Metabolic Fingerprinting of Murine L929 Fibroblasts as a Cell-Based Tumour Suppressor Model System for Methionine Restriction
Abstract
1. Introduction
2. Results
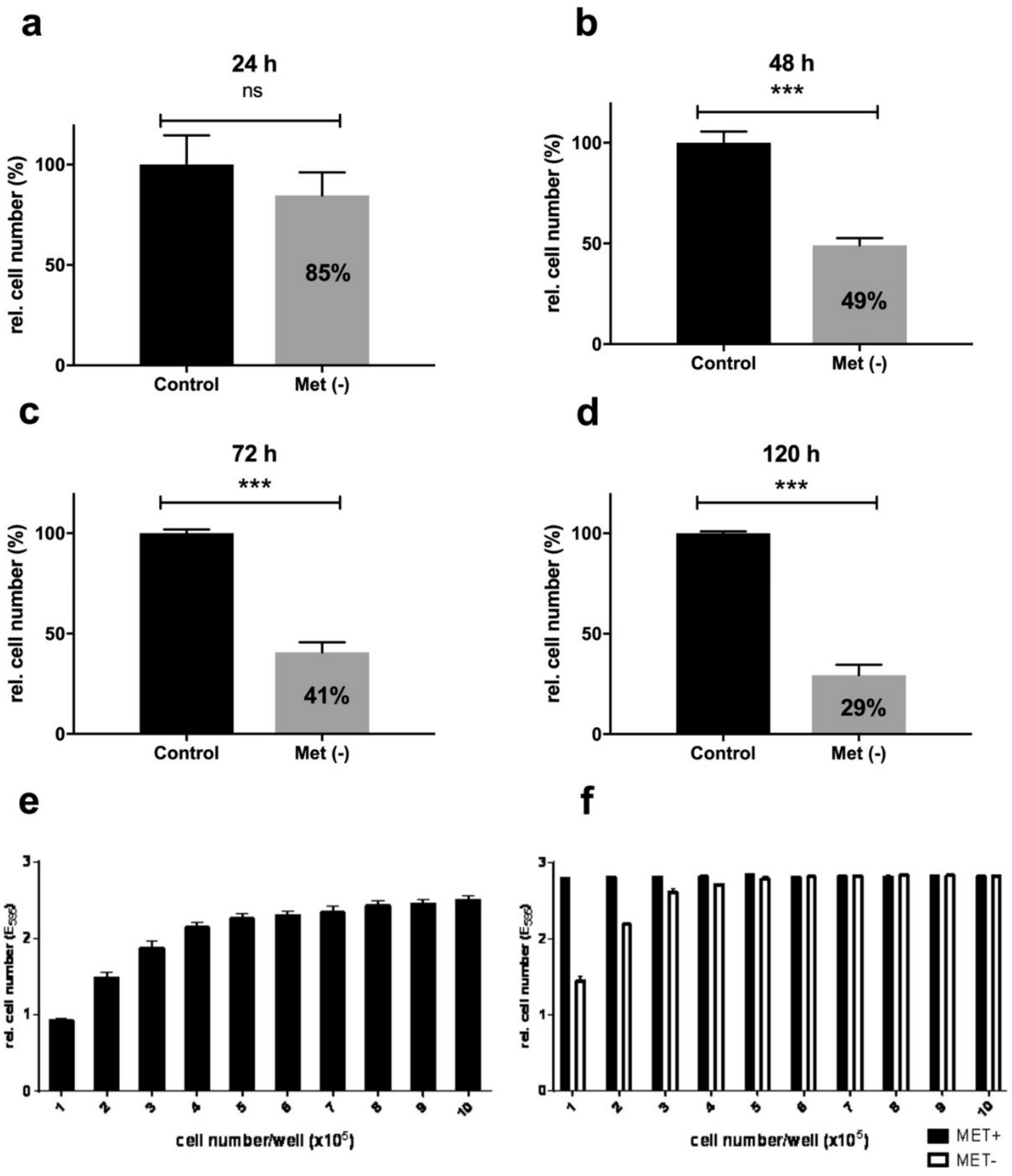
2.1. Methionine Restriction Inhibits Effective L929 Cell Proliferation
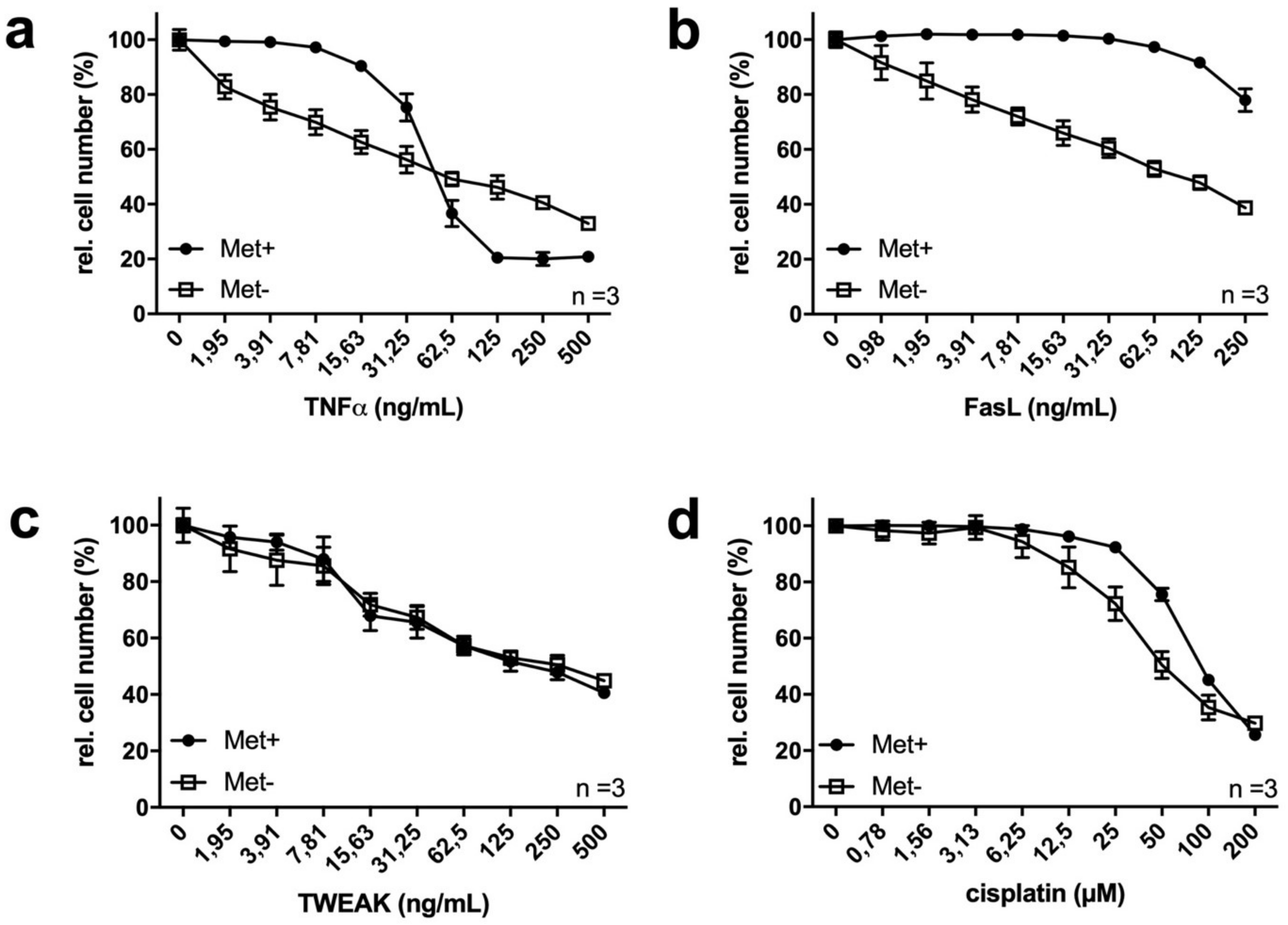
2.2. MetR Changes TNFα Ligand and Cytostatic Sensitivity in L929 Cells
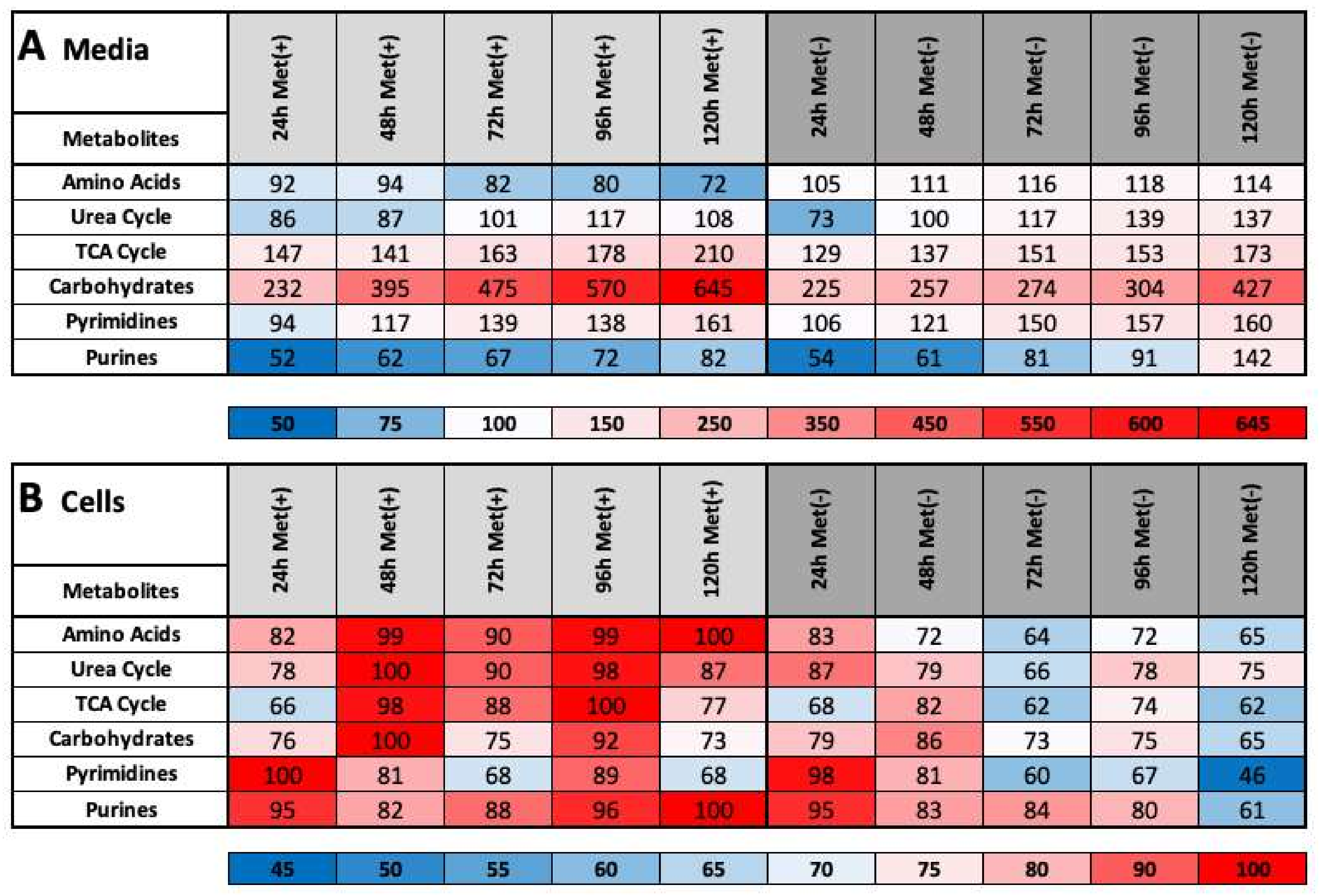
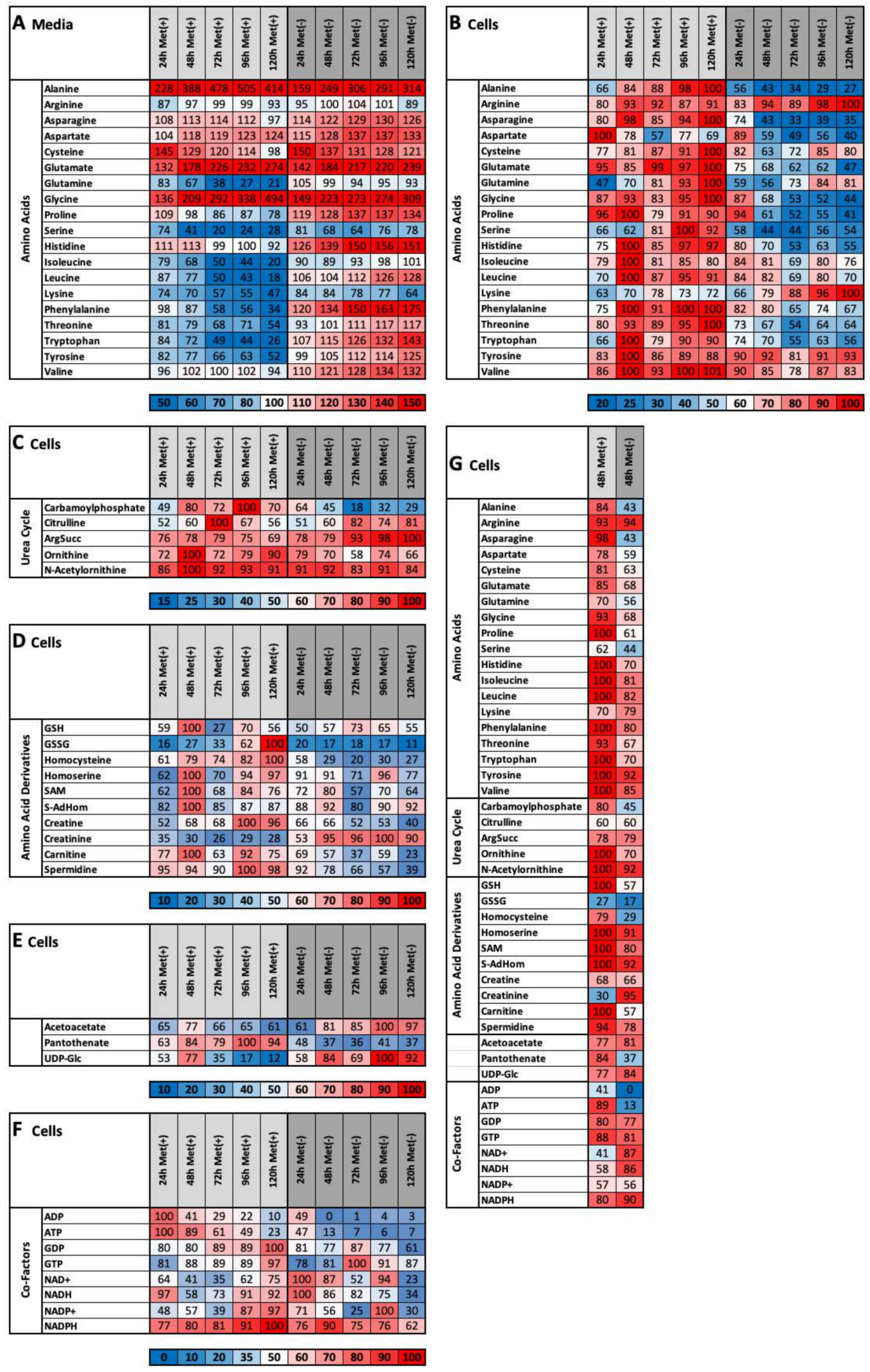
2.3. MetR Induces Metabolic Reprogramming in L929 Cells
- (1)
- important metabolic pathways
- (2)
- metabolites that are directly dependent on methionine
- (3)
- metabolites that are indirectly dependent on methionine
- (4)
- energy currencies (ATP, NADH, etc.)
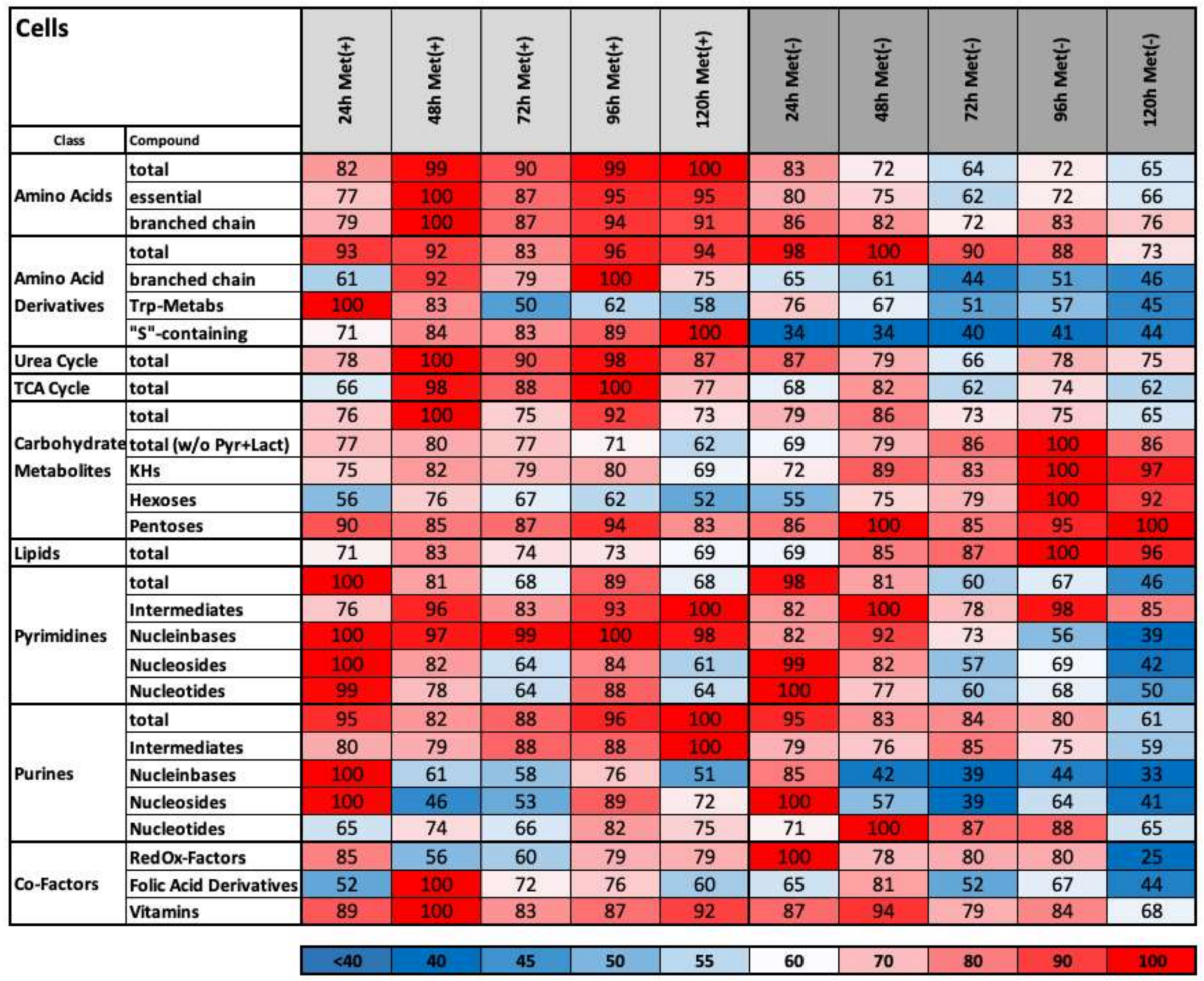
2.4. Methionine Restriction Induces a Characteristic Metabolic Fingerprint in L929 Cells
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Recombinant Protein Expression
4.3. Crystal Violet Staining (CytoTox Assay)
4.4. Liquid Chromatography/Mass Spectrometry
4.4.1. Cells
4.4.2. LC parameters
4.4.3. MS Parameters
4.4.4. Raw Data Analysis and Value Generation (In Short):
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Allemani, C.; Matsuda, T.; Di Carlo, V.; Harewood, R.; Matz, M.; Niksic, M.; Bonaventure, A.; Valkov, M.; Johnson, C.J.; Esteve, J.; et al. Global surveillance of trends in cancer survival 2000-14 (CONCORD-3): Analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet 2018, 391, 1023–1075. [Google Scholar] [CrossRef]
- Wong, M.C.S.; Lao, X.Q.; Ho, K.F.; Goggins, W.B.; Tse, S.L.A. Incidence and mortality of lung cancer: Global trends and association with socioeconomic status. Sci. Rep. 2017, 7, 14300. [Google Scholar] [CrossRef]
- Nishino, M.; Jagannathan, J.P.; Krajewski, K.M.; O’Regan, K.; Hatabu, H.; Shapiro, G.; Ramaiya, N.H. Personalized tumor response assessment in the era of molecular medicine: Cancer-specific and therapy-specific response criteria to complement pitfalls of RECIST. AJR Am. J. Roentgenol. 2012, 198, 737–745. [Google Scholar] [CrossRef]
- Tonellato, P.J.; Crawford, J.M.; Boguski, M.S.; Saffitz, J.E. A national agenda for the future of pathology in personalized medicine: Report of the proceedings of a meeting at the Banbury Conference Center on genome-era pathology, precision diagnostics, and preemptive care: A stakeholder summit. Am. J. Clin. Pathol. 2011, 135, 668–672. [Google Scholar] [CrossRef] [PubMed]
- Liberti, M.V.; Locasale, J.W. The Warburg effect: How does it benefit cancer cells? Trends Biochem. Sci. 2016, 41, 211–218. [Google Scholar] [CrossRef]
- Fontana, L.; Partridge, L. Promoting health and longevity through diet: From model organisms to humans. Cell 2015, 161, 106–118. [Google Scholar] [CrossRef] [PubMed]
- Gokarn, R.; Solon-Biet, S.M.; Cogger, V.C.; Cooney, G.J.; Wahl, D.; McMahon, A.C.; Mitchell, J.R.; Mitchell, S.J.; Hine, C.; de Cabo, R.; et al. Long-term dietary macronutrients and hepatic gene expression in aging mice. J. Gerontol. A Biol. Sci. Med. Sci. 2018, 73, 1618–1625. [Google Scholar] [CrossRef] [PubMed]
- Escobar, K.A.; Cole, N.H.; Mermier, C.M.; VanDusseldorp, T.A. Autophagy and aging: Maintaining the proteome through exercise and caloric restriction. Aging Cell 2019, 18, e12876. [Google Scholar] [CrossRef]
- Parkhitko, A.A.; Jouandin, P.; Mohr, S.E.; Perrimon, N. Methionine metabolism and methyltransferases in the regulation of aging and lifespan extension across species. Aging Cell 2019, 18, e13034. [Google Scholar] [CrossRef]
- Wanders, D.; Ghosh, S.; Stone, K.P.; Van, N.T.; Gettys, T.W. Transcriptional impact of dietary methionine restriction on systemic inflammation: Relevance to biomarkers of metabolic disease during aging. Biofactors 2014, 40, 13–26. [Google Scholar] [CrossRef] [PubMed]
- Sugimura, T.; Birnbaum, S.M.; Winitz, M.; Greenstein, J.P. Quantitative nutritional studies with water-soluble, chemically defined diets. VIII. The forced feeding of diets each lacking in one essential amino acid. Arch. Biochem. Biophys. 1959, 81, 448–455. [Google Scholar] [CrossRef]
- Ables, G.P.; Hens, J.R.; Nichenametla, S.N. Methionine restriction beyond life-span extension. Ann. N. Y. Acad. Sci. 2016, 1363, 68–79. [Google Scholar] [CrossRef] [PubMed]
- Levine, M.E.; Suarez, J.A.; Brandhorst, S.; Balasubramanian, P.; Cheng, C.W.; Madia, F.; Fontana, L.; Mirisola, M.G.; Guevara-Aguirre, J.; Wan, J.; et al. Low protein intake is associated with a major reduction in IGF-1, cancer, and overall mortality in the 65 and younger but not older population. Cell Metab. 2014, 19, 407–417. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, S.; Varshney, U. Evolution of initiator tRNAs and selection of methionine as the initiating amino acid. RNA Biol. 2016, 13, 810–819. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Liu, S.; Liu, K.; Abbasi, I.H.R.; Cai, C.; Yao, J. Molecular mechanisms relating to amino acid regulation of protein synthesis. Nutr. Res. Rev. 2019, 32, 183–191. [Google Scholar] [CrossRef]
- Lu, S.C. S-Adenosylmethionine. Int. J. Biochem. Cell Biol. 2000, 32, 391–395. [Google Scholar] [CrossRef]
- Cavuoto, P.; Fenech, M.F. A review of methionine dependency and the role of methionine restriction in cancer growth control and life-span extension. Cancer Treat. Rev. 2012, 38, 726–736. [Google Scholar] [CrossRef]
- Chaturvedi, S.; Hoffman, R.M.; Bertino, J.R. Exploiting methionine restriction for cancer treatment. Biochem. Pharmacol. 2018, 154, 170–173. [Google Scholar] [CrossRef]
- Madeo, F.; Carmona-Gutierrez, D.; Hofer, S.J.; Kroemer, G. Caloric restriction mimetics against age-associated disease: Targets, mechanisms, and therapeutic potential. Cell Metab. 2019, 29, 592–610. [Google Scholar] [CrossRef]
- Zhou, Y.; Ren, J.; Song, T.; Peng, J.; Wei, H. Methionine regulates mTORC1 via the T1R1/T1R3-PLCbeta-Ca(2+)-ERK1/2 signal transduction process in C2C12 cells. Int. J. Mol. Sci. 2016, 17, 1684. [Google Scholar] [CrossRef]
- Wang, S.Y.; Wang, W.J.; Liu, J.Q.; Song, Y.H.; Li, P.; Sun, X.F.; Cai, G.Y.; Chen, X.M. Methionine restriction delays senescence and suppresses the senescence-associated secretory phenotype in the kidney through endogenous hydrogen sulfide. Cell Cycle 2019, 18, 1573–1587. [Google Scholar] [CrossRef]
- Perrone, C.E.; Mattocks, D.A.; Plummer, J.D.; Chittur, S.V.; Mohney, R.; Vignola, K.; Orentreich, D.S.; Orentreich, N. Genomic and metabolic responses to methionine-restricted and methionine-restricted, cysteine-supplemented diets in Fischer 344 rat inguinal adipose tissue, liver and quadriceps muscle. J. Nutr. Nutr. 2012, 5, 132–157. [Google Scholar] [CrossRef] [PubMed]
- Pradas, I.; Jove, M.; Cabre, R.; Ayala, V.; Mota-Martorell, N.; Pamplona, R. Effects of aging and methionine restriction on rat kidney metabolome. Metabolites 2019, 9, 280. [Google Scholar] [CrossRef] [PubMed]
- Morris, S.M. Arginine metabolism revisited. J. Nutr. 2016, 146, 2579S–2586S. [Google Scholar] [CrossRef]
- Gil, J.; Ramirez-Torres, A.; Encarnacion-Guevara, S. Lysine acetylation and cancer: A proteomics perspective. J. Proteom. 2017, 150, 297–309. [Google Scholar] [CrossRef] [PubMed]
- Hensley, C.T.; Wasti, A.T.; DeBerardinis, R.J. Glutamine and cancer: Cell biology, physiology, and clinical opportunities. J. Clin. Investig. 2013, 123, 3678–3684. [Google Scholar] [CrossRef]
- Gillespie, Z.E.; Pickering, J.; Eskiw, C.H. Better living through chemistry: Caloric restriction (CR) and CR mimetics alter genome function to promote increased health and lifespan. Front. Genet. 2016, 7, 142. [Google Scholar] [CrossRef]
- Alves-Fernandes, D.K.; Jasiulionis, M.G. The role of SIRT1 on DNA damage response and epigenetic alterations in cancer. Int. J. Mol. Sci. 2019, 20, 3153. [Google Scholar] [CrossRef]
- Fang, E.F.; Lautrup, S.; Hou, Y.; Demarest, T.G.; Croteau, D.L.; Mattson, M.P.; Bohr, V.A. NAD(+) in aging: Molecular mechanisms and translational implications. Trends Mol. Med. 2017, 23, 899–916. [Google Scholar] [CrossRef] [PubMed]
- Wallimann, T.; Tokarska-Schlattner, M.; Schlattner, U. The creatine kinase system and pleiotropic effects of creatine. Amino Acids 2011, 40, 1271–1296. [Google Scholar] [CrossRef]
- Yang, L.; Venneti, S.; Nagrath, D. Glutaminolysis: A hallmark of cancer metabolism. Annu. Rev. Biomed. Eng. 2017, 19, 163–194. [Google Scholar] [CrossRef] [PubMed]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Morar, M.; Ealick, S.E. Structural biology of the purine biosynthetic pathway. Cell. Mol. Life Sci. 2008, 65, 3699–3724. [Google Scholar] [CrossRef] [PubMed]
- Rauert, H.; Wicovsky, A.; Muller, N.; Siegmund, D.; Spindler, V.; Waschke, J.; Kneitz, C.; Wajant, H. Membrane tumor necrosis factor (TNF) induces p100 processing via TNF receptor-2 (TNFR2). J. Biol. Chem. 2010, 285, 7394–7404. [Google Scholar] [CrossRef] [PubMed]
- Lang, I.; Fick, A.; Schafer, V.; Giner, T.; Siegmund, D.; Wajant, H. Signaling active CD95 receptor molecules trigger co-translocation of inactive CD95 molecules into lipid rafts. J. Biol. Chem. 2012, 287, 24026–24042. [Google Scholar] [CrossRef]
- Lieu, E.L.; Nguyen, T.; Rhyne, S.; Kim, J. Amino acids in cancer. Exp. Mol. Med. 2020, 52, 15–30. [Google Scholar] [CrossRef]
- Schulte, M.L.; Fu, A.; Zhao, P.; Li, J.; Geng, L.; Smith, S.T.; Kondo, J.; Coffey, R.J.; Johnson, M.O.; Rathmell, J.C.; et al. Pharmacological blockade of ASCT2-dependent glutamine transport leads to antitumor efficacy in preclinical models. Nat. Med. 2018, 24, 194–202. [Google Scholar] [CrossRef]
- Roos, C.; Wicovsky, A.; Muller, N.; Salzmann, S.; Rosenthal, T.; Kalthoff, H.; Trauzold, A.; Seher, A.; Henkler, F.; Kneitz, C.; et al. Soluble and transmembrane TNF-like weak inducer of apoptosis differentially activate the classical and noncanonical NF-kappa B pathway. J. Immunol. 2010, 185, 1593–1605. [Google Scholar] [CrossRef]
- Rajeeve, K.; Vollmuth, N.; Janaki-Raman, S.; Wulff, T.F.; Baluapuri, A.; Dejure, F.R.; Huber, C.; Fink, J.; Schmalhofer, M.; Schmitz, W.; et al. Reprogramming of host glutamine metabolism during Chlamydia trachomatis infection and its key role in peptidoglycan synthesis. Nat. Microbiol. 2020, 5, 1390–1402. [Google Scholar] [CrossRef] [PubMed]
- Bohnert, S.; Reinert, C.; Trella, S.; Schmitz, W.; Ondruschka, B.; Bohnert, M. Metabolomics in postmortem cerebrospinal fluid diagnostics: A state-of-the-art method to interpret central nervous system-related pathological processes. Int. J. Leg. Med. 2021, 135, 183–191. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schmitz, W.; Koderer, C.; El-Mesery, M.; Gubik, S.; Sampers, R.; Straub, A.; Kübler, A.C.; Seher, A. Metabolic Fingerprinting of Murine L929 Fibroblasts as a Cell-Based Tumour Suppressor Model System for Methionine Restriction. Int. J. Mol. Sci. 2021, 22, 3039. https://doi.org/10.3390/ijms22063039
Schmitz W, Koderer C, El-Mesery M, Gubik S, Sampers R, Straub A, Kübler AC, Seher A. Metabolic Fingerprinting of Murine L929 Fibroblasts as a Cell-Based Tumour Suppressor Model System for Methionine Restriction. International Journal of Molecular Sciences. 2021; 22(6):3039. https://doi.org/10.3390/ijms22063039
Chicago/Turabian StyleSchmitz, Werner, Corinna Koderer, Mohamed El-Mesery, Sebastian Gubik, Rene Sampers, Anton Straub, Alexander Christian Kübler, and Axel Seher. 2021. "Metabolic Fingerprinting of Murine L929 Fibroblasts as a Cell-Based Tumour Suppressor Model System for Methionine Restriction" International Journal of Molecular Sciences 22, no. 6: 3039. https://doi.org/10.3390/ijms22063039
APA StyleSchmitz, W., Koderer, C., El-Mesery, M., Gubik, S., Sampers, R., Straub, A., Kübler, A. C., & Seher, A. (2021). Metabolic Fingerprinting of Murine L929 Fibroblasts as a Cell-Based Tumour Suppressor Model System for Methionine Restriction. International Journal of Molecular Sciences, 22(6), 3039. https://doi.org/10.3390/ijms22063039





