Selecting Subpopulations of High-Quality Protein Conformers among Conformational Mixtures of Recombinant Bovine MMP-9 Solubilized from Inclusion Bodies
Abstract
1. Introduction
2. Results and Discussion
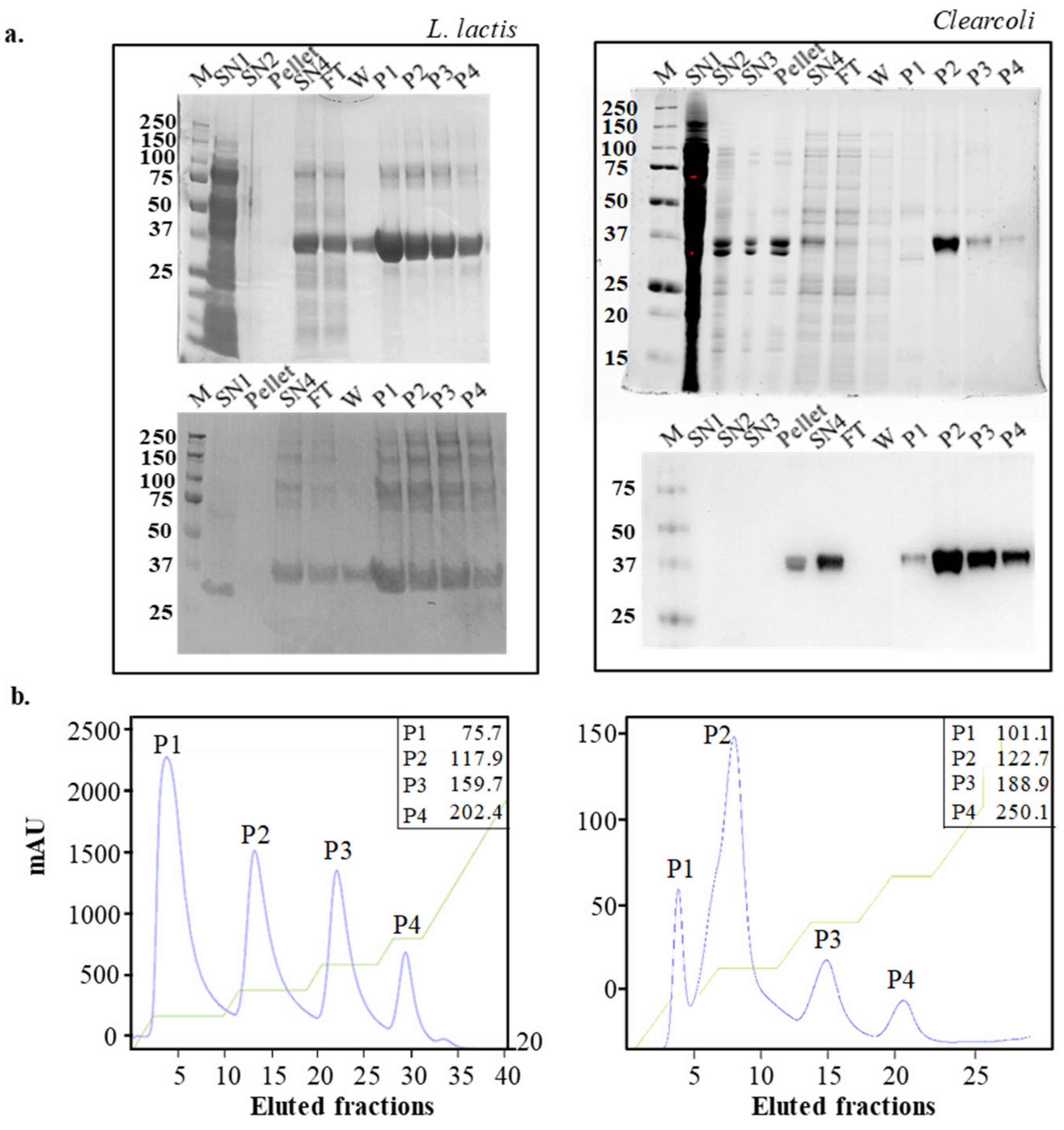
2.1. Protein Production of Soluble MMP-9 in IBs
2.2. Solubilization of Recombinant MMP-9 from IBs
2.3. Purification of Recombinant MMP-9 by Affinity Chromatography
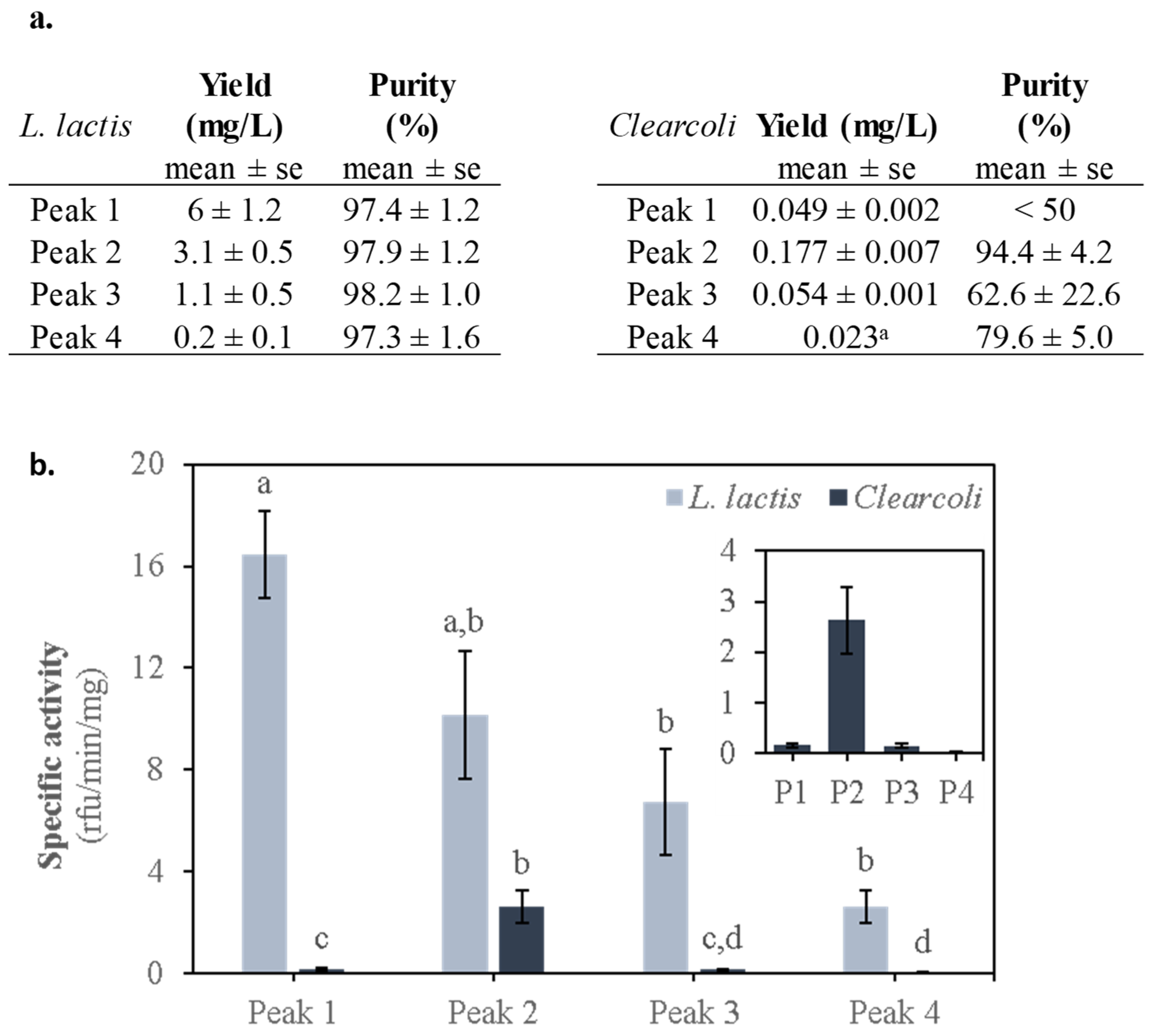
2.4. Activity of the MMP-9 Protein Peaks of L. Lactis and Clearcoli Obtained from IBs
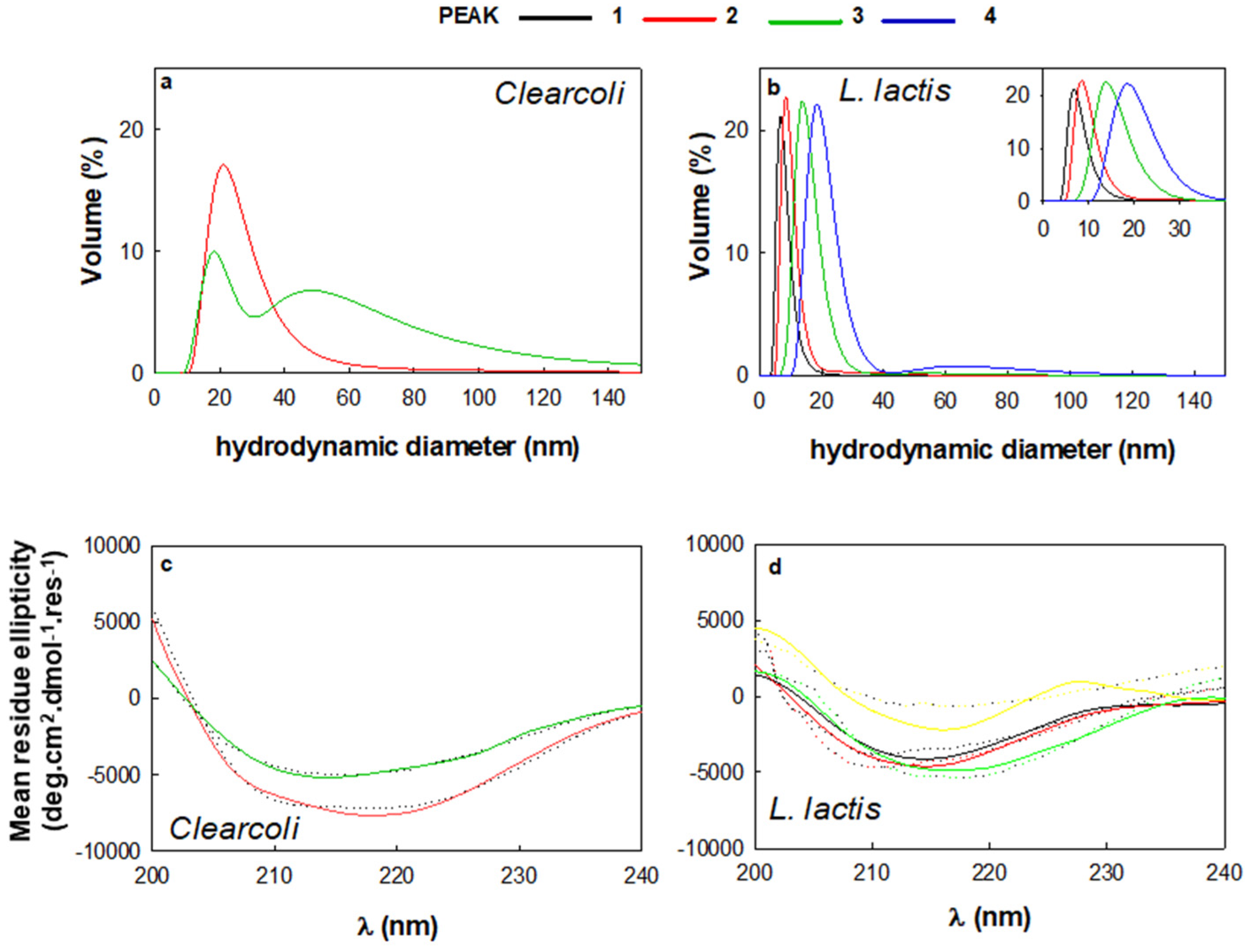
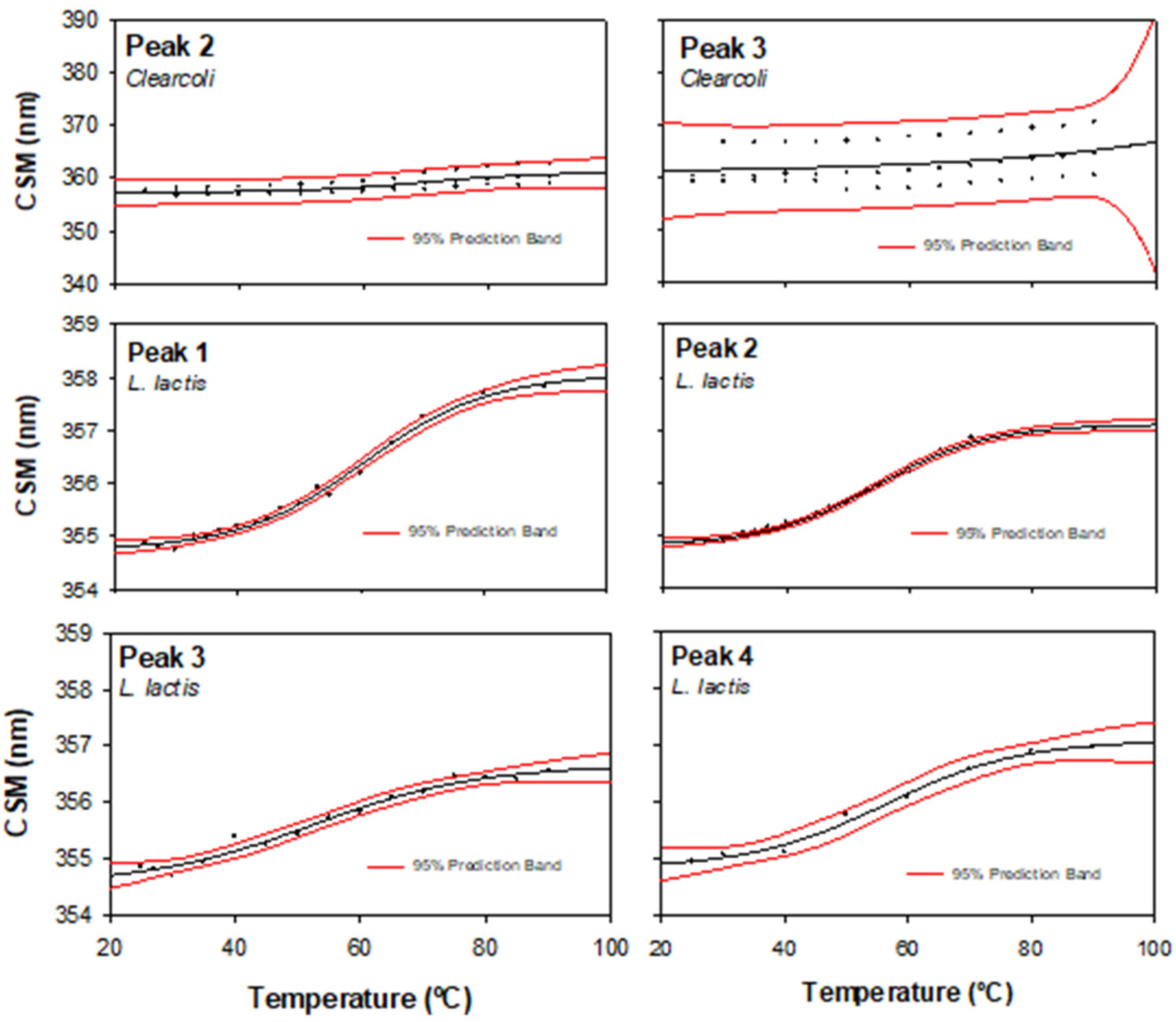
2.5. Physicochemical MMP-9 Properties
3. Materials and Methods
3.1. Bacteria Strains and Plasmids
3.2. Bacteria Strains and Plasmids
3.3. Protein Purification
3.4. Protein Detection, Yield and Purity
3.5. MMP-9 Activity Determination by DQgelatinTM Degradation Kinetics
3.6. Dynamic Light Scattering (DLS)s
3.7. Determination of Intrinsic Fluorescence
3.8. Circular Dichroism (CD)
3.9. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
Appendix A.1. Bacterial Strains and Plasmids
Appendix A.2. Protein Purification
Appendix A.3. MMP-9 Activity Determination by DQgelatinTM Degradation Kinetics
Appendix A.4. Inductively Coupled Plasma-Mass Spectrometry (ICP-MS) Analysis
References
- Ferrer-Miralles, N.; Villaverde, A. Bacterial cell factories for recombinant protein production; expanding the catalogue. Microb. Cell Fact. 2013, 12. [Google Scholar] [CrossRef]
- Sanchez-Garcia, L.; Martín, L.; Mangues, R.; Ferrer-Miralles, N.; Vázquez, E.; Villaverde, A. Recombinant pharmaceuticals from microbial cells: A 2015 update. Microb. Cell Fact. 2016, 15. [Google Scholar] [CrossRef]
- Villaverde, A.; Corchero, J.L.; Seras-Franzoso, J.; Garcia-Fruitos, E. Functional protein aggregates: Just the tip of the iceberg. Nanomedicine 2015, 10, 2881–2891. [Google Scholar] [CrossRef] [PubMed]
- De Marco, A.; Ferrer-Miralles, N.; Garcia-Fruitós, E.; Mitraki, A.; Peternel, S.; Rinas, U.; Trujillo-Roldán, M.A.; Valdez-Cruz, N.A.; Vázquez, E.; Villaverde, A. Bacterial inclusion bodies are industrially exploitable amyloids. FEMS Microbiol. Rev. 2019, 43. [Google Scholar] [CrossRef]
- Singh, S.M.; Panda, A.K. Solubilization and refolding of bacterial inclusion body proteins. J. Biosci. Bioeng. 2005, 99, 303–310. [Google Scholar] [CrossRef]
- Upadhyay, A.K.; Murmu, A.; Singh, A.; Panda, A.K. Kinetics of inclusion body formation and its correlation with the characteristics of protein aggregates in Escherichia coli. PLoS ONE 2012, 7, e33951. [Google Scholar] [CrossRef]
- Elia, F.; Cantini, F.; Chiti, F.; Dobson, C.M.; Bemporad, F. Direct Conversion of an Enzyme from Native-like to Amyloid-like Aggregates within Inclusion Bodies. Biophys. J. 2017, 112, 2540–2551. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Upadhyay, V.; Panda, A.K. Solubilization and refolding of inclusion body proteins. Methods Mol. Biol. 2015, 1258, 283–291. [Google Scholar] [CrossRef] [PubMed]
- Palmer, I.; Wingfield, P.T. Preparation and extraction of insoluble (inclusion-body) proteins from Escherichia coli. Curr. Protoc. Protein Sci. 2012, 70. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; van Oosterwijk, N.; Ali, A.M.; Adawy, A.; Anindya, A.L.; Domling, A.S.S.; Groves, M.R. A Systematic Protein Refolding Screen Method using the DGR Approach Reveals that Time and Secondary TSA are Essential Variables. Sci. Rep. 2017, 7, 9355. [Google Scholar] [CrossRef] [PubMed]
- Gifre-Renom, L.; Cano-Garrido, O.; Fàbregas, F.; Roca-Pinilla, R.; Seras-Franzoso, J.; Ferrer-Miralles, N.; Villaverde, A.; Bach, À.; Devant, M.; Arís, A.; et al. A new approach to obtain pure and active proteins from Lactococcus lactis protein aggregates. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Upadhyay, V.; Upadhyay, A.K.; Singh, S.M.; Panda, A.K. Protein recovery from inclusion bodies of Escherichia coli using mild solubilization process. Microb. Cell Fact. 2015, 14, 41. [Google Scholar] [CrossRef]
- Peternel, S.; Grdadolnik, J.; Gaberc-Porekar, V.; Komel, R. Engineering inclusion bodies for non denaturing extraction of functional proteins. Microb. Cell Fact. 2008, 7, 34. [Google Scholar] [CrossRef]
- Seras-Franzoso, J.; Peternel, S.; Cano-Garrido, O.; Villaverde, A.; Garcia-Fruitos, E. Bacterial inclusion body purification. Methods Mol. Biol. 2015, 1258, 293–305. [Google Scholar] [PubMed]
- Cano-Garrido, O.; Sánchez-Chardi, A.; Parés, S.; Giró, I.; Tatkiewicz, W.I.; Ferrer-Miralles, N.; Ratera, I.; Natalello, A.; Cubarsi, R.; Veciana, J.; et al. Functional protein-based nanomaterial produced in microorganisms recognized as safe: A new platform for biotechnology. Acta Biomater. 2016, 43, 230–239. [Google Scholar] [CrossRef] [PubMed]
- Bonnans, C.; Chou, J.; Werb, Z. Remodelling the extracellular matrix in development and disease. Nat. Rev. Mol. Cell Biol. 2014, 15, 786–801. [Google Scholar] [CrossRef]
- Page-McCaw, A.; Ewald, A.J.; Werb, Z. Matrix metalloproteinases and the regulation of tissue remodelling. Nat. Rev. Mol. Cell Biol. 2007, 8, 221–233. [Google Scholar] [CrossRef] [PubMed]
- Gifre-Renom, L.; Ugarte-Berzal, E.; Martens, E.; Boon, L.; Cano-Garrido, O.; Martínez-Núñez, E.; Luque, T.; Roca-Pinilla, R.; Conchillo-Solé, Ò.; Ferrer-Miralles, N.; et al. Recombinant protein-based nanoparticles: Elucidating their inflammatory effects in vivo and their potential as a new therapeutic format. Pharmaceutics 2020, 12, 450. [Google Scholar] [CrossRef]
- Mohseni, S.; Moghadam, T.T.; Dabirmanesh, B.; Khajeh, K. Expression, purification, refolding and in vitro recovery of active full length recombinant human gelatinase MMP-9 in Escherichia coli. Protein Expr. Purif. 2016, 126, 42–48. [Google Scholar] [CrossRef]
- Mohana-Borges, R.; Silva, J.L.; Ruiz-Sanz, J.; de Prat-Gay, G. Folding of a pressure-denatured model protein. Proc. Natl. Acad. Sci. USA 1999, 96, 7888–7893. [Google Scholar] [CrossRef] [PubMed]
- Peternel, S.; Komel, R. Isolation of biologically active nanomaterial (inclusion bodies) from bacterial cells. Microb. Cell Fact. 2010, 9, 66. [Google Scholar] [CrossRef]
- Padhiar, A.A.; Chanda, W.; Joseph, T.P.; Guo, X.; Liu, M.; Sha, L.; Batool, S.; Gao, Y.; Zhang, W.; Huang, M.; et al. Comparative study to develop a single method for retrieving wide class of recombinant proteins from classical inclusion bodies. Appl. Microbiol. Biotechnol. 2018, 102, 2363–2377. [Google Scholar] [CrossRef]
- Yamaguchi, H.; Miyazaki, M. Refolding techniques for recovering biologically active recombinant proteins from inclusion bodies. Biomolecules 2014, 4, 235–251. [Google Scholar] [CrossRef]
- Rosenblum, G.; Van den Steen, P.E.; Cohen, S.R.; Grossmann, J.G.; Frenkel, J.; Sertchook, R.; Slack, N.; Strange, R.W.; Opdenakker, G.; Sagi, I. Insights into the structure and domain flexibility of full-length pro-matrix metalloproteinase-9/gelatinase B. Structure 2007, 15, 1227–1236. [Google Scholar] [CrossRef] [PubMed]
- Vorackova, I.; Suchanova, S.; Ulbrich, P.; Diehl, W.E.; Ruml, T. Purification of proteins containing zinc finger domains using immobilized metal ion affinity chromatography. Protein Expr. Purif. 2011, 79, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Neznansky, A.; Opatowsky, Y. Expression, purification and crystallization of the phosphate-binding PstS protein from Pseudomonas aeruginosa. Acta Crystallogr. F Struct. Biol. Commun. 2014, 70, 906–910. [Google Scholar] [CrossRef] [PubMed]
- Cano-Garrido, O.; Rueda, F.L.; Sanchez-Garcia, L.; Ruiz-Avila, L.; Bosser, R.; Villaverde, A.; Garcia-Fruitos, E. Expanding the recombinant protein quality in Lactococcus lactis. Microb. Cell Fact. 2014, 13, 167. [Google Scholar] [CrossRef] [PubMed]
- Berkowitz, S.A.; Engen, J.R.; Mazzeo, J.R.; Jones, G.B. Analytical tools for characterizing biopharmaceuticals and the implications for biosimilars. Nat. Rev. Drug Discov. 2012, 11, 527–540. [Google Scholar] [CrossRef]
- Sahin, E.; Roberts, C.J. Size-exclusion chromatography with multi-angle light scattering for elucidating protein aggregation mechanisms. Methods Mol. Biol. 2012, 899, 403–423. [Google Scholar] [CrossRef]
- Wilkins, D.K.; Grimshaw, S.B.; Receveur, V.; Dobson, C.M.; Jones, J.A.; Smith, L.J. Hydrodynamic radii of native and denatured proteins measured by pulse field gradient NMR techniques. Biochemistry 1999, 38, 16424–16431. [Google Scholar] [CrossRef]
- Sanchez, J.M.; Sanchez-Garcia, L.; Pesarrodona, M.; Serna, N.; Sanchez-Chardi, A.; Unzueta, U.; Mangues, R.; Vazquez, E.; Villaverde, A. Conformational Conversion during Controlled Oligomerization into Nonamylogenic Protein Nanoparticles. Biomacromolecules 2018, 19, 3788–3797. [Google Scholar] [CrossRef]
- Houston, S.; Hof, R.; Francescutti, T.; Hawkes, A.; Boulanger, M.J.; Cameron, C.E. Bifunctional role of the Treponema pallidum extracellular matrix binding adhesin Tp0751. Infect. Immun. 2011, 79, 1386–1398. [Google Scholar] [CrossRef]
- Bujacz, G.; Alexandratos, J.; Wlodawer, A.; Merkel, G.; Andrake, M.; Katz, R.A.; Skalka, A.M. Binding of different divalent cations to the active site of avian sarcoma virus integrase and their effects on enzymatic activity. J. Biol. Chem. 1997, 272, 18161–18168. [Google Scholar] [CrossRef] [PubMed]
- Batoulis, H.; Schmidt, T.H.; Weber, P.; Schloetel, J.-G.; Kandt, C.; Lang, T. Concentration Dependent Ion-Protein Interaction Patterns Underlying Protein Oligomerization Behaviours. Sci. Rep. 2016, 6, 24131. [Google Scholar] [CrossRef]
- Salgado, E.N.; Lewis, R.A.; Mossin, S.; Rheingold, A.L.; Tezcan, F.A. Control of protein oligomerization symmetry by metal coordination: C2 and C3 symmetrical assemblies through CuII and NiII coordination. Inorg. Chem. 2009, 48, 2726–2728. [Google Scholar] [CrossRef] [PubMed]
- Malhotra, P.; Udgaonkar, J.B. How cooperative are protein folding and unfolding transitions? Protein Sci. 2016, 25, 1924–1941. [Google Scholar] [CrossRef] [PubMed]
- Otosu, T.; Ishii, K.; Tahara, T. Microsecond protein dynamics observed at the single-molecule level. Nat. Commun. 2015, 6, 7685. [Google Scholar] [CrossRef]
- Teschke, C.M.; Parent, K.N. “Let the phage do the work”: Using the phage P22 coat protein structures as a framework to understand its folding and assembly mutants. Virology 2010, 401, 119–130. [Google Scholar] [CrossRef]
- Stetefeld, J.; McKenna, S.A.; Patel, T.R. Dynamic light scattering: A practical guide and applications in biomedical sciences. Biophys. Rev. 2016, 8, 409–427. [Google Scholar] [CrossRef]
- Li, T.M.; Hook, J.W., III; Drickamer, H.G.; Weber, G. Plurality of pressure-denatured forms in chymotrypsinogen and lysozyme. Biochemistry 1976, 15, 5571–5580. [Google Scholar] [CrossRef] [PubMed]
- Ruan, K.; Weber, G. Hysteresis and conformational drift of pressure-dissociated glyceraldehydephosphate dehydrogenase. Biochemistry 1989, 28, 2144–2153. [Google Scholar] [CrossRef] [PubMed]
- Lakowicz, J.R.; Kusba, J.; Wiczk, W.; Gryczynski, I.; Szmacinski, H.; Johnson, M.L. Resolution of the conformational distribution and dynamics of a flexible molecule using frequency-domain fluorometry. Biophys. Chem 1991, 39, 79–84. [Google Scholar] [CrossRef]
- Sreerama, N.; Woody, R.W. Estimation of Protein Secondary Structure from Circular Dichroism Spectra: Comparison of CONTIN, SELCON, and CDSSTR Methods with an Expanded Reference Set. Anal. Biochem. 2000, 287, 252–260. [Google Scholar] [CrossRef]
- Provencher, S.W.; Glöckner, J. Estimation of globular protein secondary structure from circular dichroism. Biochemistry 1981, 20, 33–37. [Google Scholar] [CrossRef]
- Whitmore, L.; Wallace, B.A. Protein secondary structure analyses from circular dichroism spectroscopy: Methods and reference databases. Biopolymers 2008, 89, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Chou, H.T.; Kuo, T.Y.; Chiang, J.C.; Pei, M.J.; Yang, W.T.; Yu, H.C.; Lin, S.B.; Chen, W.J. Design and synthesis of cationic antimicrobial peptides with improved activity and selectivity against Vibrio spp. Int. J. Antimicrob. Agents 2008, 32, 130–138. [Google Scholar] [CrossRef] [PubMed]
- Giacalone, M.J.; Gentile, A.M.; Lovitt, B.T.; Berkley, N.L.; Gunderson, C.W.; Surber, M.W. Toxic protein expression in Escherichia coli using a rhamnose-based tightly regulated and tunable promoter system. Biotechniques 2006, 40, 355–364. [Google Scholar] [CrossRef] [PubMed]
| Peak 1 | Peak 2 | Peak 3 | Peak 4 | |
|---|---|---|---|---|
| Hydrodynamic diameter (nm) | ||||
| Clearcoli | n.d. | 21 ± 9 | 18.2 1 ± 5.3 | n.d. |
| (pdi = 0.6) | (pdi = 0.47) | |||
| L. lactis | 6.5 ± 2.1 | 8.04 ± 3.2 | 15.7 ± 4.1 | 18.2 ± 4.1 |
| (pdi = 0.7) | (pdi = 0.5) | (pdi = 0.7) | (pdi = 0.7) | |
| Center of spectral mass (CSM, nm) | ||||
| Clearcoli | n.d. | 357 ± 0.5 | 361 ± 9 | n.d. |
| L. lactis | 354.82 ± 0.1 | 354.89 ± 0.07 | 354.75 ± 0.2 | 354.92 ± 0.3 |
| Unfolding temperature (Tm, °C) | ||||
| Clearcoli | n.d. | 69.54 ± 8.2 | n.d. | n.d. |
| L. lactis | 60.6 ± 1.1 | 54.8 ± 0.7 | 52.4 ± 3.3 | 56.4 ± 2.5 |
| Peak 1 | Peak 2 | Peak 3 | Peak 4 | |
|---|---|---|---|---|
| Clearcoli | ||||
| Alpha helix | n.d. | 0.242 | 0.132 | n.d. |
| Beta sheet | n.d. | 0.273 | 0.336 | n.d. |
| Turns | n.d. | 0.217 | 0.213 | n.d. |
| Disordered | n.d. | 0.266 | 0.319 | n.d. |
| NRMSD | n.d. | 0.095 | 0.057 | n.d. |
| L. lactis | ||||
| Alpha helix | 0.091 | 0.125 | 0.06 | 0.039 |
| Beta sheet | 0.389 | 0.377 | 0.349 | 0.409 |
| Turns | 0.197 | 0.198 | 0.197 | 0.196 |
| Disordered | 0.323 | 0.3 | 0.394 | 0.356 |
| NRMSD | 0.491 | 0.411 | 0.176 | 0.498 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carratalá, J.V.; Gifre-Renom, L.; Roca-Pinilla, R.; Villaverde, A.; Arís, A.; Garcia-Fruitós, E.; Sánchez, J.M.; Ferrer-Miralles, N. Selecting Subpopulations of High-Quality Protein Conformers among Conformational Mixtures of Recombinant Bovine MMP-9 Solubilized from Inclusion Bodies. Int. J. Mol. Sci. 2021, 22, 3020. https://doi.org/10.3390/ijms22063020
Carratalá JV, Gifre-Renom L, Roca-Pinilla R, Villaverde A, Arís A, Garcia-Fruitós E, Sánchez JM, Ferrer-Miralles N. Selecting Subpopulations of High-Quality Protein Conformers among Conformational Mixtures of Recombinant Bovine MMP-9 Solubilized from Inclusion Bodies. International Journal of Molecular Sciences. 2021; 22(6):3020. https://doi.org/10.3390/ijms22063020
Chicago/Turabian StyleCarratalá, Jose Vicente, Laia Gifre-Renom, Ramon Roca-Pinilla, Antonio Villaverde, Anna Arís, Elena Garcia-Fruitós, Julieta María Sánchez, and Neus Ferrer-Miralles. 2021. "Selecting Subpopulations of High-Quality Protein Conformers among Conformational Mixtures of Recombinant Bovine MMP-9 Solubilized from Inclusion Bodies" International Journal of Molecular Sciences 22, no. 6: 3020. https://doi.org/10.3390/ijms22063020
APA StyleCarratalá, J. V., Gifre-Renom, L., Roca-Pinilla, R., Villaverde, A., Arís, A., Garcia-Fruitós, E., Sánchez, J. M., & Ferrer-Miralles, N. (2021). Selecting Subpopulations of High-Quality Protein Conformers among Conformational Mixtures of Recombinant Bovine MMP-9 Solubilized from Inclusion Bodies. International Journal of Molecular Sciences, 22(6), 3020. https://doi.org/10.3390/ijms22063020







