angeloni, D. Human Pathophysiological Adaptations to the Space Environment. Front. Physiol. 2017, 8, 547. [Google Scholar] [CrossRef] [PubMed]
- Pletser, V. Gravity, Weight and Their Absence; Springer: Singapore, 2018; pp. 67–84. [Google Scholar]
- Chancellor, J.C.; Scott, G.B.; Sutton, J.P. Space Radiation: The Number One Risk to Astronaut Health Beyond Low Earth Orbit. Life 2014, 4, 491–510. [Google Scholar] [CrossRef] [PubMed]
- Boerma, M.; Nelson, G.A.; Sridharan, V.; Mao, X.W.; Koturbash, I.; Hauer-Jensen, M. Space Radiation and Cardiovascular Disease Risk. World J. Cardiol. 2015, 7, 882–888. [Google Scholar] [CrossRef]
- Cucinotta, F.A.; Alp, M.; Sulzman, F.M.; Wang, M. Space Radiation Risks to the Central Nervous System. Life Sci. Space Res. 2014, 2, 54–69. [Google Scholar] [CrossRef]
- Limoli, C. Your Brain on Mars. Radiat. Res. 2015, 184, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Council, N.R. Managing Space Radiation Risk in the New Era of Space Exploration; National Academies Press: Washington, MD, USA, 2008. [Google Scholar]
- NCRP. Information Needed to Make Radiation Protection Recommendations for Space Missions Beyond Low-Earth Orbit; NCRP Report No. 153; NCRP: Bethesda, MD, USA, 2006. [Google Scholar]
- Parihar, V.K.; Allen, B.; Tran, K.K.; Macaraeg, T.G.; Chu, E.M.; Kwok, S.F.; Chmielewski, N.N.; Craver, B.M.; Baulch, J.E.; Acharya, M.M.; et al. What Happens to Your Brain on the Way to Mars. Sci. Adv. 2015, 1, e1400256. [Google Scholar] [CrossRef] [PubMed]
- Parihar, V.K.; Allen, B.D.; Caressi, C.; Kwok, S.; Chu, E.; Tran, K.K.; Chmielewski, N.N.; Giedzinski, E.; Acharya, M.M.; Britten, R.A.; et al. Cosmic Radiation Exposure and Persistent Cognitive Dysfunction. Sci. Rep. 2016, 6, 34774. [Google Scholar] [CrossRef]
- Pecaut, M.J.; Mao, X.W.; Bellinger, D.L.; Jonscher, K.R.; Stodieck, L.S.; Ferguson, V.L.; Bateman, T.A.; Mohney, R.P.; Gridley, D.S. Is Spaceflight-Induced Immune Dysfunction Linked to Systemic Changes in Metabolism. PLoS ONE 2017, 12, e0174174. [Google Scholar] [CrossRef]
- Sonnenfeld, G.; Butel, J.S.; Shearer, W.T. Effects of the Space Flight Environment on the Immune System. Rev. Environ. Health 2003, 18, 1–17. [Google Scholar] [CrossRef]
- Sonnenfeld, G. The Immune System in Space, Including Earth-Based Benefits of Space-Based Research. Curr. Pharm. Biotechnol. 2005, 6, 343–349. [Google Scholar] [CrossRef]
- Casero, D.; Gill, K.; Sridharan, V.; Koturbash, I.; Nelson, G.; Hauer-Jensen, M.; Boerma, M.; Braun, J.; Cheema, A.K. Space-Type Radiation Induces Multimodal Responses in the Mouse Gut Microbiome and Metabolome. Microbiome 2017, 5, 105. [Google Scholar] [CrossRef] [PubMed]
- Garrett-Bakelman, F.E.; Darshi, M.; Green, S.J.; Gur, R.C.; Lin, L.; Macias, B.R.; McKenna, M.J.; Meydan, C.; Mishra, T.; Nasrini, J.; et al. The Nasa Twins Study: A Multidimensional Analysis of a Year-Long Human Spaceflight. Science 2019, 364. [Google Scholar] [CrossRef]
- Laiakis, E.C.; Trani, D.; Moon, B.H.; Strawn, S.J.; Fornace, A.J., Jr. Metabolomic Profiling of Urine Samples From Mice Exposed to Protons Reveals Radiation Quality and Dose Specific Differences. Radiat. Res. 2015, 183, 382–390. [Google Scholar] [CrossRef] [PubMed]
- Miousse, I.R.; Skinner, C.M.; Sridharan, V.; Seawright, J.W.; Singh, P.; Landes, R.D.; Cheema, A.K.; Hauer-Jensen, M.; Boerma, M.; Koturbash, I. Changes in One-Carbon Metabolism and DNA Methylation in the Hearts of Mice Exposed to Space Environment-Relevant Doses of Oxygen Ions (16O). Life Sci. Space Res. 2019, 22, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, M.; Rajagopal, M.; Gill, K.; Li, Y.; Bansal, S.; Sridharan, V.; Tyburski, J.B.; Boerma, M.; Cheema, A.K. Identification of Plasma Lipidome Changes Associated With Low Dose Space-Type Radiation Exposure in a Murine Model. Metabolites 2020, 10, 252. [Google Scholar] [CrossRef]
- Li, H.H.; Wang, Y.W.; Chen, R.; Zhou, B.; Ashwell, J.D.; Fornace, A.J., Jr. Ionizing Radiation Impairs T Cell Activation by Affecting Metabolic Reprogramming. Int. J. Biol. Sci. 2015, 11, 726–736. [Google Scholar] [CrossRef] [PubMed]
- Agha, N.H.; Mehta, S.K.; Rooney, B.V.; Laughlin, M.S.; Markofski, M.M.; Pierson, D.L.; Katsanis, E.; Crucian, B.E.; Simpson, R.J. Exercise as a Countermeasure for Latent Viral Reactivation During Long Duration Space Flight. FASEB J. 2020, 34, 2869–2881. [Google Scholar] [CrossRef]
- Rooney, B.V.; Crucian, B.E.; Pierson, D.L.; Laudenslager, M.L.; Mehta, S.K. Herpes Virus Reactivation in Astronauts During Spaceflight and Its Application on Earth. Front. Microbiol. 2019, 10, 16. [Google Scholar] [CrossRef]
- Cao, D.; Song, J.; Ling, S.; Niu, S.; Lu, L.; Cui, Z.; Li, Y.; Hao, S.; Zhong, G.; Qi, Z.; et al. Hematopoietic Stem Cells and Lineage Cells Undergo Dynamic Alterations Under Microgravity and Recovery Conditions. FASEB J. 2019, 33, 6904–6918. [Google Scholar] [CrossRef]
- Crucian, B.E.; Choukèr, A.; Simpson, R.J.; Mehta, S.; Marshall, G.; Smith, S.M.; Zwart, S.R.; Heer, M.; Ponomarev, S.; Whitmire, A.; et al. Immune System Dysregulation During Spaceflight: Potential Countermeasures for Deep Space Exploration Missions. Front. Immunol. 2018, 9, 1437. [Google Scholar] [CrossRef]
- Moreno-Villanueva, M.; Feiveson, A.H.; Krieger, S.; Kay Brinda, A.; von Scheven, G.; Bürkle, A.; Crucian, B.; Wu, H. Synergistic Effects of Weightlessness, Isoproterenol, and Radiation on DNA Damage Response and Cytokine Production in Immune Cells. Int. J. Mol. Sci. 2018, 19, 3689. [Google Scholar] [CrossRef]
- Paul, A.M.; Cheng-Campbell, M.; Blaber, E.A.; Anand, S.; Bhattacharya, S.; Zwart, S.R.; Crucian, B.E.; Smith, S.M.; Meller, R.; Grabham, P. Beyond Low-Earth Orbit: Characterizing Immune and microRNA Differentials Following Simulated Deep Spaceflight Conditions in Mice. iScience 2020, 23, 101747. [Google Scholar] [CrossRef]
- Tascher, G.; Gerbaix, M.; Maes, P.; Chazarin, B.; Ghislin, S.; Antropova, E.; Vassilieva, G.; Ouzren-Zarhloul, N.; Gauquelin-Koch, G.; Vico, L.; et al. Analysis of Femurs From Mice Embarked on Board Bion-M1 Biosatellite Reveals a Decrease in Immune Cell Development, Including B Cells, After 1 wk of Recovery on Earth. FASEB J. 2019, 33, 3772–3783. [Google Scholar] [CrossRef] [PubMed]
- Bronte, V.; Pittet, M.J. The Spleen in Local and Systemic Regulation of Immunity. Immunity 2013, 39, 806–818. [Google Scholar] [CrossRef] [PubMed]
- Rizvi, A.; Pecaut, M.J.; Slater, J.M.; Subramaniam, S.; Gridley, D.S. Low-Dose γ-Rays Modify CD4(+) T Cell Signalling Response to Simulated Solar Particle Event Protons in a Mouse Model. Int. J. Radiat. Biol. 2011, 87, 24–35. [Google Scholar] [CrossRef]
- Cheema, A.K.; Mehta, K.Y.; Rajagopal, M.U.; Wise, S.Y.; Fatanmi, O.O.; Singh, V.K. Metabolomic Studies of Tissue Injury in Nonhuman Primates Exposed to Gamma-Radiation. Int. J. Mol. Sci. 2019, 20, 3360. [Google Scholar] [CrossRef]
- Jang, W.G.; Park, J.Y.; Lee, J.; Bang, E.; Kim, S.R.; Lee, E.K.; Yun, H.J.; Kang, C.M.; Hwang, G.S. Investigation of Relative Metabolic Changes in the Organs and Plasma of Rats Exposed to X-Ray Radiation Using HR-MAS (1)H NMR and Solution (1)H NMR. NMR Biomed. 2016, 29, 507–518. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Hu, M.; Liu, M.; Hu, J.Z. 1h Nmr Metabolomics Study of Spleen from C57BL/6 Mice Exposed to Gamma Radiation. Metabolomics 2016, 6, 1–11. [Google Scholar]
- Weil, M.M.; Bedford, J.S.; Bielefeldt-Ohmann, H.; Ray, F.A.; Genik, P.C.; Ehrhart, E.J.; Fallgren, C.M.; Hailu, F.; Battaglia, C.L.; Charles, B.; et al. Incidence of Acute Myeloid Leukemia and Hepatocellular Carcinoma in Mice Irradiated With 1 Gev/nucleon (56)Fe Ions. Radiat. Res. 2009, 172, 213–219. [Google Scholar] [CrossRef]
- Weil, M.M.; Ray, F.A.; Genik, P.C.; Yu, Y.; McCarthy, M.; Fallgren, C.M.; Ullrich, R.L. Effects of 28Si Ions, 56Fe Ions, and Protons on the Induction of Murine Acute Myeloid Leukemia and Hepatocellular Carcinoma. PLoS ONE 2014, 9, e104819. [Google Scholar]
- Nia, A.M.; Khanipov, K.; Barnette, B.L.; Ullrich, R.L.; Golovko, G.; Emmett, M.R. Comparative RNA-Seq Transcriptome Analyses Reveal Dynamic Time-Dependent Effects of 56fe, 16o, and 28si Irradiation on the Induction of Murine Hepatocellular Carcinoma. BMC Genom. 2020, 21, 453. [Google Scholar] [CrossRef]
- Nia, A.M.; Chen, T.; Barnette, B.L.; Khanipov, K.; Ullrich, R.L.; Bhavnani, S.K.; Emmett, M.R. Efficient Identification of Multiple Pathways: RNA-Seq Analysis of Livers From 56Fe Ion Irradiated Mice. BMC Bioinform. 2020, 21, 118. [Google Scholar] [CrossRef]
- Astarita, G.; Dhungana, S.; Shrestha, B.; Laiakis, E.C. Metabolomic Approaches to Study the Tumor Microenvironment. Methods Enzym. 2020, 636, 93–108. [Google Scholar]
- Smith, C.A.; O′Maille, G.; Want, E.J.; Qin, C.; Trauger, S.A.; Brandon, T.R.; Custodio, D.E.; Abagyan, R.; Siuzdak, G. Metlin: A Metabolite Mass Spectral Database. Ther. Drug Monit. 2005, 27, 747–751. [Google Scholar] [CrossRef]
- Wishart, D.S.; Jewison, T.; Guo, A.C.; Wilson, M.; Knox, C.; Liu, Y.; Djoumbou, Y.; Mandal, R.; Aziat, F.; Dong, E.; et al. Hmdb 3.0--the Human Metabolome Database in 2013. Nucleic Acids Res. 2013, 41, D801–D807. [Google Scholar] [CrossRef]
- Sud, M.; Fahy, E.; Cotter, D.; Brown, A.; Dennis, E.A.; Glass, C.K.; Merrill, A.H.; Murphy, R.C.; Raetz, C.R.; Russell, D.W.; et al. Lmsd: Lipid Maps Structure Database. Nucleic Acids Res. 2007, 35, D527–D532. [Google Scholar] [CrossRef]
- Chong, J.; Xia, J. Using Metaboanalyst 4.0 for Metabolomics Data Analysis, Interpretation, and Integration with Other Omics Data. Methods Mol. Biol. 2020, 2104, 337–360. [Google Scholar]
- Gao, J.; Tarcea, V.G.; Karnovsky, A.; Mirel, B.R.; Weymouth, T.E.; Beecher, C.W.; Cavalcoli, J.D.; Athey, B.D.; Omenn, G.S.; Burant, C.F.; et al. Metscape: A Cytoscape Plug-in for Visualizing and Interpreting Metabolomic Data in the Context of Human Metabolic Networks. Bioinformatics 2010, 26, 971–973. [Google Scholar] [CrossRef]
- Simonsen, L.C.; Slaba, T.C.; Guida, P.; Rusek, A. Nasa′s First Ground-Based Galactic Cosmic Ray Simulator: Enabling a New Era in Space Radiobiology Research. PLoS Biol. 2020, 18, e3000669. [Google Scholar] [CrossRef]
- Pedley, A.M.; Benkovic, S.J. A New View into the Regulation of Purine Metabolism: The Purinosome. Trends Biochem. Sci. 2017, 42, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Keenan, R.T. The Biology of Urate. Semin. Arthritis Rheum. 2020, 50, S2–S10. [Google Scholar] [CrossRef]
- Stoecklein, V.M.; Osuka, A.; Ishikawa, S.; Lederer, M.R.; Wanke-Jellinek, L.; Lederer, J.A. Radiation Exposure Induces Inflammasome Pathway Activation in Immune Cells. J. Immunol. 2015, 194, 1178–1189. [Google Scholar] [CrossRef]
- Chang, P.Y.; Kanazawa, N.; Lutze-Mann, L.; Winegar, R. HZE Particle Radiation Induces Tissue-Specific and p53-Dependent Mutagenesis in Transgenic Animals. Phys. Med. 2001, 17, 189–191. [Google Scholar]
- Yamaoka, K.; Niki, E.; Takahashi, M.; Iriyama, K. Effects of Low Dose X-Ray Irradiation on Purine Metabolism in Mouse Splenocytes. Physiol. Chem. Phys. Med. NMR 1997, 29, 1–9. [Google Scholar]
- Meyts, I.; Aksentijevich, I. Deficiency of Adenosine Deaminase 2 (DADA2): Updates on the Phenotype, Genetics, Pathogenesis, and Treatment. J. Clin. Immunol. 2018, 38, 569–578. [Google Scholar] [CrossRef]
- Pell, V.R.; Chouchani, E.T.; Frezza, C.; Murphy, M.P.; Krieg, T. Succinate Metabolism: A New Therapeutic Target for Myocardial Reperfusion Injury. Cardiovasc. Res. 2016, 111, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Mills, E.L.; Kelly, B.; Logan, A.; Costa, A.S.H.; Varma, M.; Bryant, C.E.; Tourlomousis, P.; Däbritz, J.H.M.; Gottlieb, E.; Latorre, I.; et al. Succinate Dehydrogenase Supports Metabolic Repurposing of Mitochondria to Drive Inflammatory Macrophages. Cell 2016, 167, 457–470.e13. [Google Scholar] [CrossRef]
- Haas, R.; Cucchi, D.; Smith, J.; Pucino, V.; Macdougall, C.E.; Mauro, C. Intermediates of Metabolism: From Bystanders to Signalling Molecules. Trends Biochem. Sci. 2016, 41, 460–471. [Google Scholar] [CrossRef]
- Dalla Pozza, E.; Dando, I.; Pacchiana, R.; Liboi, E.; Scupoli, M.T.; Donadelli, M.; Palmieri, M. Regulation of Succinate Dehydrogenase and Role of Succinate in Cancer. Semin. Cell Dev. Biol. 2020, 98, 4–14. [Google Scholar] [CrossRef]
- Ryan, D.G.; Murphy, M.P.; Frezza, C.; Prag, H.A.; Chouchani, E.T.; O′Neill, L.A.; Mills, E.L. Coupling Krebs Cycle Metabolites to Signalling in Immunity and Cancer. Nat. Metab. 2019, 1, 16–33. [Google Scholar] [CrossRef]
- Esser-von Bieren, J. Immune-Regulation and -Functions of Eicosanoid Lipid Mediators. Biol. Chem. 2017, 398, 1177–1191. [Google Scholar] [CrossRef]
- Laiakis, E.C.; Pannkuk, E.L.; Chauthe, S.K.; Wang, Y.W.; Lian, M.; Mak, T.D.; Barker, C.A.; Astarita, G.; Fornace, A.J., Jr. A Serum Small Molecule Biosignature of Radiation Exposure from Total Body Irradiated Patients. J. Proteome Res. 2017, 16, 3805–3815. [Google Scholar] [CrossRef] [PubMed]
- da Silveira, W.A.; Fazelinia, H.; Rosenthal, S.B.; Laiakis, E.C.; Kim, M.S.; Meydan, C.; Kidane, Y.; Rathi, K.S.; Smith, S.M.; Stear, B.; et al. Comprehensive Multi-Omics Analysis Reveals Mitochondrial Stress as a Central Biological Hub for Spaceflight Impact. Cell 2020, 183, 1185–1201.e20. [Google Scholar] [CrossRef] [PubMed]
- Jonscher, K.R.; Alfonso-Garcia, A.; Suhalim, J.L.; Orlicky, D.J.; Potma, E.O.; Ferguson, V.L.; Bouxsein, M.L.; Bateman, T.A.; Stodieck, L.S.; Levi, M.; et al. Spaceflight Activates Lipotoxic Pathways in Mouse Liver. PLoS ONE 2016, 11, e0152877. [Google Scholar]
- Espinosa-Jeffrey, A.; Nguyen, K.; Kumar, S.; Toshimasa, O.; Hirose, R.; Reue, K.; Vergnes, L.; Kinchen, J.; Vellis, J. Simulated Microgravity Enhances Oligodendrocyte Mitochondrial Function and Lipid Metabolism. J. Neurosci. Res. 2016, 94, 1434–1450. [Google Scholar] [CrossRef] [PubMed]
- Rabin, B.M.; Poulose, S.M.; Carrihill-Knoll, K.L.; Ramirez, F.; Bielinski, D.F.; Heroux, N.; Shukitt-Hale, B. Acute Effects of Exposure to (56)Fe and (16)O Particles on Learning and Memory. Radiat. Res. 2015, 184, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Gridley, D.S.; Pecaut, M.J.; Nelson, G.A. Total-Body Irradiation with High-Let Particles: Acute and Chronic Effects on the Immune System. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2002, 282, R677–R688. [Google Scholar] [CrossRef]
- Hoehn, D.; Pujol-Canadell, M.; Young, E.F.; Serban, G.; Shuryak, I.; Maerki, J.; Xu, Z.; Chowdhury, M.; Luna, A.M.; Vlada, G.; et al. Effects of High- and Low-Let Radiation on Human Hematopoietic System Reconstituted in Immunodeficient Mice. Radiat. Res. 2019, 191, 162–175. [Google Scholar] [CrossRef]
- Baba, S.; Smith, T.; Hellmann, J.; Bhatnagar, A.; Carter, K.; Vanhoover, A.; Caruso, J. Space Flight Diet-Induced Deficiency and Response to Gravity-Free Resistive Exercise. Nutrients 2020, 12, 2400. [Google Scholar] [CrossRef]
- Ray, S.; Gebre, S.; Fogle, H.; Berrios, D.C.; Tran, P.B.; Galazka, J.M.; Costes, S.V. Genelab: Omics Database for Spaceflight Experiments. Bioinformatics 2019, 35, 1753–1759. [Google Scholar] [CrossRef]
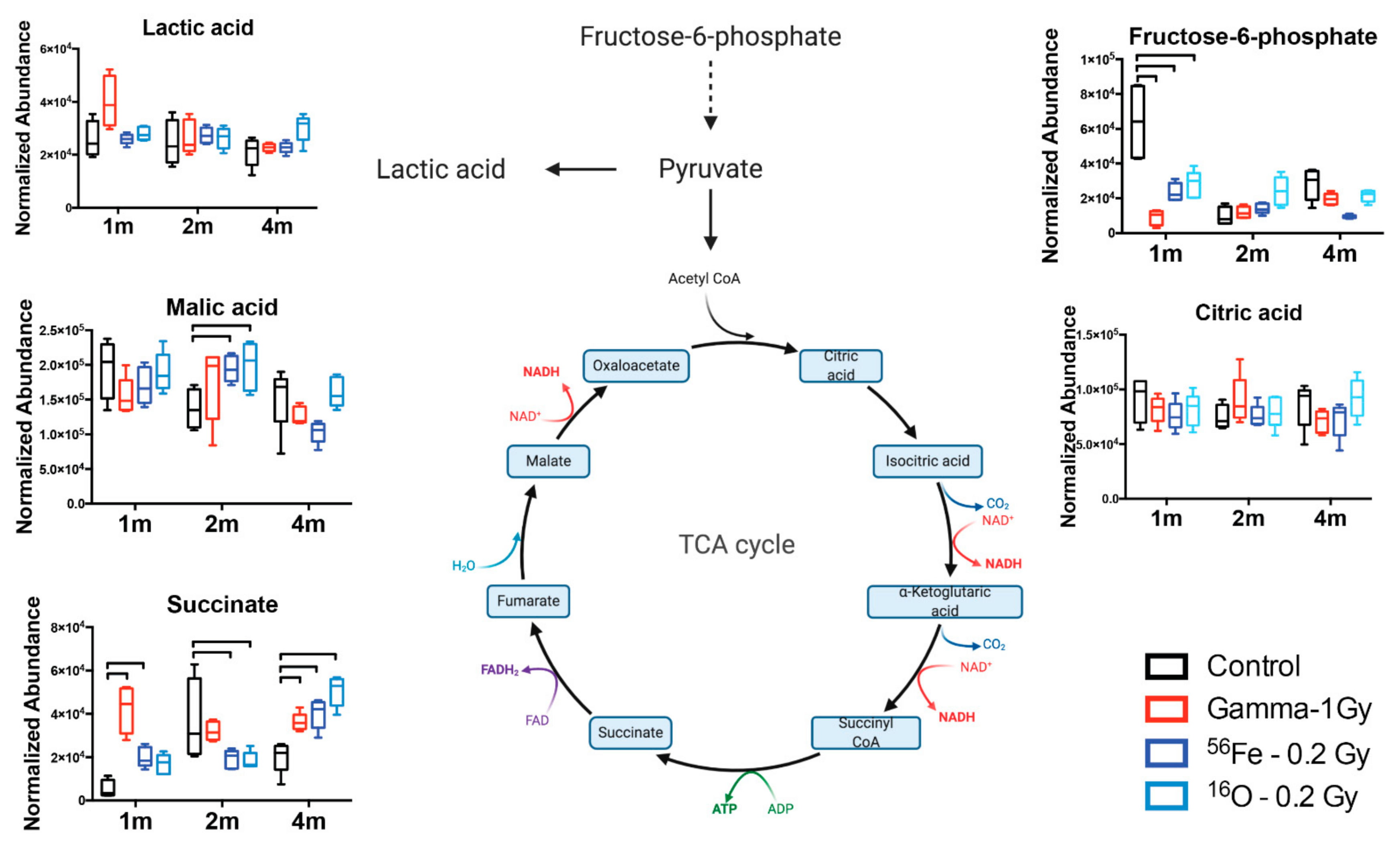
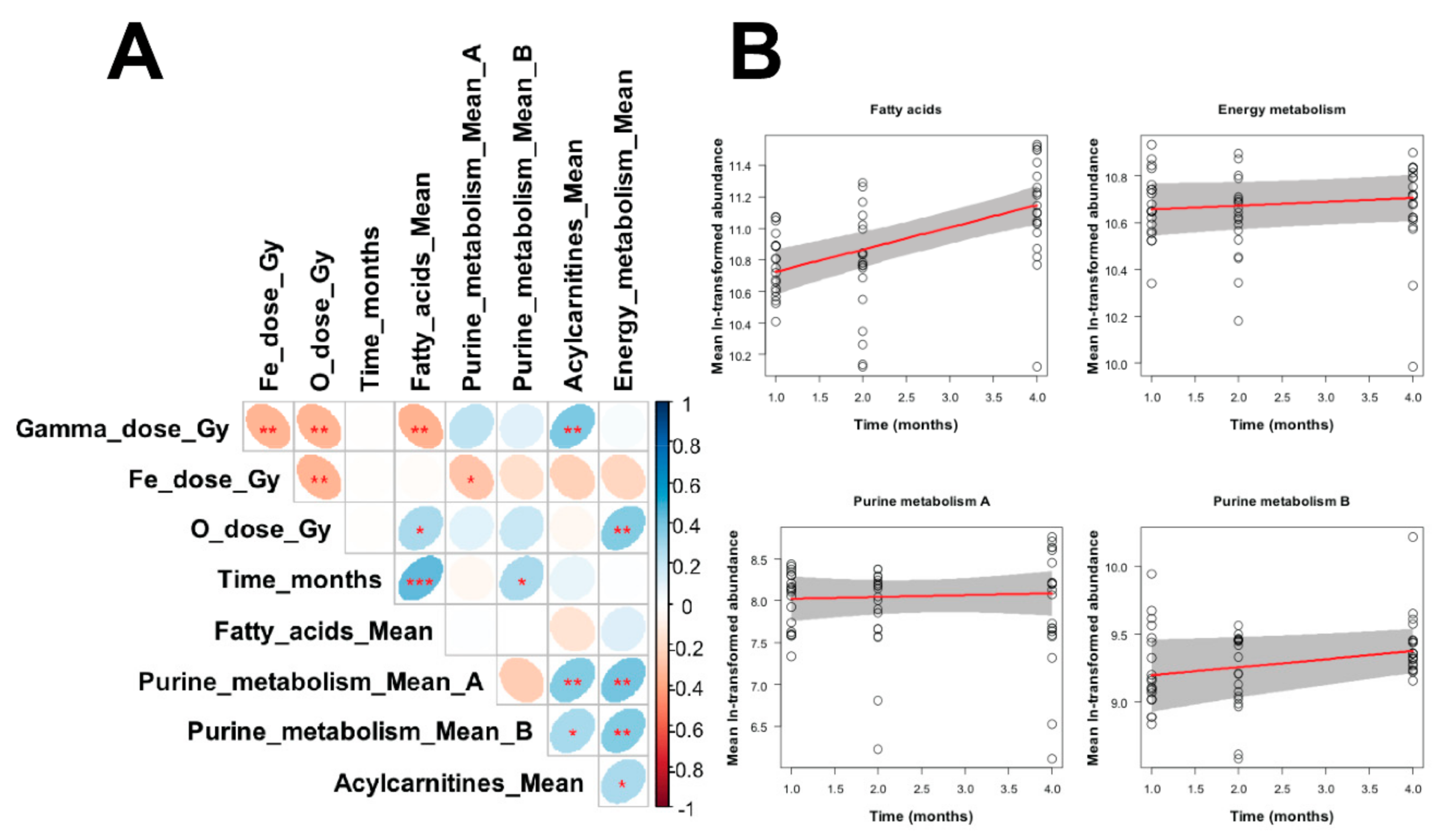
| Adduct | Metabolite | m/z | Ret. Time | Ppm Error | Two way ANOVA p Value (Time Factor) | Organic Compound Class |
|---|---|---|---|---|---|---|
| [M + H] + | Docosahexaenoic acid | 329.2476 | 6.97 | 0.35 | 0.0003 | Polyunsaturated fatty acid (PUFA) |
| [M + H] + | Phenylalanine | 166.0866 | 0.49 | 2.21 | 0.0338 | Amino acid |
| [M + H] + | Guanosine monophosphate (GMP) | 364.0653 | 0.44 | 0.04 | 0.3411 | Purine ribonucleoside monophosphate |
| [M + NH4] + | Xanthine | 170.0658 | 0.33 | 9.33 | 0.4095 | Purine derivative |
| [M + H] + | Oleoylcarnitine | 426.3578 | 5.84 | 0.19 | 0.0107 | Acylcarnitine |
| [M + H] + | Guanine | 152.0571 | 0.44 | 3.03 | 0.0876 | Purine derivative |
| [M + Na] + | Glycerophosphocholine | 280.0925 | 0.38 | 2.23 | <0.0001 | Choline derivative |
| [M + H] + | Acetylcarnitine | 204.1235 | 0.41 | 2.35 | 0.01 | Acylcarnitine |
| [M + H] + | Octanoylcarnitine | 288.2172 | 3.5 | 0.98 | 0.2444 | Acylcarnitine |
| [M + H] + | Arginine | 175.1194 | 0.4 | 2.54 | 0.1893 | Amino acid |
| [M + H] + | Carnitine | 162.1128 | 0.41 | 2.32 | 0.1449 | Non-essential amino acid, vitamin |
| [M + H] + | N6,N6,N6-Trimethyl-L-lysine | 189.1601 | 0.35 | 1.71 | 0.8555 | Amino acid derivative |
| [M + H] + | Taurine | 126.0223 | 0.45 | 3.26 | 0.0043 | Amino acid derivative |
| [M − H] − | Taurine | 124.0061 | 0.38 | 9.85 | 0.0438 | |
| [M + H] + | Cytidine | 244.0924 | 0.43 | 1.38 | 0.1413 | Nucleoside |
| [M + H] + | Alpha-linolenic acid | 279.2322 | 6.7 | 1.53 | 0.0114 | PUFA |
| [M + Na] + | Inosine | 291.0689 | 0.43 | 3.8 | 0.0007 | Purine nucleoside |
| [M + Na] + | Hypoxanthine | 159.0282 | 0.41 | 3.83 | 0.0048 | Purine derivative |
| [M + Na] + | Adenosine monophosphate | 370.0512 | 0.41 | 3.04 | 0.0448 | Purine ribonucleoside monophosphate |
| [M + H] + | Hexanoylcarnitine | 260.1854 | 2.69 | 0.77 | 0.1674 | Acylcarnitine |
| [M + H] + | Nicotinamide (Niacinamide) | 123.0555 | 0.43 | 1.69 | 0.0041 | Pyridine derivative, vitamin |
| [M + H] + | Pipecolic acid | 130.0865 | 0.35 | 2.64 | 0.3599 | Amino acid derivative |
| [M + H] + | Docosapentaenoic acid | 331.2647 | 7.2 | 4.77 | <0.0001 | PUFA |
| [M + H] + | Glutathione reduced | 308.0912 | 0.43 | 0.64 | 0.0003 | Amino acid derivative |
| [M − H] − | Glutathione reduced | 306.0756 | 0.41 | 2.95 | <0.0001 | |
| [M − H] − | Citric acid | 191.0184 | 0.41 | 6.81 | 0.8517 | Weak organic acid |
| [M − H] − | Erythronic acid | 135.0291 | 0.41 | 5.32 | 0.1656 | Sugar acid |
| [M − H] − | Eicosapentaenoic acid | 301.2162 | 6.63 | 3.49 | 0.1158 | PUFA |
| [M − H] − | Histidine | 154.0612 | 0.37 | 6.14 | 0.0419 | Amino acid |
| [M − H] − | Lactic acid | 89.0241 | 0.44 | 2.52 | 0.005 | Organic acid |
| [M − H] − | Uric acid | 167.02 | 0.41 | 6.12 | 0.0283 | Purine derivative |
| [M − H] − | Adenosine diphosphate (ADP) | 426.0229 | 0.45 | 1.8 | <0.0001 | Purine ribonucleoside diphosphate |
| [M − H] − | Fructose-6-phosphate | 259.0217 | 0.43 | 2.57 | <0.0001 | Hexose phosphate |
| [M − H] − | Adenine | 134.0459 | 0.38 | 10.3 | 0.5518 | Purine base |
| [M − H] − | Succinate | 117.0186 | 0.45 | 5.35 | <0.0001 | Dicarboxylic acid |
| [M − H] − | Serine | 104.0347 | 0.37 | 0.88 | 0.0027 | Amino acid |
| [M − H] − | Uridine monophosphate (UMP) | 323.028 | 0.43 | 1.71 | 0.7291 | Pyrimidine ribonucleoside monophosphate |
| [M − H] − | Ribulose-5-phosphate | 229.0113 | 0.43 | 2.09 | 0.014 | Pentose phosphate |
| [M − H] − | Malic acid | 133.0133 | 0.41 | 6.79 | 0.0002 | Dicarboxylic acid |
| [M − H] − | Arachidonic acid | 303.2317 | 7.07 | 4.1 | 0.0605 | PUFA |
| Metabolite Grouping | Robust Linear Regression Parameters | |||
|---|---|---|---|---|
| Meaning | Best-fit Value | Standard Error | p Value | |
| Fatty acids | Intercept | 10.585 | 0.095 | <2 × 10−16 |
| Slope for gamma ray dose (Gy−1) | −0.243 | 0.106 | 0.025 | |
| Slope for O ion dose (Gy−1) | 0.88 | 0.572 | 0.13 | |
| Slope for Fe ion dose (Gy−1) | −0.375 | 0.379 | 0.326 | |
| Slope for time (months−1) | 0.141 | 0.028 | 4.5 × 10−6 | |
| Purine metabolism A * | Intercept | 7.997 | 0.185 | <2 × 10−16 |
| Slope for gamma ray dose (Gy−1) | 0.392 | 0.169 | 0.024 | |
| Slope for O ion dose (Gy−1) | 1.311 | 0.954 | 0.175 | |
| Slope for Fe ion dose (Gy−1) | −0.147 | 0.679 | 0.829 | |
| Slope for time (months−1) | 0.023 | 0.062 | 0.712 | |
| Purine metabolism B # | Intercept | 9.136 | 0.159 | <2 × 10−16 |
| Slope for gamma ray dose (Gy−1) | 0.136 | 0.115 | 0.241 | |
| Slope for O ion dose (Gy−1) | 0.771 | 0.666 | 0.253 | |
| Slope for Fe ion dose (Gy−1) | −0.22 | 0.587 | 0.709 | |
| Slope for time (months−1) | 0.06 | 0.028 | 0.035 | |
| Acylcarnitines | Intercept | 10.914 | 0.192 | <2 × 10−16 |
| Slope for gamma ray dose (Gy−1) | 0.319 | 0.162 | 0.054 | |
| Slope for O ion dose (Gy−1) | 0.154 | 0.95 | 0.872 | |
| Slope for Fe ion dose (Gy−1) | −0.122 | 0.866 | 0.888 | |
| Slope for time (months−1) | 0.025 | 0.044 | 0.578 | |
| Energy metabolism | Intercept | 10.639 | 0.066 | <2 × 10−16 |
| Slope for gamma ray dose (Gy−1) | −0.017 | 0.063 | 0.784 | |
| Slope for O ion dose (Gy−1) | 0.502 | 0.296 | 0.096 | |
| Slope for Fe ion dose (Gy−1) | −0.423 | 0.316 | 0.186 | |
| Slope for time (months−1) | 0.017 | 0.014 | 0.245 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Laiakis, E.C.; Shuryak, I.; Deziel, A.; Wang, Y.-W.; Barnette, B.L.; Yu, Y.; Ullrich, R.L.; Fornace, A.J., Jr.; Emmett, M.R. Effects of Low Dose Space Radiation Exposures on the Splenic Metabolome. Int. J. Mol. Sci. 2021, 22, 3070. https://doi.org/10.3390/ijms22063070
Laiakis EC, Shuryak I, Deziel A, Wang Y-W, Barnette BL, Yu Y, Ullrich RL, Fornace AJ Jr., Emmett MR. Effects of Low Dose Space Radiation Exposures on the Splenic Metabolome. International Journal of Molecular Sciences. 2021; 22(6):3070. https://doi.org/10.3390/ijms22063070
Chicago/Turabian StyleLaiakis, Evagelia C., Igor Shuryak, Annabella Deziel, Yi-Wen Wang, Brooke L. Barnette, Yongjia Yu, Robert L. Ullrich, Albert J. Fornace, Jr., and Mark R. Emmett. 2021. "Effects of Low Dose Space Radiation Exposures on the Splenic Metabolome" International Journal of Molecular Sciences 22, no. 6: 3070. https://doi.org/10.3390/ijms22063070
APA StyleLaiakis, E. C., Shuryak, I., Deziel, A., Wang, Y.-W., Barnette, B. L., Yu, Y., Ullrich, R. L., Fornace, A. J., Jr., & Emmett, M. R. (2021). Effects of Low Dose Space Radiation Exposures on the Splenic Metabolome. International Journal of Molecular Sciences, 22(6), 3070. https://doi.org/10.3390/ijms22063070








