Abstract
The regenerative capacity of cardiomyocytes is insufficient to functionally recover damaged tissue, and as such, ischaemic heart disease forms the largest proportion of cardiovascular associated deaths. Human-induced pluripotent stem cells (hiPSCs) have enormous potential for developing patient specific cardiomyocytes for modelling heart disease, patient-based cardiac toxicity testing and potentially replacement therapy. However, traditional protocols for hiPSC-derived cardiomyocytes yield mixed populations of atrial, ventricular and nodal-like cells with immature cardiac properties. New insights gleaned from embryonic heart development have progressed the precise production of subtype-specific hiPSC-derived cardiomyocytes; however, their physiological immaturity severely limits their utility as model systems and their use for drug screening and cell therapy. The long-entrenched challenges in this field are being addressed by innovative bioengingeering technologies that incorporate biophysical, biochemical and more recently biomimetic electrical cues, with the latter having the potential to be used to both direct hiPSC differentiation and augment maturation and the function of derived cardiomyocytes and cardiac tissues by mimicking endogenous electric fields.
1. Introduction
Cardiovascular disease is the primary cause of death worldwide. In 2016, the World Health Organisation estimated that 17.9 million people died from cardiovascular disease, corresponding to 31% of deaths worldwide [1,2]. Ischaemic heart disease represents the largest proportion of cardiovascular related deaths; in 2015, approximately 8.92 million deaths globally were due to ischaemic myocardial infarctions [3]. This is particularly concerning considering that whilst cardiomyocytes are renewed in the adult heart under particular in vivo physiological or pathological conditions, their regenerative rate is limited [4,5]. More specifically, cardiomyocytes are regenerated at a rate of approximately 1% per year in healthy 25-year-old adults, progressively declining over the life span to approximately 0.45% annually in 75-year-old humans. Therefore, less than 50% of cardiomyocytes renew during an average life span [6,7]. Consequently, the rate of regeneration is insufficient to functionally restore damaged tissue via native processes necessary to overcome substantial myocardial damage and loss of cardiomyocytes associated with ischaemia [6,8].
Thus, emerging regenerative medicine therapies involving either the stimulation of endogenous regenerative processes or cell transplantation therapies have drawn increasing attention as cardiomyocyte replacement strategies [9]. Human-induced pluripotent stem cells (hiPSCs) offer much promise for developing patient-specific hiPSC-derived cardiomyocytes for modelling heart disease, pharmacology testing and regenerative replacement therapy [10,11,12] (Figure 1). However, the application of cardiomyocytes derived from hiPSCs is limited as current protocols result in mixed populations of atrial, ventricular and nodal-like cells with immature cardiac properties [13]. More specifically, hiPSC-derived cardiomyocytes display foetal-like properties in terms of morphology, electrophysiology, calcium handling, metabolism and gene expression compared to adult cardiomyocytes. Furthermore, hiPSC-derived cardiomyocyte culture methods do not provide 100% differentiation, with non-cardiomyocyte and undifferentiated cells contaminating cell hiPSC-derived cardiomyocyte cultures (Table 1) [14]. Such issues are also apparent in other leading applications for pluripotent stem cell derivatives. These include the production of oligodendrocytes, A9 dopaminergic neurons, pancreatic β-islet cells and retinal pigment epithelium to treat spinal cord injury, Parkinson’s disease, diabetes and age-related macular degeneration, respectively [15,16,17,18,19,20]. Overcoming the challenges of impurity and immaturity of iPSC-derived products is pertinent to applying hiPSC technology for regenerative medicine regardless of the disease target. Evidently, precise production of mature, subtype-specific cardiomyocytes without contaminating cells will drastically improve the translational applications of hiPSCs for disease modelling, drug screening, toxicological studies and cell therapy [21,22,23].
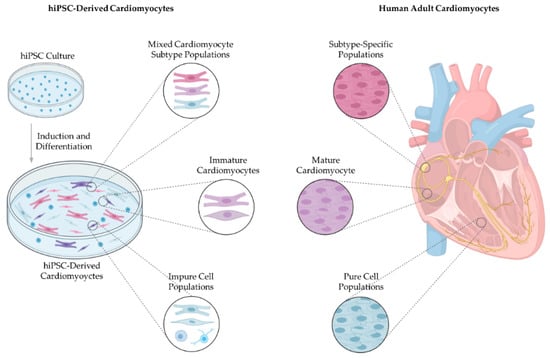
Figure 1.
Bioengineered cardiac tissue compared to native adult cardiac tissue. Current hiPSC cardiomyocyte differentiation protocols yield mixed cardiomyocyte subtype populations with immature structure and function, and cell cultures are contaminated with non-cardiomyocytes and undifferentiated cells. Developing a hiPSC-derived cardiomyocyte differentiation process driving subtype-specific, mature and pure cell cultures will be instrumental in extending the clinical application of hiPSC-derived cardiomyocytes. hiPSCs, human-induced pluripotent stem cells.
2. Clinical Need for Bioengineered Cardiomyocytes
The adult mammalian heart is not capable of repairing damaged tissue via native processes as cardiomyocytes have an insufficient regeneration rate [8]. Thus, cardiomyocytes are particularly vulnerable to multiple pathologies acute or chronic, such as apoptosis, necrosis and oncosis, which increase the risk of heart failure [24]. Dead cardiomyocytes accumulate at infarcted areas of the heart, subsequently activating host immune inflammatory responses, triggering the upregulation of cytokines and concomitant infiltration of neutrophils and macrophages to the area of necrotised tissue [25,26]. Consequently, matrix metalloproteases, a family of 25 enzymes, are activated, subsequently leading to the degradation of the surrounding coronary vasculature and the excessive extracellular matrix (ECM) turnover [27,28]. The collagen matrix becomes weakened, causing ventricular myocyte wall thinning and ventricular dilation as a result of mural realignment of myocyte bundles [29]. Fibrillary collagen deposited by fibroblast-like cells at injured sites results in the formation of scar tissue [30]. During this remodelling process, the collagen composition shifts from predominantly type I collagen to type III collagen. Hence, scar tissue with absent or reduced contraction capabilities and impulse propagation replaces regions of necrotic tissue arising from cardiomyocyte damage. The remodelling process, introducing scar tissue, becomes maladaptive due to a loss of contractile force and impulse propagation [31,32]. Hypertrophy of the remaining tissue occurs as a compensatory mechanism for the reduced cardiac function. The resultant cardiac overload is a progressive process leading to cardiac fibrosis and myocyte sliding displacement [33,34]. The end-stage of this degenerative process is heart failure, occurring when the metabolic demands of the body exceed the pumping ability of the heart. Maladaptive remodelling of the heart ultimately increases the risk of irreversible heart failure [35].
Notwithstanding significant success and progress in the treatment of myocardial infarction via pharmacological therapies, percutaneous coronary intervention or conventional coronary artery bypass graft surgery, the progression to heart failure remains high—approximately 15–30% [36,37]. Unfortunately, cardiomyocytes continue to degrade and lose contractile ability over time. Hence, patients with end-stage heart failure require more advanced treatments involving cardiac transplantation and mechanical assist devices [38]. The gold standard treatment for all causes of heart failure has been heart transplantation; however, it has a high mortality rate with a documented 15–20% mortality rate within the first year proceeding transplant [39,40]. Cardiac transplantation is also limited by the restricted availability of donor organs, potential cardiac allograft vasculopathy, malignancy and detrimental side effects associated with immunosuppression such as kidney disease, hypertension, diabetes mellitus and obesity [41]. Given the high incidence and severity of heart failure, more advanced treatments such as regenerative medicine using hiPSCs which aim to remuscularise damaged heart tissue are under investigation as potential cures for conditions associated with cardiomyocyte ischaemia [42]. Such treatments involve the implantation of exogenous hiPSC-derived cardiomyocytes to the damaged area for replacement of scar tissue with contractile tissue that is integrated electrically with the host cardiomyocytes [43].
Additionally, stem cell therapy is also being investigated as a therapeutic approach to overcome disturbances within the cardiac conduction system (CCS), particularly for pacemaker cells. The production of functional nodal cells holds great promise for advancing the treatment of cardiac disorders associated with the dysfunction or failure of sinoatrial (SA) nodal cells, collectively known as sick sinus syndrome [44,45]. The collective term sick sinus syndrome comprises sinus bradycardia, bradycardia-tachycardia syndrome, chronic atrial tachyarrhythmias, sinus pauses, sinoatrial node (SAN) exit block and inappropriate responses of heart rate to autonomic regulation [46]. Disturbances of the pacemaker cells can arise due to developmental and congenital defects, acquired injury or ischeamia or less commonly due to inherited diseases associated with alterations to the pacemaking function. The function of the SAN progressively declines with age, potentially resulting in age-related fibrosis or Lenegre–Lev syndrome [47]. Acute tissue destruction due to ischaemia or injury can occur suddenly, for example, during valve replacement surgery [48,49]. Moreover, disturbances to the cardiac conduction system may be congenital, such as congenital malformations including cardiac isomerisation and congenital heart blocks [50]. Furthermore, disturbances may arise during embryogenesis, resulting in errors in the function of the SAN and ectopic pacemakers, leading to conditions such as ventricular pre-excitation or Wolff–Parkinson–White syndrome [51]. Typically, symptomatic sick sinus syndrome requires the implantation of an electronic pacemaker [52].
There are several major drawbacks of electronic pacemakers with traditional methods of energy harvesting. For example, complications associated with technical failure of implanted systems and limited battery durability require monitoring and maintenance and increase the probability of recurrent surgery for battery and/or electrode replacement [53]. Moreover, traditional cardiac pacemakers fail to respond to the autonomic modulatory system; thereby, the heart rate fails to adapt to the demands of exercise and emotion [54]. Furthermore, the introduction of foreign material to the body is associated with infection. The incidence of pacemaker-associated infection varies from 0.1% to 20%, with 25% of pacemaker-associated infections occurring within two months of implantation. Alarmingly, cardiac pacemaker-related infection complications are associated with a mortality of up to 70%. The risk of pacemaker infection increases upon successive pacemaker related surgeries and in individuals with co-morbidities that are predisposed to pacemaker infection, such as malignancy, corticosteroid use, diabetes mellitus and skin diseases [55].
Cell-based therapies aim to address the hitherto mentioned limitations, potentially providing a long-term cure for SA nodal dysfunction. Additionally, hiPSC-derived cardiomyocytes offer opportunities to understand human heart development, cardiac disease modelling and investigation of the effects of therapeutic interventions.
3. Subtype-Specific Cardiomyocytes
The heart has two predominant categories of cardiomyocytes, i.e., contractile cardiomyocytes and non-contractile cardiomyocytes, which together preserve the heart’s mechanical, electrical and biochemical activity [56]. Contractile cardiomyocytes, the principal cardiomyocytes, comprise the myocardium, forming sheets of overlapping spiral patterns that attach to the fibrous skeleton of the heart [57,58]. Contractile cardiomyocytes have a distinct gene expression and cooperate with various cells of the heart, including endothelial cells, endocardial cells and epicardial-derived fibroblasts to perform the heart’s pumping action, subsequently projecting blood throughout the hearts chambers and into the peripheral circulatory system [59] (Figure 2B). The mechanical pumping is initiated by an electrophysiological stimulus to generate coordinated chronological electrical action potentials that result in muscle contraction [60].
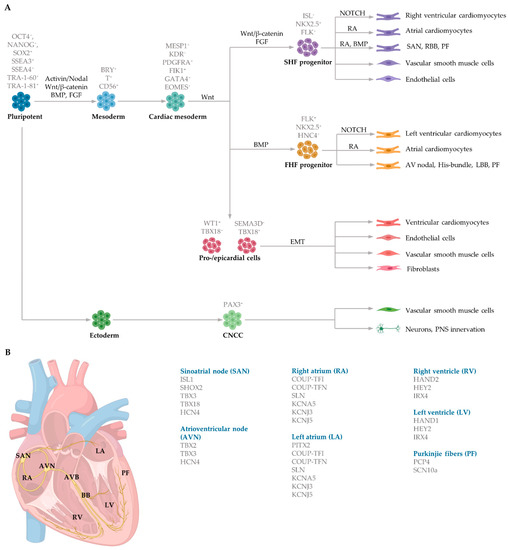
Figure 2.
Schematic of embryonic cardiac commitment highlighting (A) key signalling pathways regulating each differentiation stage during cardiogenesis, including cell markers expressed during each stage, and (B) gene expression in distinct regions of the human heart. Mesoderm induction is induced in response to Activin/Nodal, Wnt/β-catenin, BMP, and FGF signalling. Cardiac specification is regulated by MESP1, which coordinates the migration of early cardiovascular progenitors (ISL+) to the FHF and the SHF. The FHF progenitors generate the left ventricle, portions of the atria and conducting cells. The SHF progenitors contribute to the right ventricle, portions of the atria, conducting cells, vascular smooth muscle cells and endothelial cells. Epicardial cells undergo epithelial-to-mesenchymal transition to form fibroblasts, endothelial cells, vascular smooth muscle cells and parts of the ventricle. Ectoderm progenitors also generate vascular smooth muscle and neurons responsible for autonomic innervation. EMT, epithelial-to-mesenchymal transition. CNCC, cranial neural crest cells. RA, retinoic acid.
Electrophysiological initiation and propagation are made possible by specialised non-contractile cells, the second category of cardiomyocytes present in the heart [61]. The CCS collectively refers to non-contractile cardiac cells, comprised of several nodes and conducting cells responsible for generating and transducing electrical signals to control the rate and rhythm of heart muscle contraction [62,63] (Figure 2B). These include the SAN, atrioventricular node (AVN), atrioventricular bundle, right and left bundle branches (BB) and Purkinje fibres (collectively termed the subendocardial system) [64]. Automaticity is present within cells of the SAN, AVN, atrioventricular bundle, BB and Purkinje fibres [65]. However, SA nodal cells have a relatively faster depolarisation rate compared to other components of the CCS; therefore, the pacemaker activity of the SAN overrides automaticity of other structures and sets the heart’s pace [66,67]. Although, in cases of SAN failure, the automaticity of the AVN becomes dominant, acting to secure repetitive ventricular contraction [68]. Additionally, Purkinje fibres can, to a lesser extent, act as pacemakers; this is particularly true in cases of atrioventricular block [69,70]. Gap junctions electrically connect neighbouring cardiomyocytes, allowing the transmission of electrical energy between the components of the CCS and to the rest of the heart [71]. As a result, the heart contracts as a synchronised unit independent of neural input [72]. The concomitant electrophysiological activity of the CCS and subsequent rhythmic contraction of contractile cardiomyocytes subserve the function of the heart; namely, to distribute perfused blood to the body and remove cellular waste products [73].
3.1. Cardiomyocyte Embryonic Specification and Differentiation
The heart is the first functional organ to form during embryogenesis [74]. Human cardiovascular system development begins during gastrulation at the end of the second week of gestation when the developing embryo’s metabolic demands exceed that supplied from placental diffusion alone [75,76]. Following gastrulation, the mammalian heart begins to develop from progenitor cells within the anterior lateral plate mesoderm [77]. On approximately day 15 of gestation, the cardiac progenitor cells within the thoracic region condense into two bilaterally located endocardial tubes on either side of the trilaminar embryo.
Mesoderm induction is regulated by several distinct and overlapping signalling pathways, including Activin and Nodal, both members of the Transforming growth factor beta (TGFβ) family, alongside the canonical (β-catenin-dependent) Wingless (Wnt) signalling pathway (Figure 2A). Bone morphogenetic proteins (BMP), also members of the TGFβ family, and fibroblast growth factors (FGF) further aid in mesoderm determination in embryos [78,79]. The transcription factors homeobox protein NKX2.5 (NK2 transcription factor related, locus 5) and mesoderm posterior basic helix-loop-helix transcription factor 1 (MESP1) play a vital role in initiating cardiac commitment. NKX2.5 collaborates with the zinc finger protein transcription factors belonging to the GATA family to induce cardiomyocyte induction. Cardiac progenitors, cardiac mesoderm cells and cardiomyocytes in the left atria and ventricle express NKX2.5 [80,81]. Mesoderm induction commences in response to the secretion of growth factors from the adjacent endoderm germ layer. These factors include BMP4 and nodal signalling. The activation of the nodal signalling pathway in the proximal region of the primitive ectoderm initiates the expression of Bmp4 within the adjacent extraembryonic ectoderm. BMP4 in the extraembryonic ectoderm then stimulates Wnt3 expression in the epiblast, triggering the upregulation of mesoendodermal markers such as Brachyury (Bry) and Eomesodermin (EOMES), contributing to both the definitive endoderm and the cardiac mesoderm induction [82]. EOMES-positive cells then activate MESP1 to control early cardiac progenitor commitment. MESP1 is the primary regulator of multipotent cardiac progenitor specification, regulating the induction of cardiac and mesoderm genes to activate the migration of mesodermal cells to the anterior lateral plate mesoderm via the primitive streak [79,83]. MESP1 activates Dickkopf Wnt signalling pathway inhibitor 1 (DKK1), thereby inhibiting Wnt and promoting cardiac differentiation and maturation [84,85]. Interestingly, Wnt/β-catenin signalling during cardiogenesis is biphasic. Prior to gastrulation, activation of Wnt/β-catenin signalling promotes cardiac lineage specification, whereas activation during gastrulation inhibits cardiomyocyte differentiation [86,87].
During cardiac crescent formation, two distinguishable cell populations exist: cells within the splanchnic mesoderm and cells in the epithelial structure of the crescent [77]. These different populations of progenitor cells, referred to as the first heart field (FHF) and second heart field (SHF), produce different regions of the developing heart. MESP1 controls both the distinct populations of the FHF and SHF during early gastrulation which give rise to the precursors of the subtype-specific cardiomyocytes. Early cardiac progenitors, positive for the marker ISL1, induce the production of FHF and SHF progenitor cells. FHF progenitors (ISL1- NKX2.5+), which control myocardium specification, are regulated by BMP and contribute to portions of the atria and left ventricle and the atrioventricular bundle conduction cells [88]. The SHF progenitors (ISL1 + NKX2.5+) that regulate both myocardium and endocardium specification are controlled by FGF and WNT and give rise to portions of the atria, right ventricle and SAN conduction cells [89,90] (Figure 2A).
Cardiac mesoderm progenitors coalesce, forming two paired primordia heart tubes that are composed of both endocardial and myocardial lineage. Subsequently, FHF- and SHF-derived cardiac progenitor cells proliferate within the developing structure [91]. At approximately day 21 of gestation, the bilaterally paired tubes fuse, forming a primitive linear tube. Blood flows into the tube at the caudal side and outflows at the cranial side [74]. During early heart development, dominant pacemaker activity arises at the intake region at the caudal end of the primitive tube, referred to as the sinus venosus. The embryonic cells within the sinus venosus are characterised by the expression of HCN4, encoding the potassium/sodium hyperpolarized-activated cyclic nucleotide-gated channel 4 responsible for the hyperpolarisation-activated current essential for pacemaking ability [92]. The heart tube is elongated as FHF- and SHF-derived progenitor cardiac cells rapidly proliferate and differentiate into cardiomyocytes in response to NOTCH and retinoic acid signalling from the endocardium and epicardium, respectively. Retinoic acid, the active derivative of vitamin A, modifies the expression of critical genes to induce cardiomyocyte differentiation by the induction of fibroblast growth factor signalling [93]. NOTCH coordinates cardiac specification by interaction with other signalling pathways, including WNT and BMP [94].
3.2. Generation of Subtype-Specific Cardiomyocytes from Human Pluripotent Stem Cells
There are a variety of protocols enabling the generation of human cardiomyocytes from iPSCs [95]. Such approaches include the use of growth factors, small molecules, genetic manipulation and biophysical cues within both monolayer and three-dimensional (3D) cultures [96]. However, these studies typically yield mixed populations of atrial, ventricular and pacemaker-like cardiomyocytes [97,98,99]. The mixed subtypes of cardiomyocytes may distort innate cellular physiology, confound disease phenotypes and jeopardize the therapeutic effects of transplanted cardiomyocytes for cardiac tissue regeneration [92]. Accordingly, at the forefront of cardiac engineering is the ability to generate subtype-specific cardiomyocytes for the precise fabrication of atrial, ventricular and pacemaker-like hiPSC-derived cardiomyocytes. Recent research has identified that subtype-specific cardiomyocytes from hiPSCs can be generated by imitating embryonic heart chamber development [100,101]. These strategies have focused on the manipulation of the critical developmental signalling pathways of cardiogenesis, in particular, WNT, NOTCH and retinoic acid signalling pathways (Table 1) [22].
NOTCH signaling is vital for ventricular cardiomyocyte specification and morphogenesis. Moreover, NOTCH regulates coronary vessel specification and is a primary modulator of arteriovenous specification during vessel development [102]. The NOTCH signalling pathway consists of two families of membrane-bound ligands, Jagged1 and 2, and Delta-like (DLL)1, 3 and 4, that bind to four transmembrane-receptors, NOTCH 1 to 4, of an adjacent cell [103]. Ligand interaction with these receptors initiates a cascade of proteolytic cleavages, including the proteolytic cleavage-by-γ-secretase leading to the NOTCH intracellular domain release from the cell-membrane and subsequent translocation to the nucleus [104]. In the nucleus, the NOTCH intracellular domain reacts with the recombining binding protein suppressor of hairless protein, resulting in the expulsion of a histone deacetylase corepressor complex, thus giving rise to an activation complex [105]. Further, Mastermind-like protein 1 and histone acetyltransferase are recruited to the site, activating the transcription of NOTCH target genes, such as the basic helix–loop–helix transcriptional repressors HEY1 (Hairy/enhancer-of-split related with YRPW motif protein 1) and HEY2 (Hairy/enhancer-of-split related with YRPW motif protein 2). The activation of these NOTCH target genes results in ventricular chamber development and spatial arrangement of cardiomyocytes within the myocardium [94].
Recent studies have demonstrated that retinoic acid signalling can promote atrial-specific differentiation of hiPSCs through the upregulation of the transcription factor COUP-TFII [106]. Conversely, the lack of retinoic acid is associated with ventricular-specific differentiation [100]. In particular, retinoic acid aids in atrial/ventricular-specific hiPSC differentiation during a restrictive temporal window proceeding mesoderm commitment [22,23]. Moreover, retinoic acid-mediated chamber specific differentiation of hiPSCs depends on the distinct mesoderm progenitor population. Different concentrations of BMP4 and Activin A induce two mesoderm populations characterised by unique cell surface markers, i.e., CD235a+ CYP26A1+ and RALDH2+ ALDH+, which generate ventricular and atrial cardiomyocytes, respectively. During hiPSC differentiation, retinoic acid signalling will efficiently give rise to atrial cardiomyocytes in RALDH2+ ALDH+ mesoderm progenitors. Comparatively, a lack of retinoic acid signalling will generate ventricular cardiomyocytes in CD235a+ CYP26A1+ progenitors [22]. Further, retinoic acid signalling combined with WNT and BMP guides the differentiation of vascular smooth muscle and cardiac fibroblast lineage from hiPSCs. Moreover, the combined treatment of RA and BMP signalling directs hiPSC differentiation towards pacemaker-like cardiomyocytes [107]. Interestingly, the inhibition of COUP-TFII signalling ablates the repressing effect on NOTCH signalling, thereby indirectly promoting ventricular differentiation [106]. Inhibition of retinoic acid signalling indirectly promotes cardiac progenitors toward a ventricular-like subtype [100]. Additionally, combined treatment with BMP4, Activin A and the WNT signalling pathway inhibitor is highly efficient at stimulating hiPSC ventricular subtype specification in differentiating cardiomyocytes [108].
The production of pacemaker-like cardiomyocytes from hiPSCs has also been achieved by mirroring embryonic pacemaker lineage specification. Sinoatrial cells originate from TBX18+ NKX2.5− mesoderm progenitors, distinguishing them from chamber-specific cardiomyocytes and other components of the CCS [109]. Pacemaker-like cardiomyocyte induction using transient stage-specific exposure to FGF and BMP signalling with concomitant inhibition of Wnt signalling produced relatively pure populations of pacemaker-like cardiomyocytes [110]. However, more recently, Ren, et al. [111] demonstrated that canonical Wnt signalling directs pacemaker-like specification of differentiating hiPSC-derived cardiomyocytes. Wnt signalling promoted enhanced expression of pacemaker-specific genes such as SHOX2, HCN4, ISL1, TBX3 and TBX18 with a concomitant reduction in NKX2.5 expression. Wnt-activated cardiomyocytes demonstrated less negative resting membrane potentials corresponding with native pacemaker electrophysiological properties, thereby functionally supporting genetic alterations. Moreover, the Wnt-activated hiPSC-derived pacemaker-like cardiomyocytes augmented pacing of bioengineered “mini heart” models. Similarly, Protze, Liu, Nussinovitch, Ohana, Backx, Gepstein and Keller [107] also used a transgene-independent method to produce pacemaker-like cardiomyocytes from hiPSCs and human embryonic stem cells by stage-specific alteration of developmental signal pathways. More specifically, FGF signalling was inhibited at the developmental stage corresponding to cardiac mesoderm induction, and BMP and retinoic acid signalling pathways were activated. Hence, the development of NKX2.5+ cardiomyocytes was eradicated, thereby directing cardiomyocyte differentiation towards a pacemaker specific lineage. Electrophysiological analysis demonstrated that the majority (90%) of pacemaker-specific cardiomyocytes had characteristic pacemaker action potentials representative of native pacemaker cardiomyocytes, suggesting typical pacemaker ion current profiles. Remarkably, when transplanted into rat hearts, the bioengineered cardiomyocytes functioned as biological pacemakers, capable of altering the pace of the host heart. Transgenic approaches to generate pacemaker-like cardiomyocytes from hiPSCs include the forced expression of SHOX2, TBX3 and TGF-beta activated kinase [112,113,114].
Alternatively, the native cellular interactions can be recapitulated in vivo to aid subtype-specific differentiation to provide an alternative protocol to achieve pacemaker-like specific cardiomyocytes. Schweizer, Darche, Ullrich, Geschwill, Greber, Rivinius, Seyler, Müller-Decker, Draguhn, Utikal, Koenen, Katus and Thomas [52] generated hiPSC pacemaker-specific cardiomyocytes by initially co-culturing hiPSCs with visceral endoderm-like (END-2) cells for ten to twelve days to form beating cell aggregates. The beating cell aggregates were then isolated and transferred to foetal bovine serum-enriched medium for further culturing for six weeks. Interestingly, the expressions of HCN1, HNC4, NCX1, Cav1.2 and Cav1.3 within the beating clusters were similar to, or higher than those of human sinoatrial nodal cells. Action potential measurements of cells isolated from clusters demonstrated predominantly nodal-type (63.4%) with less atrial-type (36.6%) action potential configurations and no ventricular action potential morphologies. However, the relative expression of connexin demonstrated enhanced pacemaker lineage induction, as demonstrated by a fourfold increase in Cx45 levels following differentiation. Furthermore, the beating clusters demonstrated constant and synchronised pacing of neonatal rat ventricular myocytes in co-culture experiments.
Whilst these findings aid in generating subtype-specific cardiomyocytes derived from hiPSCs, the clinical application of these methods remain limited as they yield mixed cardiomyocyte subtype-populations. Moreover, the aforementioned protocols produce cardiomyoctes with relatively immature structure and function than their native counterparts. Further refinement of the generation of subtype-specific cardiomyocytes promises to propel the translational applications of patient-derived hiPSCs for disease modelling, drug discovery/toxicity and regenerative medicine.

Table 1.
Overview of protocols for the generation of subtype-specific cardiomyocytes involving the manipulation of cardiac development signalling pathways.
Table 1.
Overview of protocols for the generation of subtype-specific cardiomyocytes involving the manipulation of cardiac development signalling pathways.
| Subtype-Specific Cardiomyocyte Differentiation Protocol | Main Finding |
|---|---|
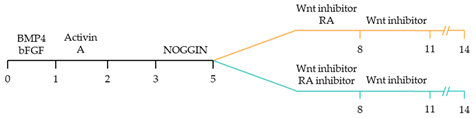 | ~51–65% cardiomyocyte differentiation efficiency ~94% atrial-specific cardiomyocyte differentiation ~83% ventricular-specific cardiomyocyte differentiation [100]. |
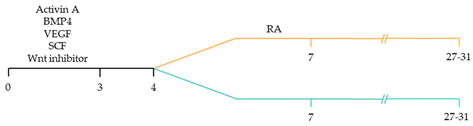 | ~51% cardiomyocyte differentiation efficiency ~85% atrial-specific cardiomyocyte differentiation ~80% ventricular-specific cardiomyocyte differentiation [115]. |
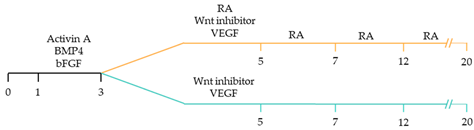 | ~70–98% cardiomyocyte differentiation efficiency Upregulation of atrial-specific genes following RA treatment Upregulation of ventricular-specific genes without RA treatment [22]. |
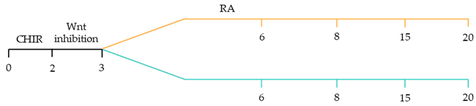 | ~87–90.5% cardiomyocyte differentiation efficiency ~94.7% atrial-specific cardiomyocyte differentiation ~77.8% ventricular-specific cardiomyocyte differentiation [23]. |
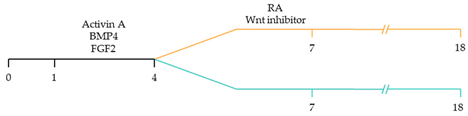 | ~93–94% cardiomyocyte differentiation efficiency RA treatment increased atrial specific genes, and tissue had a similar pharmacological response to muscarinic agonist as native atrial tissue [116]. |
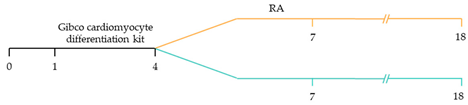 | ~85% atrial-specific cardiomyocyte differentiation ~86% ventricular-specific cardiomyocyte differentiation [117]. |
 | ~98% cardiomyocyte differentiation efficiency ~91.5% efficiency of ventricular-specific differentiation with 87% cardiac differentiation efficiency [118]. |
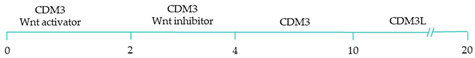 | 80–95% cardiac differentiation efficiency, with 50-60% of cardiomyocytes displaying ventricular specific APs and ventricular-specific genes [119]. |
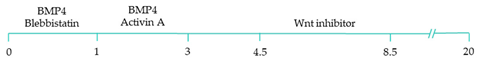 | 100% of cardiomyocytes displayed ventricular-like action potential parameters, with 89.42% cardiac differentiation efficiency [108]. |
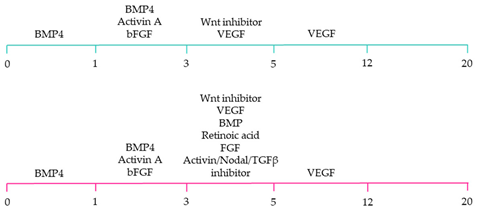 | ~80% cardiomyocyte differentiation efficiency. ~90% ventricular-specific differentiation ~90% pacemaker-specific differentiation. hiPSC-derived sinoatrial nodal cells demonstrated pacing ability when transplanted into rat hearts [107]. |
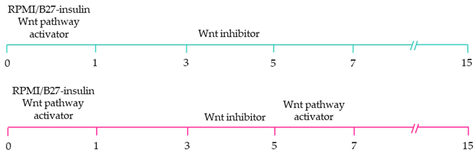 | ~85% cardiomyocyte differentiation efficiency. Wnt-activated hiPSC-cardiomyocytes expressed significantly elevated expression of pacemaker-related genes with pacemaker-like APs [111]. |
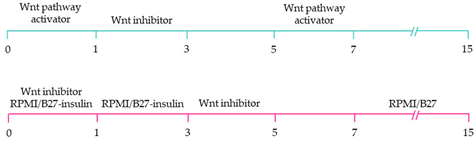 | ~60% cardiomyocyte differentiation efficiency. Wnt signalling promoted pacemaker lineage, whereas Wnt inhibition yielded chamber-specific cardiomyocytes [101]. |
Yellow, blue and pink represent atrial, ventricular and pacemaker-like cardiomyocyte differentiation, respectively. CDM3: chemically defined medium, 3 components: RPMI 1640, L-ascorbic 2-phosphate, and rice-derived recombinant human albumin. CDM3L: CDM3 without D-glucose and supplemented with sodium DL-lactate.
4. Maturity of Bioengineered Cardiomyocytes
Bioengineered hiPSC-derived cardiomyocytes display many of the characteristics of their native counterparts, rendering them a clinically relevant model for studying the mechanisms underlying cardiac diseases and, as a corollary, designing new pharmacological therapies. hiPSCs-derived cardiomyocytes are employed to model and trial therapeutic strategies for cardiac arrhythmias [120,121], cardiomyopathies [122,123] and metabolic disorders [124,125]. Although heart tissue was first cultured almost a century ago, most current protocols generate foetal-like cardiomyocytes that are relatively immature compared to adult cardiomyocytes, thereby limiting their clinical applications [126]. Inconsistencies with adult cardiomyocytes and the subsequent impact associated with immature characteristics (summarised in Table 2) include foetal-like morphology, poor expression of several contractile-protein and ion channel encoding genes contributing to immature electrophysiological and contractile properties. Moreover, hiPSC-derived cardiomyocytes have an adrenergic response and metabolism that are representative of foetal cardiomyocytes rather than mature adult cardiomyocytes [127]. The relative immaturity of hiPSC-derived cardiomyocytes hinders disease modelling of cardiomyopathies including metabolic syndromes, sarcomeric syndromes such as mutations of the sarcomere protein titin (TTN), disorders affecting cardiomyocyte contractility and cell cycle re-entry in mature cardiomyocytes in response to mitogens [125,128,129,130].

Table 2.
Different properties of foetal, hiPSC-derived and adult cardiomyocytes.
4.1. Morphology
Adult cardiomyocyte morphology provides the structural framework necessary for the execution of the functional electrophysiological and contractile properties of the cell. For example, morphological parameters including cell geometry, distribution of gap junctions and the interstitial space directly impact cell conductance [163]. In vivo cardiomyocyte hypertrophy associated with maturation following birth is linked to an alteration in the distribution of gap junctions and cell adhesion molecules. As native cardiomyocytes grow, gap junctions and cell adhesion molecules concentrate at intercalated discs located at the lateral border of cells. As such, larger, more mature cardiomyocytes have an increased maximal action potential upstroke velocity and enhanced impulse propagation velocity than smaller cardiomyocytes [164]. Current hiPSC differentiation protocols lack the biochemical and biophysical cues required to recapitulate native cardiomyocyte hypertrophy. Consequently, cultured hiPSC-derived cardiomyocytes are circular to oblong shaped, measuring 5-fold and 10-fold shorter in length and width, respectively, even after prolonged culture [132]. A reduced length-to-width ratio reduces the presence of lengthened myofibrils with laterally registered sarcomeres, thereby reducing contractility efficiency [165].
4.2. Electrophysiology
hiPSC-derived cardiomyocytes contract when exposed to a force of 0.08–4 mN/mm2, conduction velocity of approximately 10–20 cm/s and an upstroke velocity of 10–50 V/s. By contrast, in native adult cardiomyocytes, these parameters are significantly higher: 40–80 mN/mm2, 60 cm/s and 150–350 V/s, respectively [166]. Furthermore, hiPSC-derived cardiomyocyte cultures are typically mixed subtype populations and hence display diverse action potential phenotypes [167]. Adult atrial, ventricular and pacemaker-specific cardiomyocytes display different cellular electrophysiology due to differences in the function and expression of membrane-bound ion channels [168]. The ion channels expressed on subtype-specific cardiomyocytes have different conductances of ions, resulting in markedly different action potentials, including spontaneous diastolic depolarisation in SAN cardiomyocytes, a more negative resting membrane potential in ventricular cardiomyocytes and a less pronounced plateau in atrial cardiomyocytes.
Contractile cardiomyocytes have a stable resting membrane proceeding depolarisation, produced by the inward rectifying potassium channel (IK1), whereas pacemaker cells possess intrinsic automaticity, enabled by an unstable membrane potential that begins at about −60 mV and gradually drifts towards the threshold, resulting in continuous cyclic depolarisation [168]. In pacemaker cells, hyperpolarisation proceeding an action potential opens slow sodium channels and closes potassium channels. The decrease in the conductance of the IK1 results in relatively lower potassium resting permeability. Once the membrane potential rises to −50 mV, the potassium/sodium hyperpolarisation-activated cyclic nucleotide-gated channel 4 (HCN4) is activated. The hyperpolarisation-activated, slow inward current of sodium, referred to as the “funny” current (If), alters the balance between potassium loss and sodium entry [169]. The sodium channel responsible for rapid depolarization (INa); Nav1.5, encoded by Scn5a, is relatively low in the SAN compared to atrial and ventricular myocytes; however, SA nodal cells are characterised by a high expression of HCN4. Resultantly, the inner membrane potential is less negative, enabling slow depolarisation, therefore, reaching the threshold sooner. High expression of Nav1.5 in contractile cardiomyocytes contributes to an inward sodium current triggering rapid depolarisation. The maximum upstroke velocities for depolarisation are comparatively lower in atrial cardiomyocytes, varying between 150 and 300 V/s, compared to 300 and 400 V/s for that of ventricular cardiomyocytes [170]. The reduced upstroke velocity in atrial myocytes is attributed to the relatively depolarised resting potential compared to ventricular myocytes, thereby lowering the rapid sodium ion influx entry into the sarcolemma [171]. The succeeding plateau phase occurs as the potential approaches +30 mV, initiating concomitant voltage-gated sodium channels closure and active efflux of sodium from the cell. Simultaneously, voltage-gated L-type calcium channels (long-lasting calcium channels; ICaL) open, resulting in activation of ryanodine receptor type- 2 channels on the sarcoplasmic reticulum, triggering a surge in calcium concentration. The resulting entry of extracellular calcium stimulates the release of calcium from sarcoplasmic reticular stores, thereby inducing excitation–contraction coupling [172]. Compared with the spike and dome shape of the ventricular action potential, atrial action potentials exhibit a more triangular morphology with a less prominent plateau phase [173,174]. Therefore, negative voltage of the triangular atrial plateau phase increases the driving force of calcium influx, resulting in an increased inward calcium current compared to the ventricular counterpart. The ICaL are solely responsible for the atrial action potential plateau phase as there is no T-type calcium (transient opening calcium channel: ICaT) current present [170]. The ICaT forms the last portion of the pacemaker potential and is characterised by small conductances that transiently open. Atrial cardiomyocytes have a reduced inward-rectifying potassium current (approximately five- to sixfold less) compared to their ventricular counterpart, likely related to a comparatively more depolarised atrial resting membrane potential between –65 and −80 mV and the triangular, more negative plateau phase [175]. Kir2.1 is the primary molecular correlate of the IK1. As such, ventricular cardiomyocytes have a lower expression of Kir2.1 in comparison to atrial cardiomyocytes. Moreover, atrial cardiomyocytes possess several currents that are mostly absent in the ventricle, such as the acetylcholine-activated inward-rectifying potassium current (IKACh) and the ultra-rapid delayed-rectifier potassium current (IKur) [176] (Figure 3).
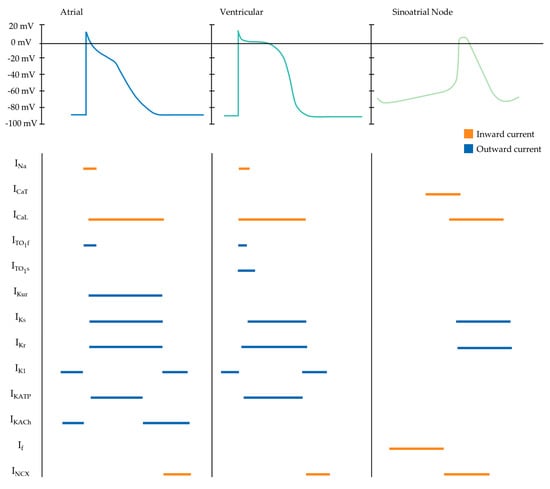
Figure 3.
Schematic depicting atrial, ventricular and sinoatrial nodal action potential waveforms and their underlying ionic currents. Horizontal orange and blue lines represent inward and outward cellular currents, respectively. INa, inward sodium. ICaT, T-type calcium. ICaL, L-type calcium. Ito1f, fast transient outward. Ito1s, slow transient outward. IKur, ultra-rapid potassium delayed rectifier. IKs, slow potassium delayed rectifier. IKr, rapid potassium delayed rectifier. IK1, inward rectifier. IKATP, ADP-activated potassium channel. IKACh, muscarinic-gated potassium channel. If, “funny” current. INCX, sodium/potassium exchange current.
Electrophysiological immaturity and mixed subtypes of hiPSC-derived cardiomyocytes can interfere with therapeutic applications including efficient disease modelling and drug trials. For example, hiPSC-derived cardiomyocytes often lack the inward rectifying potassium current that is critical for a stable resting membrane potential, therefore rendering them insufficient for modelling long QT syndrome [177].
Gap junction channels mediate intercellular cardiac electrical communication, allowing the transmission of coordinated electrical energy throughout the heart. Gap junctions play a critical role in the intercellular conduction velocities within the various cardiomyocyte populations; the different connexin subunit (Cx) expression among the subtype populations reflects this difference. For example, pacemaker activity within the SAN requires a high intercellular resistance (low conductivity) [66]. In the SAN, Cx45, Cx30.2 and Cx40 subunits mediate cell-to-cell coupling. These subunits form gap junctions with overall greater electrical resistance (<40 pS), which aids to shelter the SA nodal cells from the hyperpolarising current of the adjacent atrial cardiomyocytes by providing electrical isolation [178,179]. In contrast, the fast-conducting atrial and ventricular myocytes express predominantly Cx40 and Cx43 subunits that form gap junction channels of relative high conductance (low intercellular resistance) (>70 pS). The high conductance connections facilitate the rapid conduction essential for mechanical contraction [180].
Furthermore, the components of the intercalated disc complex such as N-cadherin-mediated adherens junctions, desmosomes, Nav1.5 and Cx43 polarise to the lateral ends of mature native cardiomyocytes. In vivo, the polarisation occurs gradually as the circumferentially distributed components polarise towards the long axis of the cell; however, the mechanisms mediating this process remain unknown [181]. The redistribution of these components is absent in standard cultured hiPSC-derived cardiomyocytes [182], although induced polarisation has been demonstrated by spatial restriction via micropatterned laminin surfaces [183], cyclic mechanical stretching [154] and co-culturing with bone marrow stromal cells or fibroblasts [184]. Together, it is critical to orientate the cytoarchitecture for efficient electromechanical coupling of cardiomyocytes to recapitulate native tissue and allow for the transmission of direction contraction across large distances. Cultured cardiomyocytes typically display mixed gap junction profiles, disorganized myofibrils and diffuse intercellular junctions with insufficient similarity to native adult cardiomyocytes. This further impedes the biomedical applications of hiPSC-derived cardiomyocytes.
4.3. Calcium Handling
Native adult cardiomyocytes develop advanced molecular machinery including T-tubules (transverse tubules) and sarcoplasmic reticulum within the sarcomere’s Z-line to modulate fast excitation–contraction coupling and calcium-induced calcium release. There are critical differences between these sophisticated structures within the various cardiomyocyte subtypes, resulting in a significantly faster atrial muscle contraction than ventricular myocyte contraction. Following calcium release from the sarcoplasmic reticulum and the initiation of contraction, reuptake of calcium into the sarcoplasmic reticulum occurs via the sarco(endo)plasmic reticulum calcium ATPase (SERCA2a), the activation of which is modulated by phospholamban (PLN) and sarcolipin (SLN). Interestingly, atrial cardiomyocytes have a higher expression of SERCA2a and lower PLN levels than ventricular cardiomyocytes [185]. However, atrial cardiomyocytes have a higher expression of SLN. Additionally, atrial cardiomyocytes also have reduced expression of ryanodine receptor type- 2 (RYR2) channels present on their sarcoplasmic reticulum and a weaker expression of the sarcoplasmic reticulum calcium buffer, calsequestrin (CASQ2), compared with that of the ventricular counterpart.
Further, ventricular myocytes express a complex system of t-tubules, consequently bringing sarcolemmal components deep to the cell interior. As a result, ventricular RYR2 channels clusters are coupled to the surface membrane via the t-tubules, resulting in a centripetal calcium wave propagation [60]. However, atrial cardiomyocytes typically lack or express only rudimentary t-tubule components. Accordingly, most atrial RYR2 channels clusters are not associated with any surface membrane contacts. Therefore, in atrial myocytes, RYR2channels closest to the sarcolemma are activated first following sequential activation of adjacent RYR2channels [186]. Together, these differences result in subtype-specific calcium handling properties of cardiomyocytes.
Consequently, in vitro cardiomyocyte cultures derived from hiPSCs typically yield mixed cardiomyocyte subtype populations, with mixed calcium handling properties consequently compromising the clinical relevance and potential therapeutic applications. Additionally, in hiPSC-derived-cardiomyocytes, sarcoplasmic reticula are foetal-like and disorganised with low expression of SERCA, and T-tubules are absent. Accordingly, hiPSC-derived cardiomyocytes depend on l-type channels for calcium release, thereby slowing fast excitation–contraction coupling. The electrophysiology immaturity further hinders the clinical relevance, in particular for disease modelling and pharmacological testing of conditions associated with dysregulated calcium handling such as catecholaminergic polymorphic ventricular tachycardia, triggered and re-entrant tachyarrhythmias and heart failure [187].
4.4. Metabolism
Metabolically, adult cardiomyocytes depend on fatty acids as opposed to glycolytic substrates as the preferred metabolic source. Throughout embryonic development, cardiomyocytes derive approximately 80% of energy from glycolysis following a shift to fatty acid β-oxidation during early postnatal maturation. As native cardiomyocytes mature, the mitochondrial volume increases by up to 40% and aligns between myofibrils and sarcolemma to enhance ATP production and distribution, thereby enhancing the oxidative capacity and promoting the switch in metabolic substrate [188,189,190]. Comparatively, hiPSC-derived cardiomyocytes rely predominately on glycolysis rather than fatty acid β-oxidation, although they inherently possess the capacity to utilise fatty acids [191,192].
4.5. Gene Expression
Classifying the genes associated with human cardiomyocyte maturation remains an enduring process. Nevertheless, the general expression patterns of maturation-related genes recognised in mice and humans are predominantly alike [193,194]. Mature cardiac tissue expresses high levels of genes such as SERCA2 (sarcoplasmic reticulum ATPase), ITPR3 (inositol-1,4,5-triphosphate), KCNH2 (potassium voltage-gated channel, CAV3 (caveolin 3) and RYR2 (ryanodine receptor 2). Additionally, indicators of cardiomyocyte maturation are also CASQ2 (calsequestrin 2), COX6A2 (cytochrome oxidase), S100A1 (S100 calcium binding protein A1), SCN5A (sodium voltage-gated channel alpha subunit 5) and MYOM2/3 (myomesin-2/3) [195]; these mature cardiac tissue markers are downregulated in hiPSC-derived cardiomyocytes [191,196,197,198,199]. Moreover, there are several discrepancies between the essential structural gene expressions of native adult cardiac tissue and hiPSC-derived cardiac tissue (summarised in Table 2). Adult cardiomyocytes display high levels of the β-isoform of the cardiac myosin heavy chain sarcomere gene (MYH7), whereas hiPSC-derived cardiomyocytes typically display elevated levels of the α-isoform (MYH6). Typically, the isoform switch occurs during the fetal to adult transition. Titin (TTN) has three predominant isoforms; N2B, N2BA and fetal cardiac titin (FCT). Adult cardiac tissue highly expresses the N2A isoform. In contrast, the N2BA isoform is predominant in hiPSC-derived cardiomyocytes. Troponin I (TnI) also has three significant isoforms including slow skeletal (ssTnI), fast skeletal (fsTnI), and cardiac (cTnI) encoded by TNNI1, TNNI2, and TNNI3, respectively. Adult cardiomyocytes display enhanced cTnI, while, the primary isoform remains ssTnI in hiPSC-derived cardiomyocytes [166,200].
Driving the maturation of hiPSC-derived cardiomyocytes is pivotal to enhance the capabilities and potential applications of hiPSC-derived cardiomyocytes. Accurate production of subtype-specific cardiomyocytes promises to progress hiPSC heart disease modelling, cardiac toxicity testing and translational medical application.
5. Approaches for Driving Pluripotent Stem Cell-Derived Cardiomyocyte Maturity
Native cardiomyocyte maturation involves the activation and integration of an orchestra of cellular signalling events. Bioengineering mature cardiomyocytes involves activating the mechanisms responsible for initiating the cellular events responsible for maturation [201]. Various approaches have been explored to accelerate maturation to overcome the immature phenotypes of hiPSC-derived cardiomyocytes such as prolonged culturing, biophysical and biochemical stimuli, genetic/epigenetic manipulation [164,202], in vivo maturation, bioengineered culture environments including 3D culturing techniques and electromechanical conditioning (Figure 4).
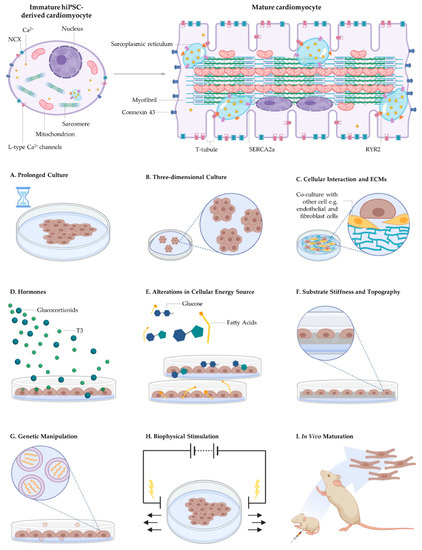
Figure 4.
Engineered microenvironments used to augment the maturity of human-induced pluripotent stem cell-derived cardiomyocytes. Current strategies include (A) prolonged culture period, (B) three-dimensional culture periods, (C) co-culture with fibroblasts, endothelial cells and ECMs, and biochemical factors such as (D) hormones including glucocorticoids and T3 and (E) fatty acids rather than glucose for energy production. Approaches also include (F) substrate stiffness and topography, (G) genetic manipulation, (H) biophysical stimulation including mechanical and electrical stimulation and (I) in vivo maturation. However, the mechanisms by which these signals drive cardiomyocyte maturation remain largely unknown.
Prolonged cell culture was the first approach implemented to augment hiPSC-derived cardiomyocyte maturity. Long-term cell culture for forty days demonstrated augmented cellular hypertrophy, enhanced contractility, calcium handling and increased expression of ventricular-specific genes including increased CX43 expression, TnI isoform transition and MHC isoform switch [203]. Culturing for up to one year further improved sarcomeric organization; however, T-tubule development remained insufficient, indicating that prolonged cell culture alone is inadequate to mature hiPSC-derived cardiomyocytes [191]. Whilst prolonging the culture period is a simple method that facilitates maturation [135,204,205], it is neither economically nor logistically ideal, particularly in terms of up-scaling for clinical use. Further, long-term culturing is highly variable in terms of the level of maturation achieved within cultures. As such, various approaches aiming to amplify maturation within a constrained time frame are being explored.
5.1. Biophysical Cues
Biophysical cues such as nanogrooved substrates and micropatterned, adhesive rectangles in addition with more physiologically relevant substrate stiffness aid to recapitulate the structural features of native cardiomyocytes [131,197,206]. The use of micropatterned matrigel rectangles (2000 µm2 rectangles with length:width aspect ratios of 5:1–7:1) [131] and microgrooved scaffolds [197] force hiPSC-derived cardiomyocytes to adopt an elongated morphology with enhanced directionality representative of native anisotropic, rod-like shaped mature adult cardiomyocytes. The anisotropic, rod-like shape of native cardiomyocytes facilitates myofibril alignment and cell contractility. Consequently, patterning hiPSC-derived cardiomyocytes to adopt a rod-shaped morphology enhances sarcomere alignment and organisation, thereby enhancing calcium handling and subsequent force generation.
The elastic modulus of the ECM progressively increases from <10 kPa in neonatal hearts to approximately 25 kPa in adult cardiac tissue due to collagen accumulation, thereby facilitating cardiomyocyte maturation [131,207] Culturing hiPSC-derived cardiomyocytes on surfaces within physiological matrix stiffness induces enhanced sarcomere alignment, calcium handling and contractility compared with softer matrices [208,209]. Moreover, the myocardial ECM is anisotropic and fibrillar and produces essential nanotopical cues to guide cardiomyocyte alignment and assembly. These extracellular matrix features were recreated in vitro using nanofibrous scaffolds with physiologically relevant porosity to support cell infiltration and promote tissue morphogenesis, leading to enhanced native-like myocardial tissue genesis and chamber-level contractile function [210]. Decellularised myocardial matrix is an emerging strategy used to recapitulate the adult myocardial biophysical cues to guide hiPSC-derived cardiomyocyte alignment. hiPSC-derived cardiomyocytes seeded onto thin slices of decellularised porcine myocardial matrix exhibited enhanced structural and functional maturity compared to standard culture that was maintained for more than 200 days [211].
Although cardiomyocytes contribute to approximately 75% of myocardial volume, they account for only around 30% of the total cell number present in the heart. Non-cardiomyocytes such as vascular endothelial cells, cardiac fibroblasts, smooth muscle cells, immune cells and neural cells comprise the remaining portion [212]. Non-cardiomyocytes directly improve native cardiomyocyte maturation through paracrine signalling and cell-to-cell interactions [213,214,215,216]. Recreating the cardiac cell heterogeneity in vitro contributes to enhanced maturation of hiPSC-derived cardiomyocytes. Co-culturing with endothelial cells has demonstrated enhanced hypertrophy, increased expression of ion channel and sarcomere genes and an improved chronotropic response compared to monoculture hiPSC-derived cardiomyocytes [217]. Furthermore, co-culturing hiPSCs with mesenchymal stem cells improves the structural, electrophysiological and metabolic maturity of cardiomyocytes [142]. Interestingly, hiPSC-derived cardiomyocytes cultured with cardiac fibroblasts and cardiac endothelial cells within constructed, scaffold-free, three-dimensional microtissues displayed augmented electrophysiological and contractile maturity, improved sarcomeric structures with T-tubules and enhanced mitochondrial respiration compared to control tissue without multiple cardiac cell types [218].
Further research is required to establish the impact of smooth muscle cells and peripheral neurons on hiPSC-derived cardiomyocyte maturity. In summary, the aforementioned research demonstrates that the maturity of hiPSC-derived cardiomyocytes is enhanced in vitro by mirroring the native cell shape, physiological stiffness and intercellular crosstalk.
5.2. Biochemical Cues
Biochemical cues to improve hiPSC-cardiomyocyte differentiation and maturation include alterations in the cellular energy source to induce mainly fatty acid β-oxidation and the addition of hormones (thyroid hormone triiodothyronine (T3) and glucocorticoids) [151,219,220]. An indicator of cardiomyocyte maturation is the postnatal metabolic switch from glycolysis to fatty acid synthesis initiating hypertrophic growth and cell cycle withdrawal, thereby accelerating cardiomyocyte maturation [188,190]. The switch in adenosine triphosphate production can be mimicked in hiPSC-derived cardiomyocyte development to upregulate adult-like gene expression patterns. Supplementation of hiPSC-derived cardiomyocyte cultures with fatty acids to promote β-oxidation prevents non-cardiomyocytes induction and enriches hiPSC-derived cardiomyocytes. Substituting glucose for galactose or fatty acids, in particular, oleic acid, palmate, carnitine and linoleic acid, improved hiPSC-derived cardiomyocyte maturation. Promoting fatty acid oxidation improved morphological, structural and functional maturation as well as mitochondrial number [156,221]. Conversely, culturing hiPSC-derived cardiomyocytes with glucose enriched media promoted nucleotide biosynthesis, leading to a reduction of cardiac glucose uptake and increased nucleotide deprivation, significantly reducing cardiomyocyte maturation [222].
Alterations in metabolic hormonal signalling coincide with cardiomyocyte development and cardiovascular maturation. The serum levels of T3 surge dramatically postnatally, evoking hypertrophic growth, polyploidisation, the induction of SERCA expression and the isoform switch of myosin heavy chain and TTN [223]. The omnipresent effects of T3 are translatable to in vitro cultures for improved maturation, with T3-treated hiPSC-derived cardiomyocytes demonstrating hypertrophy, elongated morphology, increased sarcomere length, mitochondrial biogenesis and increased mitochondrial respiration, improved calcium handling and contractile properties [224]. Similarly, glucocorticoid signalling is also critical for healthy heart development, including maturation of the fetal heart. Endogenous glucocorticoid synthesis accelerates during the antepartum period, with serum levels remaining high following birth to promote cardiomyocyte maturation [225]. Glucocorticoids bind to nuclear glucocorticoid receptors (encoded by Nr3c1) expressed on cardiomyocytes to stimulate myofibril organisation and maturation [226]. Co-treatment of dexamethasone, a glucocorticoid analog, in addition with T3, further augments hiPSC-derived cardiomyocyte maturation as demonstrated by extensive T-tubule networking, improved sarcomere organisation and calcium kinetics [151]. Conversely, mutations of Nr3c1 impede cardiomyocyte maturation as indicated by sarcomere disorganisation, disruption of myocyte alignment and impaired calcium handling [145,226]. To date, research highlights the necessity of hormonal signalling and points to investigating further molecules and combinations such as angiotensin II, insulin-like growth factor and neuregulin 1β that may also have a role in cardiomyocyte maturation [227,228,229]. Together, hormone signalling and metabolic alteration can significantly enhance cardiomyocyte maturation in vitro by inducing cell cycle exit, hypertrophy, enhanced electrophysiological function and cell cycle withdrawal. Further research is required to elucidate the mechanisms behind these beneficial effects completely; however, preliminary research indicates several critical pathways such as protein synthesis, mitochondrial biogenesis and metabolic sensing and signalling.
5.3. Three-Dimensional Cell Culturing
Recently, 3D culture systems (with or without scaffolds) have become a physiologically relevant alternative to monolayer cultures for the maturation of hiPSC-derived cardiomyocytes. Whilst convention monolayer cultures provide simplicity and scalability, they fail to recapitulate the native extracellular microenvironment and tissue architecture, limiting cellular interaction with apical–basal polarity [230]. 3D cell cultures aim to overcome the intrinsic limitations of monolayer cell cultures by better resembling native cardiac tissue architecture [231]. In 3D cell cultures, hiPSC-derived cardiomyocytes develop improved physiological maturity than monolayer cultures, including enhanced myofibrillar organization and sarcolemma alignment, leading to enhanced calcium handling with concomitant improved contractility and electrophysiology. Importantly, hiPSC-derived cardiomyocytes mature more rapidly compared to monolayer cell cultures [96,192,232,233,234,235].
The use of 3D tissue that better resembles native cardiac tissue potentially allows for enhanced accuracy in disease modelling and drug trials to recapitulate disease phenotypes that monolayer cultures fail to represent [128,232]. While 3D cardiac tissue is a promising method for augmenting hiPSC-derived cardiomyocyte maturation, it is insufficient to produce adult-like cardiomyocytes alone. Consequently, the engineering of mature hiPSC-derived cardiomyocytes for clinical applications requires the combination of 3D cell culture with the aforementioned maturation-promoting techniques such as co-culturing [146,236,237], hormonal signalling [145,238] and electrical stimulation [136,153].
5.4. In Vivo Maturation
Interestingly, hiPSC-derived cardiomyocytes rapidly mature once implanted in vivo; however, the mechanisms responsible for the augmented maturity remain unknown [239,240]. Proceeding immediate cell death following transplantation, the in vivo environment provides the essential cues for rapid cardiac differentiation and maturation of hiPSCs. Whilst there remains dissent regarding the optimal developmental stage and health status of the host heart upon transplantation, hiPSCs mature when transplanted into a neonatal, an adult, or adult heart after infarction [239,240,241]. In vivo transplanted hiPSC-derived cardiomyocytes developed adult-like cardiomyocyte morphology, physiology and gene expression, including regular T-tubules consistent with adult calcium transients and sarcomere shortening within one month following transplantation to early postnatal rat hearts. These results indicate that hiPSC-derived cardiomyocyte maturation is accelerated within a host heart, guided by essential environmental cues, including extracellular factors [240]. In summary, in vivo studies indicate that hiPSC-derived cardiomyocytes have the capacity for extensive maturation and development of adult cardiomyocytes properties when placed into their native environment. The mechanisms governing the accelerated maturation remain unknown but are likely to include biochemical and biophysical cues, electromechanical conditioning, metabolic fuels and neurohormones in addition to potentially unknown cues.
The ability of hiPSC-derived cardiomyocytes to differentiate and mature into near-adult phenotypes following in vivo transplantation highlights that under standard cell culture conditions, critical signals from the native environment are absent. The previously mentioned methods in this section have demonstrated augmented maturity of hiPSC-derived cardiomyocytes. However, no research to date has demonstrated complete cardiomyocyte maturation in terms of structure, metabolism and electrophysiology. hiPSC-derived cardiomyocytes matured using only these methods generated cells that did not entirely mirror native adult cardiomyocytes. Hence, such approaches require the combination with further, more complex techniques. Electrical and mechanical stimulation has emerged as a potent method to augment hiPSC maturation [153,242]. Research has indicated that 3D cell culture with electromechanical conditioning and in vivo maturation via transplantation into an animal heart have tremendous success in maturing hiPSC-derived cardiomyocytes with the former presenting the more clinically amenable option [127].
6. Electrical Stimulation for Tissue Engineering
The overarching objective of tissue engineering and regenerative medicine is to produce tissue that is both structurally and functionally equivalent to in vivo tissue [243]. Biochemical and biophysical stimuli, including chemical mechanical, magnetic and material-based (e.g., scaffolds, topography) cues regulate native cellular activity, including cell development and maintenance (e.g., scaffolds, topography) cues [244]. Employment of these biochemical and biophysical stimuli for in vitro cell culturing has advanced tissue engineering, aiding the production of more physiologically relevant tissue [245]. More recently, electrical stimulation has been utilised to augment and influence cell behaviour in vitro. Cells and tissue within the human body are naturally bioelectric, responding to and generating electric fields for normal physiological development, function, maintenance, repair and regeneration [246]. Therein, bioelectric potentials control and regulate cell proliferation, differentiation, migration, gene expression, apoptosis and morphology. The bioelectric potentials act as long-range intercellular signals, varying depending on their physiological state, employing different signals to elicit different outcomes in cells and organs [247,248]. Thus, it is important that we understand and consider applying electrical stimuli to enhance the quality of engineered tissue that is clinically amenable for regenerative medicine [249].
Electroceuticals or bioelectronics encompasses medical devices that utilise electrical stimulation to alter and modify biological tissue. Such devices have an extensive history in human medicine with devices such as cochlear implants for auditory function, pacemakers and implantable defibrillators for cardiac arrhythmias and neuromodulation with spinal cord stimulation for chronic pain [250,251,252]. Further, the field of electroceuticals has expanded to include electrical stimulation of the vagus nerve and deep brain stimulation [253,254]. Moreover, electroceuticals may apply to a broader range of diseases, including but not limited to asthma, chronic obstructive airway, diabetes and cardiovascular and gastrointestinal diseases [255]. The use of electrical stimulation in vitro is emerging as a tool for modulating human cell behaviour for tissue engineering, modelling and regenerative medicine. The application of endogenous electrical fields to in vitro tissue is thought to better recapitulate the native environment, thereby improving control over cell proliferation, differentiation adhesion and migration [256,257,258].
Electrical Stimulation for Bioengineering Mature Cardiac Tissue
Electrical stimulation may augment the maturity of hiPSC-derived cardiac tissue [127]. Endogenous electric fields play a critical role during normal foetal development, aiding in embryogenesis and subsequent maturation, with disturbances to these native endogenous electric fields leading to abnormal embryonic development [259]. Hence, the biomimetic paradigm has emerged, involving the application of biophysical cues, including electrical stimulation to differentiating engineered cardiac tissue to mirror the signals present in vivo to aid in producing clinically amenable bioengineered cardiomyocytes [260,261]. This led to the application of pulsatile electric fields to enhance the maturity of neonatal cardiomyocytes within 2D cell cultures. Remarkably, this preliminary work demonstrated enhanced cell alignment and sarcomere structure compared to non-stimulated controls [262]. Such findings encouraged the application of electrical stimulation of various duration, timing and frequency for enhanced differentiation of hiPSC-derived cardiomyocytes.
Interestingly, research indicates that incremental rises in stimulation intensity over the differentiation period enhance maturation compared to fixed-rate maturation. This was demonstrated within a 3D cardiac tissue platform exposed to electrical stimulation that aimed to mature hiPSC-derived cardiomyocytes [153]. hiPSCs seeded into a collagen gel surrounding a template suture in a polydimethylsiloxane microfabricated well were exposed to two different electrical stimulation protocols. The protocols involved progressive daily increments in the stimulation rate from 1 to 3 Hz (low-frequency group) and from 1 to 6 Hz (high-frequency group) [153]. Cardiac tissue subjected to electrical stimulation demonstrated augmented myofibril ultrastructural organization, additional H-zones per sarcomere, an increased number of desmosomes per membrane length and enhanced electrophysiological and calcium handling properties compatible with functional sarcoplasmic reticulum compared to non-stimulated controls. Research by Hirt, Boeddinghaus, Mitchell, Schaaf, Bornchen, Muller, Schulz, Hubner, Stenzig, Stoehr, Neuber, Eder, Luther, Hansen and Eschenhagen [136] compared two pacing protocols: 0.5 Hz for the entire stimulation period (low-frequency group) or 2 Hz in the first week and 1.5 Hz thereafter (high-frequency group). The engineered heart tissue generated from hiPSC-derived cardiomyocytes was exposed to symmetric biphasic pulses (2 ms in both polarities) of 2 V/cm via carbon electrodes. Interestingly, the high-frequency group demonstrated an augmented force of spontaneous cardiomyocyte beating. Moreover, electrically stimulated cardiac tissues had a higher cytoplasm-to-nucleus ratio with a more sophisticated muscular network composed of longitudinally oriented cardiomyocytes with an augmented contractile mass relative to the cell-free matrix in comparison to unstimulated controls. As such, these results indicate that decreasing electrical pacing frequency during cardiomyocyte differentiation enhances the structural and functional properties of hiPSC-derived cardiomyocytes for the engineering of mature cardiac tissue.
In addition to enhancing maturation, exogenous electrical stimulation can be applied to enhance cardiomyocyte subtype-specific differentiation of hiPSCs. This was initially demonstrated with human iPS(Foreskin)- 2 cells that were differentiated to cardiomyocytes by initially forming embryoid bodies (EBs) for five days [97]. The EBs were then exposed to acute biphasic current pulses at 65 mV/mm or 200 mV/mm at 1 Hz frequency and 1 ms pulse width for 5 min. The EBs were electrically stimulated in suspension via gold electrodes coated with platinum. Remarkably, electrical stimulation augmented the proportion of contracting EBs and upregulated the cardiac gene expression of ACTC1, TNNT2, MYH7, TBX5 and MYL7. Moreover, they found that electrical stimulation at 200 mV/mm for 5 min enhanced ventricular cardiomyocyte differentiation rather than atrial cardiomyocyte differentiation. Furthermore, electrically stimulated EBs showed a concentration-dependent increase in contraction frequency in response to isoproterenol, indicating increased β-adrenoceptor sensitivity, suggesting augmented maturity of electrically stimulated cardiomyocytes. Interestingly, the cardiogenic effect of brief electrical stimulation was not observed in a different iPSC line, CERA007c6, demonstrating that the effects of electrical stimulation vary depending on the cell line used [97].
Electrical stimulation-induced ventricular cardiomyocyte-specific differentiation was further demonstrated with EBs electrically stimulated continuously for seven days by square signals at 5 V/cm for 2 ms at 0.5, 1 or 2 Hz via carbon rods [263]. Electrically stimulated cardiomyocytes displayed augmented maturity as demonstrated by upregulated markers of cardiac differentiation (Nkx2.5), structural development (MYH6 and IGF1R), electrical development (KCNJ2) and cardiac chamber specification (NPPA, MYL2 and ANKRD1) in comparison to controls. Interestingly, electrical stimulation influenced the type of connexin expression. More specifically, the electrical stimulation enhanced the expression of GJA1 (Cx43) and CJA5 (Cx40), the predominant gap junctions present in chamber-specific cardiomyocytes and secondary ventricular cells, respectively, in comparison to controls. Moreover, the cardiomyocyte beating rate was influenced by the frequency at which they were stimulated, indicating that electrical stimulation can regulate the cardiomyocyte beating rate. The acquired beating frequency from the electrical stimulation was transferable to neighbouring cardiomyocytes and was maintained after the stimulation was discontinued.
Enhanced ventricular–cardiomyocyte differentiation was also demonstrated following electrical field conditioning of 2 Hz, beginning on day 7 post cell seeding with a weekly 1 Hz ramp-up protocol until 6 Hz was reached and maintained for one week. The stimulation frequency was then decreased to 3 Hz and maintained for up to six months [264]. The electrical field conditioning was combined with direct hiPSC-derived atrial [22] and ventricular [118] differentiation protocols. In addition to the upregulation of ventricular specific proteins such as MLC2v, electrical conditioning enhanced sarcomeric organisation, matured cellular metabolism and chamber-specific drug responses [264]. Such results indicate that electrical stimulation may enhance cardiomyocyte maturation whilst simultaneously directing differentiation towards a specific lineage. An approach that simultaneously matures and drives subtype-specific differentiation of hiPSCs-derived cardiomyocytes provides a major step towards industrial biomedical applications of engineered cardiac tissue [166].
More recent evidence highlights that electrical stimulation-augmented cardiac differentiation of hiPSCs occurs mechanistically through the activation of the calcium/PKC/ERK pathway [265]. Electric fields were produced by via a C-Pace electric stimulator consisting of two electrodes emitting bipolar stimuli of 1 or 1.5 V/cm with a biphasic square pulse (5 ms) at 5 Hz frequency for 1–30 days. Remarkably, the functional maturity of the hiPSCs was significantly augmented by electrical stimulation as demonstrated by calcium indicators, intracellular calcium levels and enhanced expression of cardiac-specific genes.
The aforementioned studies using electrical stimulation are limited by traditional tethered, penetrating wired systems, creating an inherent risk for infection and inflammation. Additionally, electrical stimulation devices may not be financially viable for industrial scalability [201]. However, the striking degree of maturity attained through combined electrical stimulation and 3D culturing techniques provides a promising approach for overcoming the immaturity of hiPSC-derived cardiomyocytes. However, the mechanisms by which electrical pacing augments cardiomyocyte maturation and induces subtype-specific differentiation are yet to be completely understood.
7. Discussion and Future Perspectives
Current differentiation protocols by and large yield mixed subtype populations of foetal-like cardiomyocytes, representing a significant obstacle to translational applications of hPSC-derived cardiomyocytes, including disease modelling, cell therapy and drug discovery. Whilst recently there has been much effort directed towards cardiomyocyte subtype-specific differentiation, the reliability and therefore reproducibility of methods are yet to be verified. Consistent production of ventricular, atrial and pacemaker cardiomyocytes is necessary for their application in pre-clinical and clinical testing towards translational applications. In particular, hiPSC-derived atrial-like cardiomyocytes offer a human physiologically relevant model for atrial arrhythmias and for identifying novel atrial-selective pharmacology candidates. Highly enriched ventricular cardiomyocytes would be ideal for remuscularisation of the ventricular wall to restore cardiac function following myocardial infarction. A pure ventricular cell culture, devoid of contaminating atrial and pacemaker cardiomyocytes, would help to abrogate ventricular arrhythmias observed in animal models after transplantation of grafts comprised of mixed cardiomyocyte subtypes.
Biological pacemakers have theoretical advantages compared to artificial pacemakers by providing battery independency and autonomic responsiveness. Biological pacemakers are particularly promising for infants and children with heart conditions, due to their potential adaptation during adolescence. Although effective protocols for deriving homogenous populations of atrial, ventricular- and nodal-like cardiomyocytes remain a continuing challenge, current difficulties may be circumvented through critical stage-dependent activation and inhibition of crucial development pathways and the enrichment of cardiac progenitors committed to specific subtype lineages.
Maturation of hiPSC-derived cardiomyocytes is also a major issue for the field. While specific maturation strategies (such as biochemical and biophysical cues) provide incremental advances in cardiomyocyte maturity, adult cardiomyocyte phenotypes are yet to be achieved. Collective research indicates an interdependence between specific maturation strategies with an optimal combinatory approach; in particular, including the application of electrical stimulation of 3D tissue. However, the integration of environmental cues and biophysical stimulation is yet to be optimised, and the molecular mechanisms facilitating maturation remain poorly understood.
In conclusion, future research should focus on driving cost-effective and reproducible hPSC cardiac maturation and subtype-specific differentiation, specifically through combinatorial approaches that coordinate the intensity and timing of interventions to recapitulate the native cardiac developmental process. By doing so, it is anticipated that cardiac regenerative medicine will quickly become a reality, having broad translational applications for improved therapeutics.
Author Contributions
Conceptualization, E.C.J., E.T.-C., J.M.C. Writing—original draft preparation, E.C.J.; writing—review and editing, J.M.C., E.T.-C., E.C.J. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Australian Research Council (ARC) Centre of Excellence Scheme (CE140100012). ECJ was funded by an Australian Government Research Training Program (RTP) Scholarship.
Conflicts of Interest
The authors declare no conflict of interest.
References
- WHO. Cardiovascular Diseases (CVD) Fact Sheet. Available online: https://www.who.int/en/news-room/fact-sheets/detail/cardiovascular-diseases-(cvds) (accessed on 10 January 2021).
- WHO. World Health Statistics 2020: Monitoring Health for the SDGs, Sustainable Development Goals; World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]
- Roth, G.A.; Johnson, C.; Abajobir, A.; Abd-Allah, F.; Abera, S.F.; Abyu, G.; Ahmed, M.; Aksut, B.; Alam, T.; Alam, K.; et al. Global, Regional, and National Burden of Cardiovascular Diseases for 10 Causes, 1990 to 2015. J. Am. Coll. Cardiol. 2017, 70, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Finnemore, A.; Groves, A. Physiology of the fetal and transitional circulation. Semin. Fetal Neonatal Med. 2015, 20, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Kiuchi, S.; Usami, A.; Shimoyama, T.; Otsuka, F.; Yamaguchi, S.; Nakamura, T.; Suzuki, S.; Ono, K. Cardiac Pacemaker Cells Generate Cardiomyocytes from Fibroblasts in Long-Term Cultures. Sci. Rep. 2019, 9, 15174. [Google Scholar] [CrossRef]
- Bergmann, O.; Bhardwaj, R.D.; Bernard, S.; Zdunek, S.; Barnabé-Heider, F.; Walsh, S.; Zupicich, J.; Alkass, K.; Buchholz, B.A.; Druid, H.; et al. Evidence for Cardiomyocyte Renewal in Humans. Science 2009, 324, 98. [Google Scholar] [CrossRef] [PubMed]
- Bergmann, O.; Zdunek, S.; Felker, A.; Salehpour, M.; Alkass, K.; Bernard, S.; Sjostrom, S.L.; Szewczykowska, M.; Jackowska, T.; Dos Remedios, C.; et al. Dynamics of Cell Generation and Turnover in the Human Heart. Cell 2015, 161, 1566–1575. [Google Scholar] [CrossRef]
- Curtis, M.W.; Russell, B. Cardiac tissue engineering. J. Cardiovasc. Nurs. 2009, 24, 87–92. [Google Scholar] [CrossRef]
- Tohyama, S.; Fukuda, K. Future Treatment of Heart Failure Using Human iPSC-Derived Cardiomyocytes. In Etiology and Morphogenesis of Congenital Heart Disease: From Gene Function and Cellular Interaction to Morphology; Springer: Tokyo, Japan, 2016; pp. 25–31. [Google Scholar]
- Sacchetto, C.; Vitiello, L.; de Windt, L.J.; Rampazzo, A.; Calore, M. Modeling Cardiovascular Diseases with hiPSC-Derived Cardiomyocytes in 2D and 3D Cultures. Int. J. Mol. Sci. 2020, 21, 3404. [Google Scholar] [CrossRef]
- Paik, D.T.; Chandy, M.; Wu, J.C. Patient and Disease–Specific Induced Pluripotent Stem Cells for Discovery of Personalized Cardiovascular Drugs and Therapeutics. Pharmacol. Rev. 2020, 72, 320. [Google Scholar] [CrossRef]
- Sadahiro, T. Cardiac regeneration with pluripotent stem cell-derived cardiomyocytes and direct cardiac reprogramming. Regen. Ther. 2019, 11, 95–100. [Google Scholar] [CrossRef]
- Van den Heuvel, N.H.; van Veen, T.A.; Lim, B.; Jonsson, M.K. Lessons from the heart: Mirroring electrophysiological characteristics during cardiac development to in vitro differentiation of stem cell derived cardiomyocytes. J. Mol. Cell. Cardiol. 2014, 67, 12–25. [Google Scholar] [CrossRef] [PubMed]
- Kishino, Y.; Fujita, J.; Tohyama, S.; Okada, M.; Tanosaki, S.; Someya, S.; Fukuda, K. Toward the realization of cardiac regenerative medicine using pluripotent stem cells. Inflamm. Regen. 2020, 40, 1. [Google Scholar] [CrossRef]
- Mahajani, S.; Raina, A.; Fokken, C.; Kügler, S.; Bähr, M. Homogenous generation of dopaminergic neurons from multiple hiPSC lines by transient expression of transcription factors. Cell Death Dis. 2019, 10, 898. [Google Scholar] [CrossRef]
- Balboa, D.; Saarimäki-Vire, J.; Otonkoski, T. Concise Review: Human Pluripotent Stem Cells for the Modeling of Pancreatic β-Cell Pathology. Stem Cells 2019, 37, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Nagoshi, N.; Okano, H.; Nakamura, M. Regenerative therapy for spinal cord injury using iPSC technology. Inflamm. Regen. 2020, 40, 40. [Google Scholar] [CrossRef]
- Kawabata, S.; Takano, M.; Numasawa-Kuroiwa, Y.; Itakura, G.; Kobayashi, Y.; Nishiyama, Y.; Sugai, K.; Nishimura, S.; Iwai, H.; Isoda, M.; et al. Grafted Human iPS Cell-Derived Oligodendrocyte Precursor Cells Contribute to Robust Remyelination of Demyelinated Axons after Spinal Cord Injury. Stem Cell Rep. 2016, 6, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Hu, J.; He, T.-C. iPSC-based treatment of age-related macular degeneration (AMD): The path to success requires more than blind faith. Genes Dis. 2017, 4, 41–42. [Google Scholar] [CrossRef] [PubMed]
- Ye, K.; Takemoto, Y.; Ito, A.; Onda, M.; Morimoto, N.; Mandai, M.; Takahashi, M.; Kato, R.; Osakada, F. Reproducible production and image-based quality evaluation of retinal pigment epithelium sheets from human induced pluripotent stem cells. Sci. Rep. 2020, 10, 14387. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.-T.; Shao, N.-Y.; Garg, V. Subtype-specific cardiomyocytes for precision medicine: Where are we now? Stem Cells 2020, 38, 822–833. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Protze, S.I.; Laksman, Z.; Backx, P.H.; Keller, G.M. Human Pluripotent Stem Cell-Derived Atrial and Ventricular Cardiomyocytes Develop from Distinct Mesoderm Populations. Cell Stem Cell 2017, 21, 179–194. [Google Scholar] [CrossRef]
- Cyganek, L.; Tiburcy, M.; Sekeres, K.; Gerstenberg, K.; Bohnenberger, H.; Lenz, C.; Henze, S.; Stauske, M.; Salinas, G.; Zimmermann, W.H.; et al. Deep phenotyping of human induced pluripotent stem cell-derived atrial and ventricular cardiomyocytes. JCI Insight 2018, 3. [Google Scholar] [CrossRef]
- Nguyen, A.H.; Marsh, P.; Schmiess-Heine, L.; Burke, P.J.; Lee, A.; Lee, J.; Cao, H. Cardiac tissue engineering: State-of-the-art methods and outlook. J. Biol. Eng. 2019, 13, 57. [Google Scholar] [CrossRef] [PubMed]
- Zarrouk-Mahjoub, S.; Zaghdoudi, M.; Amira, Z.; Chebi, H.; Khabouchi, N.; Finsterer, J.; Mechmeche, R.; Ghazouani, E. Pro- and anti-inflammatory cytokines in post-infarction left ventricular remodeling. Int. J. Cardiol. 2016, 221, 632–636. [Google Scholar] [CrossRef]
- Puhl, S.-L.; Steffens, S. Neutrophils in Post-myocardial Infarction Inflammation: Damage vs. Resolution? Front. Cardiovasc. Med. 2019, 6. [Google Scholar] [CrossRef] [PubMed]
- DeLeon-Pennell, K.Y.; Meschiari, C.A.; Jung, M.; Lindsey, M.L. Matrix Metalloproteinases in Myocardial Infarction and Heart Failure. Prog. Mol. Biol. Transl. Sci. 2017, 147, 75–100. [Google Scholar] [CrossRef] [PubMed]
- Lindsey, M.L. Assigning matrix metalloproteinase roles in ischaemic cardiac remodelling. Nat. Rev. Cardiol. 2018, 15, 471–479. [Google Scholar] [CrossRef]
- Mann, D.L.; Bogaev, R.; Buckberg, G.D. Cardiac remodelling and myocardial recovery: Lost in translation? Eur. J. Heart Fail. 2010, 12, 789–796. [Google Scholar] [CrossRef]
- Frangogiannis, N.G. The Functional Pluralism of Fibroblasts in the Infarcted Myocardium. Circ. Res. 2016, 119, 1049–1051. [Google Scholar] [CrossRef]
- Heallen, T.R.; Martin, J.F. Heart repair via cardiomyocyte-secreted vesicles. Nat. Biomed. Eng. 2018, 2, 271–272. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Wang, Z. Current Understanding of the Biomechanics of Ventricular Tissues in Heart Failure. Bioengineering 2019, 7, 2. [Google Scholar] [CrossRef]
- Schiattarella, G.G.; Hill, J.A. Inhibition of hypertrophy is a good therapeutic strategy in ventricular pressure overload. Circulation 2015, 131, 1435–1447. [Google Scholar] [CrossRef]
- Kong, P.; Christia, P.; Frangogiannis, N.G. The pathogenesis of cardiac fibrosis. Cell Mol. Life Sci. 2014, 71, 549–574. [Google Scholar] [CrossRef] [PubMed]
- Arnal-Pastor, M.; Chachques, J.C.; Pradas, M.M.; VallÈs-Lluch, A. Biomaterials for Cardiac Tissue Engineering. In Regenerative Medicine and Tissue Engineering; Intechopen: London, UK, 2013. [Google Scholar]
- Stone, G.W.; Sabik, J.F.; Serruys, P.W.; Simonton, C.A.; Généreux, P.; Puskas, J.; Kandzari, D.E.; Morice, M.-C.; Lembo, N.; Brown, W.M.; et al. Everolimus-Eluting Stents or Bypass Surgery for Left Main Coronary Artery Disease. N. Engl. J. Med. 2016, 375, 2223–2235. [Google Scholar] [CrossRef] [PubMed]
- Mäkikallio, T.; Holm, N.R.; Lindsay, M.; Spence, M.S.; Erglis, A.; Menown, I.B.; Trovik, T.; Eskola, M.; Romppanen, H.; Kellerth, T.; et al. Percutaneous coronary angioplasty versus coronary artery bypass grafting in treatment of unprotected left main stenosis (NOBLE): A prospective, randomised, open-label, non-inferiority trial. Lancet 2016, 388, 2743–2752. [Google Scholar] [CrossRef]
- Miller, L.; Birks, E.; Guglin, M.; Lamba, H.; Frazier, O.H. Use of Ventricular Assist Devices and Heart Transplantation for Advanced Heart Failure. Circ. Res. 2019, 124, 1658–1678. [Google Scholar] [CrossRef]
- Yamakawa, M.; Kyo, S.; Yamakawa, S.; Ono, M.; Kinugawa, K.; Nishimura, T. Destination therapy: The new gold standard treatment for heart failure patients with left ventricular assist devices. Gen. Thorac Cardiovasc. Surg. 2013, 61, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Anyanwu, A.; Treasure, T. Prognosis after heart transplantation: Transplants alone cannot be the solution for end stage heart failure. BMJ 2003, 326, 509–510. [Google Scholar] [CrossRef]
- Tonsho, M.; Michel, S.; Ahmed, Z.; Alessandrini, A.; Madsen, J.C. Heart transplantation: Challenges facing the field. Cold Spring Harb. Perspect. Med. 2014, 4, a015636. [Google Scholar] [CrossRef]
- Elgendy, I.Y.; Mahtta, D.; Pepine, C.J. Medical Therapy for Heart Failure Caused by Ischemic Heart Disease. Circ. Res. 2019, 124, 1520–1535. [Google Scholar] [CrossRef]
- Tehzeeb, J.; Manzoor, A.; Ahmed, M.M. Is Stem Cell Therapy an Answer to Heart Failure: A Literature Search. Cureus 2019, 11, e5959. [Google Scholar] [CrossRef]
- Alonso, A.; Jensen, P.N.; Lopez, F.L.; Chen, L.Y.; Psaty, B.M.; Folsom, A.R.; Heckbert, S.R. Association of sick sinus syndrome with incident cardiovascular disease and mortality: The Atherosclerosis Risk in Communities study and Cardiovascular Health Study. PLoS ONE 2014, 9, e109662. [Google Scholar] [CrossRef]
- Laflamme, M.A.; Chen, K.Y.; Naumova, A.V.; Muskheli, V.; Fugate, J.A.; Dupras, S.K.; Reinecke, H.; Xu, C.; Hassanipour, M.; Police, S.; et al. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat. Biotechnol. 2007, 25, 1015–1024. [Google Scholar] [CrossRef] [PubMed]
- Dobrzynski, H.; Boyett Mark, R.; Anderson Robert, H. New Insights into Pacemaker Activity. Circulation 2007, 115, 1921–1932. [Google Scholar] [CrossRef] [PubMed]
- Mittnacht, A.; Reich, D.L.; Rhee, A.J.; Kaplan, J.A. Chapter 2—Cardiac Diseases. In Anesthesia and Uncommon Diseases, 6th ed.; Fleisher, L.A., Ed.; W.B. Saunders: Philadelphia, PA, USA, 2012; pp. 28–74. [Google Scholar]
- Jabbour, F.; Kanmanthareddy, A. Sinus Node Dysfunction; StatPearls Publishing LLC.: Treasure Island, FL, USA, 2020. [Google Scholar]
- Van Eif, V.W.W.; Devalla, H.D.; Boink, G.J.J.; Christoffels, V.M. Transcriptional regulation of the cardiac conduction system. Nat. Rev. Cardiol. 2018, 15, 617–630. [Google Scholar] [CrossRef] [PubMed]
- Smits, J.P.P.; Veldkamp, M.W.; Wilde, A.A.M. Mechanisms of inherited cardiac conduction disease. EP Eur. 2005, 7, 122–137. [Google Scholar] [CrossRef] [PubMed]
- Gollob, M.H.; Green, M.S.; Tang, A.S.; Gollob, T.; Karibe, A.; Ali Hassan, A.S.; Ahmad, F.; Lozado, R.; Shah, G.; Fananapazir, L.; et al. Identification of a gene responsible for familial Wolff-Parkinson-White syndrome. N. Engl. J. Med. 2001, 344, 1823–1831. [Google Scholar] [CrossRef] [PubMed]
- Schweizer, P.A.; Darche, F.F.; Ullrich, N.D.; Geschwill, P.; Greber, B.; Rivinius, R.; Seyler, C.; Müller-Decker, K.; Draguhn, A.; Utikal, J.; et al. Subtype-specific differentiation of cardiac pacemaker cell clusters from human induced pluripotent stem cells. Stem Cell Res. Ther. 2017, 8, 229. [Google Scholar] [CrossRef]
- Cho, H.C.; Marban, E. Biological therapies for cardiac arrhythmias: Can genes and cells replace drugs and devices? Circ. Res. 2010, 106, 674–685. [Google Scholar] [CrossRef]
- Rosen, M.R.; Brink, P.R.; Cohen, I.S.; Robinson, R.B. 26—Biological Pacing. In Cardiac Electrophysiology: From Cell to Bedside, 6th ed.; Zipes, D.P., Jalife, J., Eds.; W.B. Saunders: Philadelphia, PA, USA, 2014; pp. 253–263. [Google Scholar]
- Millar, M.; Wareham, D. Chapter 42—Infections associated with implanted medical devices. In Antibiotic and Chemotherapy, 9th ed.; Finch, R.G., Greenwood, D., Norrby, S.R., Whitley, R.J., Eds.; W.B. Saunders: London, UK, 2010; pp. 538–555. [Google Scholar]
- Feher, J. 5.5—The Cardiac Action Potential. In Quantitative Human Physiology, 2nd ed.; Feher, J., Ed.; Academic Press: Boston, MA, USA, 2012; pp. 528–536. [Google Scholar]
- Tran, D.B.; Weber, C.; Lopez, R.A. Anatomy, Thorax, Heart Muscles; StatPearls Publishing LLC.: Treasure Island, FL, USA, 2020. [Google Scholar]
- Miller, L.M.; Gal, A. Chapter 10—Cardiovascular System and Lymphatic Vessels1. In Pathologic Basis of Veterinary Disease, 6th ed.; Zachary, J.F., Ed.; Mosby: Maryland Heights, MO, USA, 2017; pp. 561–616. [Google Scholar]
- Talman, V.; Kivelä, R. Cardiomyocyte-Endothelial Cell Interactions in Cardiac Remodeling and Regeneration. Front. Cardiovasc. Med. 2018, 5, 101. [Google Scholar] [CrossRef]
- Brandenburg, S.; Arakel, E.C.; Schwappach, B.; Lehnart, S.E. The molecular and functional identities of atrial cardiomyocytes in health and disease. Biochim. Biophys. Acta BBA Mol. Cell Res. 2016, 1863, 1882–1893. [Google Scholar] [CrossRef]
- Veeraraghavan, R.; Gourdie, R.G.; Poelzing, S. Mechanisms of cardiac conduction: A history of revisions. Am. J. Physiol. Heart Circ. Physiol. 2014, 306, H619–H627. [Google Scholar] [CrossRef]
- Tessadori, F.; van Weerd, J.H.; Burkhard, S.B.; Verkerk, A.O.; de Pater, E.; Boukens, B.J.; Vink, A.; Christoffels, V.M.; Bakkers, J. Identification and functional characterization of cardiac pacemaker cells in zebrafish. PLoS ONE 2012, 7, e47644. [Google Scholar] [CrossRef]
- Park, D.S.; Fishman, G.I. Development and Function of the Cardiac Conduction System in Health and Disease. J. Cardiovasc. Dev. Dis. 2017, 4, 7. [Google Scholar] [CrossRef] [PubMed]
- Hund, T.J.; Smith, S.A.; Makara, M.A.; Mohler, P.J. Chapter 7—Cellular and Molecular Pathobiology of the Cardiac Conduction System. In Cellular and Molecular Pathobiology of Cardiovascular Disease; Willis, M.S., Homeister, J.W., Stone, J.R., Eds.; Academic Press: San Diego, CA, USA, 2014; pp. 121–134. [Google Scholar]
- Issa, Z.F.; Miller, J.M.; Zipes, D.P. 3—Electrophysiological Mechanisms of Cardiac Arrhythmias. In Clinical Arrhythmology and Electrophysiology, 3rd ed.; Issa, Z.F., Miller, J.M., Zipes, D.P., Eds.; Content Repository Only; Elsevier: Philadelphia, PA, USA, 2019; pp. 51–80. [Google Scholar]
- Mangoni, M.E.; Nargeot, J. Genesis and regulation of the heart automaticity. Physiol. Rev. 2008, 88, 919–982. [Google Scholar] [CrossRef]
- Kashou, A.H.; Basit, H.; Chhabra, L. Physiology, Sinoatrial Node (SA Node); StatPearls Publishing: Treasure Island, FL, USA, 2019. [Google Scholar]
- James, T.N. Structure and function of the sinus node, AV node and his bundle of the human heart: Part II—Function. Prog. Cardiovasc. Dis. 2003, 45, 327–360. [Google Scholar] [CrossRef]
- Miquerol, L.; Meysen, S.; Mangoni, M.; Bois, P.; van Rijen, H.V.; Abran, P.; Jongsma, H.; Nargeot, J.; Gros, D. Architectural and functional asymmetry of the His-Purkinje system of the murine heart. Cardiovasc. Res. 2004, 63, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Ideker, R.E.; Kong, W.; Pogwizd, S. Purkinje fibers and arrhythmias. Pacing Clin. Electrophysiol. 2009, 32, 283–285. [Google Scholar] [CrossRef] [PubMed]
- Stoppel, W.L.; Kaplan, D.L.; Black, L.D., 3rd. Electrical and mechanical stimulation of cardiac cells and tissue constructs. Adv. Drug Deliv. Rev. 2016, 96, 135–155. [Google Scholar] [CrossRef]
- McCuller, C.; Jessu, R.; Callahan, A.L. Physiology, Skeletal Muscle; StatPearls Publishing LLC.: Treasure Island, FL, USA, 2020. [Google Scholar]
- Carreiro, J.E. Chapter 4—The cardiovascular system. In An Osteopathic Approach to Children, 2nd ed.; Carreiro, J.E., Ed.; Churchill Livingstone: Edinburgh, UK, 2009; pp. 73–83. [Google Scholar]
- Bressan, M.; Liu, G.; Louie, J.D.; Mikawa, T. Cardiac Pacemaker Development from a Tertiary Heart Field. In Etiology and Morphogenesis of Congenital Heart Disease: From Gene Function and Cellular Interaction to Morphology; Nakanishi, T., Markwald, R.R., Baldwin, H.S., Keller, B.B., Srivastava, D., Yamagishi, H., Eds.; Springer: Tokyo, Japan, 2016; pp. 281–288. [Google Scholar]
- Kirby, M.L. Cardiac Development; Oxford University Press: Oxford, UK, 2007. [Google Scholar]
- Buijtendijk, M.F.J.; Barnett, P.; van den Hoff, M.J.B. Development of the human heart. Am. J. Med. Genet. C Semin. Med. Genet. 2020, 184, 7–22. [Google Scholar] [CrossRef]
- Meilhac, S.M.; Buckingham, M.E. The deployment of cell lineages that form the mammalian heart. Nat. Rev. Cardiol. 2018, 15, 705–724. [Google Scholar] [CrossRef]
- Kimelman, D. Mesoderm induction: From caps to chips. Nat. Rev. Genet. 2006, 7, 360–372. [Google Scholar] [CrossRef]
- Van Vliet, P.; Wu, S.M.; Zaffran, S.; Pucéat, M. Early cardiac development: A view from stem cells to embryos. Cardiovasc. Res. 2012, 96, 352–362. [Google Scholar] [CrossRef] [PubMed]
- Pikkarainen, S.; Tokola, H.; Kerkelä, R.; Ruskoaho, H. GATA transcription factors in the developing and adult heart. Cardiovasc. Res. 2004, 63, 196–207. [Google Scholar] [CrossRef] [PubMed]
- Välimäki, M.J.; Tölli, M.A.; Kinnunen, S.M.; Aro, J.; Serpi, R.; Pohjolainen, L.; Talman, V.; Poso, A.; Ruskoaho, H.J. Discovery of Small Molecules Targeting the Synergy of Cardiac Transcription Factors GATA4 and NKX2-5. J. Med. Chem. 2017, 60, 7781–7798. [Google Scholar] [CrossRef] [PubMed]
- Zaffran, S.; Frasch, M. Early signals in cardiac development. Circ. Res. 2002, 91, 457–469. [Google Scholar] [CrossRef] [PubMed]
- Bondue, A.; Lapouge, G.; Paulissen, C.; Semeraro, C.; Iacovino, M.; Kyba, M.; Blanpain, C. Mesp1 Acts as a Master Regulator of Multipotent Cardiovascular Progenitor Specification. Cell Stem Cell 2008, 3, 69–84. [Google Scholar] [CrossRef]
- Bondue, A.; Blanpain, C. Mesp1: A key regulator of cardiovascular lineage commitment. Circ. Res. 2010, 107, 1414–1427. [Google Scholar] [CrossRef]
- David, R.; Brenner, C.; Stieber, J.; Schwarz, F.; Brunner, S.; Vollmer, M.; Mentele, E.; Müller-Höcker, J.; Kitajima, S.; Lickert, H.; et al. MesP1 drives vertebrate cardiovascular differentiation through Dkk-1-mediated blockade of Wnt-signalling. Nat. Cell Biol. 2008, 10, 338–345. [Google Scholar] [CrossRef]
- Naito, A.T.; Shiojima, I.; Akazawa, H.; Hidaka, K.; Morisaki, T.; Kikuchi, A.; Komuro, I. Developmental stage-specific biphasic roles of Wnt/beta-catenin signaling in cardiomyogenesis and hematopoiesis. Proc. Natl. Acad. Sci. USA 2006, 103, 19812–19817. [Google Scholar] [CrossRef] [PubMed]
- Ueno, S.; Weidinger, G.; Osugi, T.; Kohn, A.D.; Golob, J.L.; Pabon, L.; Reinecke, H.; Moon, R.T.; Murry, C.E. Biphasic role for Wnt/beta-catenin signaling in cardiac specification in zebrafish and embryonic stem cells. Proc. Natl. Acad. Sci. USA 2007, 104, 9685–9690. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Evans, S.M.; Sun, Y. Insights into cardiac conduction system formation provided by HCN4 expression. Trends Cardiovasc. Med. 2015, 25, 1–9. [Google Scholar] [CrossRef]
- Kelly, R.G.; Buckingham, M.E.; Moorman, A.F. Heart fields and cardiac morphogenesis. Cold Spring Harb. Perspect. Med. 2014, 4, a015750. [Google Scholar] [CrossRef] [PubMed]
- Meilhac, S.M.; Lescroart, F.; Blanpain, C.; Buckingham, M.E. Cardiac cell lineages that form the heart. Cold Spring Harb. Perspect. Med. 2014, 4, a013888. [Google Scholar] [CrossRef]
- Brade, T.; Pane, L.S.; Moretti, A.; Chien, K.R.; Laugwitz, K.-L. Embryonic heart progenitors and cardiogenesis. Cold Spring Harb. Perspect. Med. 2013, 3, a013847. [Google Scholar] [CrossRef]
- Van Weerd, J.H.; Christoffels, V.M. The formation and function of the cardiac conduction system. Development 2016, 143, 197. [Google Scholar] [CrossRef]
- Niederreither, K.; Dollé, P. Retinoic acid in development: Towards an integrated view. Nat. Rev. Genet. 2008, 9, 541–553. [Google Scholar] [CrossRef]
- MacGrogan, D.; Münch, J.; de la Pompa, J.L. Notch and interacting signalling pathways in cardiac development, disease, and regeneration. Nat. Rev. Cardiol. 2018, 15, 685–704. [Google Scholar] [CrossRef]
- Matsa, E.; Burridge, P.W.; Wu, J.C. Human stem cells for modeling heart disease and for drug discovery. Sci. Transl. Med. 2014, 6, 239ps6. [Google Scholar] [CrossRef] [PubMed]
- Branco, M.A.; Cotovio, J.P.; Rodrigues, C.A.V.; Vaz, S.H.; Fernandes, T.G.; Moreira, L.M.; Cabral, J.M.S.; Diogo, M.M. Transcriptomic analysis of 3D Cardiac Differentiation of Human Induced Pluripotent Stem Cells Reveals Faster Cardiomyocyte Maturation Compared to 2D Culture. Sci. Rep. 2019, 9, 9229. [Google Scholar] [CrossRef]
- Hernandez, D.; Millard, R.; Sivakumaran, P.; Wong, R.C.; Crombie, D.E.; Hewitt, A.W.; Liang, H.; Hung, S.S.; Pebay, A.; Shepherd, R.K.; et al. Electrical Stimulation Promotes Cardiac Differentiation of Human Induced Pluripotent Stem Cells. Stem Cells Int. 2016, 2016, 1718041. [Google Scholar] [CrossRef] [PubMed]
- Zwi, L.; Caspi, O.; Arbel, G.; Huber, I.; Gepstein, A.; Park, I.H.; Gepstein, L. Cardiomyocyte differentiation of human induced pluripotent stem cells. Circulation 2009, 120, 1513–1523. [Google Scholar] [CrossRef]
- Zhang, J.; Wilson, G.F.; Soerens, A.G.; Koonce, C.H.; Yu, J.; Palecek, S.P.; Thomson, J.A.; Kamp, T.J. Functional cardiomyocytes derived from human induced pluripotent stem cells. Circ. Res. 2009, 104, e30–e41. [Google Scholar] [CrossRef]
- Zhang, Q.; Jiang, J.; Han, P.; Yuan, Q.; Zhang, J.; Zhang, X.; Xu, Y.; Cao, H.; Meng, Q.; Chen, L.; et al. Direct differentiation of atrial and ventricular myocytes from human embryonic stem cells by alternating retinoid signals. Cell Res. 2011, 21, 579–587. [Google Scholar] [CrossRef]
- Liang, W.; Han, P.; Kim, E.H.; Mak, J.; Zhang, R.; Torrente, A.G.; Goldhaber, J.I.; Marbán, E.; Cho, H.C. Canonical Wnt signaling promotes pacemaker cell specification of cardiac mesodermal cells derived from mouse and human embryonic stem cells. Stem Cells 2020, 38, 352–368. [Google Scholar] [CrossRef] [PubMed]
- Siebel, C.; Lendahl, U. Notch Signaling in Development, Tissue Homeostasis, and Disease. Physiol. Rev. 2017, 97, 1235–1294. [Google Scholar] [CrossRef] [PubMed]
- Niessen, K.; Karsan, A. Notch Signaling in Cardiac Development. Circ. Res. 2008, 102, 1169–1181. [Google Scholar] [CrossRef] [PubMed]
- Bray, S.J. Notch signalling in context. Nat. Rev. Mol. Cell Biol. 2016, 17, 722–735. [Google Scholar] [CrossRef] [PubMed]
- De la Pompa, J.L.; Epstein, J.A. Coordinating tissue interactions: Notch signaling in cardiac development and disease. Dev. Cell 2012, 22, 244–254. [Google Scholar] [CrossRef]
- Wu, S.P.; Cheng, C.M.; Lanz, R.B.; Wang, T.; Respress, J.L.; Ather, S.; Chen, W.; Tsai, S.J.; Wehrens, X.H.; Tsai, M.J.; et al. Atrial identity is determined by a COUP-TFII regulatory network. Dev. Cell 2013, 25, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Protze, S.I.; Liu, J.; Nussinovitch, U.; Ohana, L.; Backx, P.H.; Gepstein, L.; Keller, G.M. Sinoatrial node cardiomyocytes derived from human pluripotent cells function as a biological pacemaker. Nat. Biotechnol. 2017, 35, 56–68. [Google Scholar] [CrossRef]
- Karakikes, I.; Senyei, G.D.; Hansen, J.; Kong, C.-W.; Azeloglu, E.U.; Stillitano, F.; Lieu, D.K.; Wang, J.; Ren, L.; Hulot, J.-S.; et al. Small molecule-mediated directed differentiation of human embryonic stem cells toward ventricular cardiomyocytes. Stem Cells Transl. Med. 2014, 3, 18–31. [Google Scholar] [CrossRef]
- Christoffels Vincent, M.; Smits Gertien, J.; Kispert, A.; Moorman Antoon, F.M. Development of the Pacemaker Tissues of the Heart. Circ. Res. 2010, 106, 240–254. [Google Scholar] [CrossRef]
- Birket, M.J.; Ribeiro, M.C.; Verkerk, A.O.; Ward, D.; Leitoguinho, A.R.; den Hartogh, S.C.; Orlova, V.V.; Devalla, H.D.; Schwach, V.; Bellin, M.; et al. Expansion and patterning of cardiovascular progenitors derived from human pluripotent stem cells. Nat. Biotechnol. 2015, 33, 970–979. [Google Scholar] [CrossRef]
- Ren, J.; Han, P.; Ma, X.; Farah, E.N.; Bloomekatz, J.; Zeng, X.-X.I.; Zhang, R.; Swim, M.M.; Witty, A.D.; Knight, H.G.; et al. Canonical Wnt5b Signaling Directs Outlying Nkx2.5+ Mesoderm into Pacemaker Cardiomyocytes. Dev. Cell 2019, 50, 729–743. [Google Scholar] [CrossRef]
- Ionta, V.; Liang, W.; Kim, E.H.; Rafie, R.; Giacomello, A.; Marbán, E.; Cho, H.C. SHOX2 overexpression favors differentiation of embryonic stem cells into cardiac pacemaker cells, improving biological pacing ability. Stem Cell Rep. 2015, 4, 129–142. [Google Scholar] [CrossRef]
- Jung, J.J.; Husse, B.; Rimmbach, C.; Krebs, S.; Stieber, J.; Steinhoff, G.; Dendorfer, A.; Franz, W.M.; David, R. Programming and isolation of highly pure physiologically and pharmacologically functional sinus-nodal bodies from pluripotent stem cells. Stem Cell Rep. 2014, 2, 592–605. [Google Scholar] [CrossRef] [PubMed]
- Brown, K.; Legros, S.; Ortega, F.A.; Dai, Y.; Doss, M.X.; Christini, D.J.; Robinson, R.B.; Foley, A.C. Overexpression of Map3k7 activates sinoatrial node-like differentiation in mouse ES-derived cardiomyocytes. PLoS ONE 2017, 12, e0189818. [Google Scholar] [CrossRef] [PubMed]
- Devalla, H.D.; Schwach, V.; Ford, J.W.; Milnes, J.T.; El-Haou, S.; Jackson, C.; Gkatzis, K.; Elliott, D.A.; Chuva de Sousa Lopes, S.M.; Mummery, C.L.; et al. Atrial-like cardiomyocytes from human pluripotent stem cells are a robust preclinical model for assessing atrial-selective pharmacology. EMBO Mol. Med. 2015, 7, 394–410. [Google Scholar] [CrossRef] [PubMed]
- Lemme, M.; Ulmer, B.M.; Lemoine, M.D.; Zech, A.T.L.; Flenner, F.; Ravens, U.; Reichenspurner, H.; Rol-Garcia, M.; Smith, G.; Hansen, A.; et al. Atrial-like Engineered Heart Tissue: An In Vitro Model of the Human Atrium. Stem Cell Rep. 2018, 11, 1378–1390. [Google Scholar] [CrossRef] [PubMed]
- Argenziano, M.; Lambers, E.; Hong, L.; Sridhar, A.; Zhang, M.; Chalazan, B.; Menon, A.; Savio-Galimberti, E.; Wu, J.C.; Rehman, J.; et al. Electrophysiologic Characterization of Calcium Handling in Human Induced Pluripotent Stem Cell-Derived Atrial Cardiomyocytes. Stem Cell Rep. 2018, 10, 1867–1878. [Google Scholar] [CrossRef] [PubMed]
- Lian, X.; Hsiao, C.; Wilson, G.; Zhu, K.; Hazeltine, L.B.; Azarin, S.M.; Raval, K.K.; Zhang, J.; Kamp, T.J.; Palecek, S.P. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc. Natl. Acad. Sci. USA 2012, 109, E1848. [Google Scholar] [CrossRef]
- Burridge, P.W.; Matsa, E.; Shukla, P.; Lin, Z.C.; Churko, J.M.; Ebert, A.D.; Lan, F.; Diecke, S.; Huber, B.; Mordwinkin, N.M.; et al. Chemically defined generation of human cardiomyocytes. Nat. Methods 2014, 11, 855–860. [Google Scholar] [CrossRef] [PubMed]
- Maizels, L.; Huber, I.; Arbel, G.; Tijsen, A.J.; Gepstein, A.; Khoury, A.; Gepstein, L. Patient-Specific Drug Screening Using a Human Induced Pluripotent Stem Cell Model of Catecholaminergic Polymorphic Ventricular Tachycardia Type 2. Circ. Arrhythm. Electrophysiol. 2017, 10. [Google Scholar] [CrossRef] [PubMed]
- Sala, L.; Yu, Z.; Ward-van Oostwaard, D.; van Veldhoven, J.P.; Moretti, A.; Laugwitz, K.L.; Mummery, C.L.; AP, I.J.; Bellin, M. A new hERG allosteric modulator rescues genetic and drug-induced long-QT syndrome phenotypes in cardiomyocytes from isogenic pairs of patient induced pluripotent stem cells. EMBO Mol. Med. 2016, 8, 1065–1081. [Google Scholar] [CrossRef] [PubMed]
- Sun, N.; Yazawa, M.; Liu, J.; Han, L.; Sanchez-Freire, V.; Abilez, O.J.; Navarrete, E.G.; Hu, S.; Wang, L.; Lee, A.; et al. Patient-specific induced pluripotent stem cells as a model for familial dilated cardiomyopathy. Sci. Transl. Med. 2012, 4, 130ra147. [Google Scholar] [CrossRef]
- Wu, H.; Lee, J.; Vincent, L.G.; Wang, Q.; Gu, M.; Lan, F.; Churko, J.M.; Sallam, K.I.; Matsa, E.; Sharma, A.; et al. Epigenetic Regulation of Phosphodiesterases 2A and 3A Underlies Compromised β-Adrenergic Signaling in an iPSC Model of Dilated Cardiomyopathy. Cell Stem Cell 2015, 17, 89–100. [Google Scholar] [CrossRef]
- Drawnel, F.M.; Boccardo, S.; Prummer, M.; Delobel, F.; Graff, A.; Weber, M.; Gérard, R.; Badi, L.; Kam-Thong, T.; Bu, L.; et al. Disease modeling and phenotypic drug screening for diabetic cardiomyopathy using human induced pluripotent stem cells. Cell Rep. 2014, 9, 810–821. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; McCain, M.L.; Yang, L.; He, A.; Pasqualini, F.S.; Agarwal, A.; Yuan, H.; Jiang, D.; Zhang, D.; Zangi, L.; et al. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat. Med. 2014, 20, 616–623. [Google Scholar] [CrossRef] [PubMed]
- Veerman, C.C.; Kosmidis, G.; Mummery, C.L.; Casini, S.; Verkerk, A.O.; Bellin, M. Immaturity of human stem-cell-derived cardiomyocytes in culture: Fatal flaw or soluble problem? Stem Cells Dev. 2015, 24, 1035–1052. [Google Scholar] [CrossRef]
- Karbassi, E.; Fenix, A.; Marchiano, S.; Muraoka, N.; Nakamura, K.; Yang, X.; Murry, C.E. Cardiomyocyte maturation: Advances in knowledge and implications for regenerative medicine. Nat. Rev. Cardiol. 2020, 17, 341–359. [Google Scholar] [CrossRef]
- Hinson, J.T.; Chopra, A.; Nafissi, N.; Polacheck, W.J.; Benson, C.C.; Swist, S.; Gorham, J.; Yang, L.; Schafer, S.; Sheng, C.C.; et al. Titin mutations in iPS cells define sarcomere insufficiency as a cause of dilated cardiomyopathy. Science 2015, 349, 982. [Google Scholar] [CrossRef]
- Pointon, A.; Harmer, A.R.; Dale, I.L.; Abi-Gerges, N.; Bowes, J.; Pollard, C.; Garside, H. Assessment of cardiomyocyte contraction in human-induced pluripotent stem cell-derived cardiomyocytes. Toxicol. Sci. 2015, 144, 227–237. [Google Scholar] [CrossRef]
- Titmarsh, D.M.; Glass, N.R.; Mills, R.J.; Hidalgo, A.; Wolvetang, E.J.; Porrello, E.R.; Hudson, J.E.; Cooper-White, J.J. Induction of Human iPSC-Derived Cardiomyocyte Proliferation Revealed by Combinatorial Screening in High Density Microbioreactor Arrays. Sci. Rep. 2016, 6, 24637. [Google Scholar] [CrossRef]
- Ribeiro, A.J.; Ang, Y.S.; Fu, J.D.; Rivas, R.N.; Mohamed, T.M.; Higgs, G.C.; Srivastava, D.; Pruitt, B.L. Contractility of single cardiomyocytes differentiated from pluripotent stem cells depends on physiological shape and substrate stiffness. Proc. Natl. Acad. Sci. USA 2015, 112, 12705–12710. [Google Scholar] [CrossRef] [PubMed]
- Snir, M.; Kehat, I.; Gepstein, A.; Coleman, R.; Itskovitz-Eldor, J.; Livne, E.; Gepstein, L. Assessment of the ultrastructural and proliferative properties of human embryonic stem cell-derived cardiomyocytes. Am. J. Physiol. Heart Circ. Physiol. 2003, 285, H2355–H2363. [Google Scholar] [CrossRef] [PubMed]
- Mollova, M.; Bersell, K.; Walsh, S.; Savla, J.; Das, L.T.; Park, S.-Y.; Silberstein, L.E.; dos Remedios, C.G.; Graham, D.; Colan, S.; et al. Cardiomyocyte proliferation contributes to heart growth in young humans. Proc. Natl. Acad. Sci. USA 2013, 110, 1446. [Google Scholar] [CrossRef] [PubMed]
- Boateng, S.Y.; Goldspink, P.H. Assembly and maintenance of the sarcomere night and day. Cardiovasc. Res. 2008, 77, 667–675. [Google Scholar] [CrossRef] [PubMed]
- Lundy, S.D.; Zhu, W.-Z.; Regnier, M.; Laflamme, M.A. Structural and Functional Maturation of Cardiomyocytes Derived from Human Pluripotent Stem Cells. Stem Cells Dev. 2013, 22, 1991–2002. [Google Scholar] [CrossRef]
- Hirt, M.N.; Boeddinghaus, J.; Mitchell, A.; Schaaf, S.; Bornchen, C.; Muller, C.; Schulz, H.; Hubner, N.; Stenzig, J.; Stoehr, A.; et al. Functional improvement and maturation of rat and human engineered heart tissue by chronic electrical stimulation. J. Mol. Cell. Cardiol. 2014, 74, 151–161. [Google Scholar] [CrossRef]
- Mannhardt, I.; Breckwoldt, K.; Letuffe-Brenière, D.; Schaaf, S.; Schulz, H.; Neuber, C.; Benzin, A.; Werner, T.; Eder, A.; Schulze, T.; et al. Human Engineered Heart Tissue: Analysis of Contractile Force. Stem Cell Rep. 2016, 7, 29–42. [Google Scholar] [CrossRef]
- Linke Wolfgang, A.; Hamdani, N. Gigantic Business. Circ. Res. 2014, 114, 1052–1068. [Google Scholar] [CrossRef]
- Opitz, C.A.; Leake, M.C.; Makarenko, I.; Benes, V.; Linke, W.A. Developmentally regulated switching of titin size alters myofibrillar stiffness in the perinatal heart. Circ. Res. 2004, 94, 967–975. [Google Scholar] [CrossRef] [PubMed]
- Lahmers, S.; Wu, Y.; Call, D.R.; Labeit, S.; Granzier, H. Developmental control of titin isoform expression and passive stiffness in fetal and neonatal myocardium. Circ. Res. 2004, 94, 505–513. [Google Scholar] [CrossRef]
- Reiser, P.J.; Portman, M.A.; Ning, X.H.; Schomisch Moravec, C. Human cardiac myosin heavy chain isoforms in fetal and failing adult atria and ventricles. Am. J. Physiol. Heart Circ. Physiol. 2001, 280, H1814–H1820. [Google Scholar] [CrossRef]
- Yoshida, S.; Miyagawa, S.; Fukushima, S.; Kawamura, T.; Kashiyama, N.; Ohashi, F.; Toyofuku, T.; Toda, K.; Sawa, Y. Maturation of Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes by Soluble Factors from Human Mesenchymal Stem Cells. Mol. Ther. 2018, 26, 2681–2695. [Google Scholar] [CrossRef] [PubMed]
- Hailstones, D.; Barton, P.; Chan-Thomas, P.; Sasse, S.; Sutherland, C.; Hardeman, E.; Gunning, P. Differential regulation of the atrial isoforms of the myosin light chains during striated muscle development. J. Biol. Chem. 1992, 267, 23295–23300. [Google Scholar] [CrossRef]
- England, J.; Loughna, S. Heavy and light roles: Myosin in the morphogenesis of the heart. Cell. Mol. Life Sci. 2013, 70, 1221–1239. [Google Scholar] [CrossRef]
- Huang, C.Y.; Peres Moreno Maia-Joca, R.; Ong, C.S.; Wilson, I.; DiSilvestre, D.; Tomaselli, G.F.; Reich, D.H. Enhancement of human iPSC-derived cardiomyocyte maturation by chemical conditioning in a 3D environment. J. Mol. Cell. Cardiol. 2020, 138, 1–11. [Google Scholar] [CrossRef]
- Shadrin, I.Y.; Allen, B.W.; Qian, Y.; Jackman, C.P.; Carlson, A.L.; Juhas, M.E.; Bursac, N. Cardiopatch platform enables maturation and scale-up of human pluripotent stem cell-derived engineered heart tissues. Nat. Commun. 2017, 8, 1825. [Google Scholar] [CrossRef]
- Birket, M.J.; Casini, S.; Kosmidis, G.; Elliott, D.A.; Gerencser, A.A.; Baartscheer, A.; Schumacher, C.; Mastroberardino, P.G.; Elefanty, A.G.; Stanley, E.G.; et al. PGC-1α and Reactive Oxygen Species Regulate Human Embryonic Stem Cell-Derived Cardiomyocyte Function. Stem Cell Rep. 2013, 1, 560–574. [Google Scholar] [CrossRef] [PubMed]
- Piquereau, J.; Ventura-Clapier, R. Maturation of Cardiac Energy Metabolism During Perinatal Development. Front. Physiol. 2018, 9, 959. [Google Scholar] [CrossRef]
- Piquereau, J.; Caffin, F.; Novotova, M.; Lemaire, C.; Veksler, V.; Garnier, A.; Ventura-Clapier, R.; Joubert, F. Mitochondrial dynamics in the adult cardiomyocytes: Which roles for a highly specialized cell? Front. Physiol. 2013, 4. [Google Scholar] [CrossRef]
- Ulmer, B.M.; Eschenhagen, T. Human pluripotent stem cell-derived cardiomyocytes for studying energy metabolism. Biochim. Biophys. Acta Mol. Cell Res. 2020, 1867, 118471. [Google Scholar] [CrossRef] [PubMed]
- Parikh, S.S.; Blackwell, D.J.; Gomez-Hurtado, N.; Frisk, M.; Wang, L.; Kim, K.; Dahl, C.P.; Fiane, A.; Tønnessen, T.; Kryshtal, D.O.; et al. Thyroid and Glucocorticoid Hormones Promote Functional T-Tubule Development in Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes. Circ. Res. 2017, 121, 1323–1330. [Google Scholar] [CrossRef] [PubMed]
- Brette, F.; Orchard, C. T-Tubule Function in Mammalian Cardiac Myocytes. Circ. Res. 2003, 92, 1182–1192. [Google Scholar] [CrossRef] [PubMed]
- Nunes, S.S.; Miklas, J.W.; Liu, J.; Aschar-Sobbi, R.; Xiao, Y.; Zhang, B.; Jiang, J.; Masse, S.; Gagliardi, M.; Hsieh, A.; et al. Biowire: A platform for maturation of human pluripotent stem cell-derived cardiomyocytes. Nat. Methods 2013, 10, 781–787. [Google Scholar] [CrossRef]
- Salameh, A.; Wustmann, A.; Karl, S.; Blanke, K.; Apel, D.; Rojas-Gomez, D.; Franke, H.; Mohr, F.W.; Janousek, J.; Dhein, S. Cyclic mechanical stretch induces cardiomyocyte orientation and polarization of the gap junction protein connexin43. Circ. Res. 2010, 106, 1592–1602. [Google Scholar] [CrossRef]
- Vreeker, A.; van Stuijvenberg, L.; Hund, T.J.; Mohler, P.J.; Nikkels, P.G.J.; van Veen, T.A.B. Assembly of the Cardiac Intercalated Disk during Pre- and Postnatal Development of the Human Heart. PLoS ONE 2014, 9, e94722. [Google Scholar] [CrossRef] [PubMed]
- Mills, R.J.; Titmarsh, D.M.; Koenig, X.; Parker, B.L.; Ryall, J.G.; Quaife-Ryan, G.A.; Voges, H.K.; Hodson, M.P.; Ferguson, C.; Drowley, L.; et al. Functional screening in human cardiac organoids reveals a metabolic mechanism for cardiomyocyte cell cycle arrest. Proc. Natl. Acad. Sci. USA 2017, 114, E8372–E8381. [Google Scholar] [CrossRef] [PubMed]
- Herron Todd, J.; Rocha Andre Monteiro, D.; Campbell Katherine, F.; Ponce-Balbuena, D.; Willis, B.C.; Guerrero-Serna, G.; Liu, Q.; Klos, M.; Musa, H.; Zarzoso, M.; et al. Extracellular Matrix–Mediated Maturation of Human Pluripotent Stem Cell–Derived Cardiac Monolayer Structure and Electrophysiological Function. Circ. Arrhythm Electrophysiol. 2016, 9, e003638. [Google Scholar] [CrossRef]
- Harrild, D.M.; Henriquez, C.S. A Computer Model of Normal Conduction in the Human Atria. Circ. Res. 2000, 87, e25–e36. [Google Scholar] [CrossRef]
- Hwang, H.S.; Kryshtal, D.O.; Feaster, T.K.; Sánchez-Freire, V.; Zhang, J.; Kamp, T.J.; Hong, C.C.; Wu, J.C.; Knollmann, B.C. Comparable calcium handling of human iPSC-derived cardiomyocytes generated by multiple laboratories. J. Mol. Cell. Cardiol. 2015, 85, 79–88. [Google Scholar] [CrossRef]
- Veerman, C.C.; Mengarelli, I.; Lodder, E.M.; Kosmidis, G.; Bellin, M.; Zhang, M.; Dittmann, S.; Guan, K.; Wilde, A.A.M.; Schulze-Bahr, E.; et al. Switch from Fetal to Adult SCN5A Isoform in Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes Unmasks the Cellular Phenotype of a Conduction Disease-Causing Mutation. J. Am. Heart Assoc. 2017, 6. [Google Scholar] [CrossRef]
- Horváth, A.; Lemoine, M.D.; Löser, A.; Mannhardt, I.; Flenner, F.; Uzun, A.U.; Neuber, C.; Breckwoldt, K.; Hansen, A.; Girdauskas, E.; et al. Low Resting Membrane Potential and Low Inward Rectifier Potassium Currents Are Not Inherent Features of hiPSC-Derived Cardiomyocytes. Stem Cell Rep. 2018, 10, 822–833. [Google Scholar] [CrossRef]
- Ieda, M.; Tsuchihashi, T.; Ivey, K.N.; Ross, R.S.; Hong, T.-T.; Shaw, R.M.; Srivastava, D. Cardiac fibroblasts regulate myocardial proliferation through beta1 integrin signaling. Dev. Cell 2009, 16, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Spach, M.S.; Heidlage, J.F.; Barr, R.C.; Dolber, P.C. Cell size and communication: Role in structural and electrical development and remodeling of the heart. Heart Rhythm 2004, 1, 500–515. [Google Scholar] [CrossRef] [PubMed]
- Biermann, M.; Cai, W.; Lang, D.; Hermsen, J.; Profio, L.; Zhou, Y.; Czirok, A.; Isai, D.G.; Napiwocki, B.N.; Rodriguez, A.M.; et al. Epigenetic Priming of Human Pluripotent Stem Cell-Derived Cardiac Progenitor Cells Accelerates Cardiomyocyte Maturation. Stem Cells 2019, 37, 910–923. [Google Scholar] [CrossRef]
- McCain, M.L.; Parker, K.K. Mechanotransduction: The role of mechanical stress, myocyte shape, and cytoskeletal architecture on cardiac function. Pflug. Arch. 2011, 462, 89–104. [Google Scholar] [CrossRef]
- Denning, C.; Borgdorff, V.; Crutchley, J.; Firth, K.S.A.; George, V.; Kalra, S.; Kondrashov, A.; Hoang, M.D.; Mosqueira, D.; Patel, A.; et al. Cardiomyocytes from human pluripotent stem cells: From laboratory curiosity to industrial biomedical platform. Biochim. Biophys. Acta BBA Mol. Cell Res. 2016, 1863, 1728–1748. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Guo, L.; Fiene, S.J.; Anson, B.D.; Thomson, J.A.; Kamp, T.J.; Kolaja, K.L.; Swanson, B.J.; January, C.T. High purity human-induced pluripotent stem cell-derived cardiomyocytes: Electrophysiological properties of action potentials and ionic currents. Am. J. Physiol. Heart Circ. Physiol. 2011, 301, H2006–H2017. [Google Scholar] [CrossRef] [PubMed]
- Fox, A.A.; Sharma, S.; Mounsey, J.P.; Durieux, M.E.; Whitlock, R.; Bennett-Guerrero, E. Chapter 6—Molecular and Genetic Cardiovascular Medicine and Systemic Inflammation. In Kaplan’s Essentials of Cardiac Anesthesia, 2nd ed.; Kaplan, J.A., Ed.; Elsevier: Philadelphia, PA, USA, 2018; pp. 94–111. [Google Scholar]
- DeMazumder, D.; Tomaselli, G.F. Chapter 41—Molecular and Cellular Mechanisms of Cardiac Arrhythmias. In Muscle; Hill, J.A., Olson, E.N., Eds.; Academic Press: Boston, MA, USA, 2012; pp. 583–599. [Google Scholar]
- Pandit, S.V. 31—Ionic Mechanisms of Atrial Action Potentials. In Cardiac Electrophysiology: From Cell to Bedside, 7th ed.; Zipes, D.P., Jalife, J., Stevenson, W.G., Eds.; Elsevier: Philadelphia, PA, USA, 2018; pp. 293–303. [Google Scholar]
- Kay, G.N.; Shepard, R.B. 3—Stimulation and Excitation of Cardiac Tissues. In Clinical Cardiac Pacing, Defibrillation and Resynchronization Therapy, 5th ed.; Ellenbogen, K.A., Wilkoff, B.L., Kay, G.N., Lau, C.-P., Auricchio, A., Eds.; Elsevier: Philadelphia, PA, USA, 2017; pp. 61–113. [Google Scholar]
- Heijman, J.; Erfanian Abdoust, P.; Voigt, N.; Nattel, S.; Dobrev, D. Computational models of atrial cellular electrophysiology and calcium handling, and their role in atrial fibrillation. J. Physiol. 2016, 594, 537–553. [Google Scholar] [CrossRef] [PubMed]
- Amos, G.J.; Wettwer, E.; Metzger, F.; Li, Q.; Himmel, H.M.; Ravens, U. Differences between outward currents of human atrial and subepicardial ventricular myocytes. J. Physiol. 1996, 491, 31–50. [Google Scholar] [CrossRef]
- Trautwein, W.; Kassebaum, D.G.; Nelson, R.M.; Hecht, H.H. Electrophysiological study of human heart muscle. Circ. Res. 1962, 10, 306–312. [Google Scholar] [CrossRef]
- Grandi, E.; Pandit, S.V.; Voigt, N.; Workman, A.J.; Dobrev, D.; Jalife, J.; Bers, D.M. Human atrial action potential and Ca2+ model: Sinus rhythm and chronic atrial fibrillation. Circ. Res. 2011, 109, 1055–1066. [Google Scholar] [CrossRef]
- Jeevaratnam, K.; Chadda, K.R.; Huang, C.L.; Camm, A.J. Cardiac Potassium Channels: Physiological Insights for Targeted Therapy. J. Cardiovasc. Pharm. 2018, 23, 119–129. [Google Scholar] [CrossRef]
- Doss, M.X.; Di Diego, J.M.; Goodrow, R.J.; Wu, Y.; Cordeiro, J.M.; Nesterenko, V.V.; Barajas-Martínez, H.; Hu, D.; Urrutia, J.; Desai, M.; et al. Maximum Diastolic Potential of Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes Depends Critically on IKr. PLoS ONE 2012, 7, e40288. [Google Scholar] [CrossRef]
- Kreuzberg, M.M.; Willecke, K.; Bukauskas, F.F. Connexin-mediated cardiac impulse propagation: Connexin 30.2 slows atrioventricular conduction in mouse heart. Trends Cardiovasc. Med. 2006, 16, 266–272. [Google Scholar] [CrossRef] [PubMed]
- Csepe, T.A.; Zhao, J.; Hansen, B.J.; Li, N.; Sul, L.V.; Lim, P.; Wang, Y.; Simonetti, O.P.; Kilic, A.; Mohler, P.J.; et al. Human sinoatrial node structure: 3D microanatomy of sinoatrial conduction pathways. Prog. Biophys. Mol. Biol. 2016, 120, 164–178. [Google Scholar] [CrossRef]
- Dobrzynski, H.; Boyett, M.R.; Anderson, R.H. New insights into pacemaker activity: Promoting understanding of sick sinus syndrome. Circulation 2007, 115, 1921–1932. [Google Scholar] [CrossRef]
- Davis, L.M.; Rodefeld, M.E.; Green, K.; Beyer, E.C.; Saffitz, J.E. Gap Junction Protein Phenotypes of the Human Heart and Conduction System. J. Cardiovasc. Electrophysiol. 1995, 6, 813–822. [Google Scholar] [CrossRef] [PubMed]
- Scuderi, G.J.; Butcher, J. Naturally Engineered Maturation of Cardiomyocytes. Front. Cell Dev. Biol. 2017, 5, 50. [Google Scholar] [CrossRef] [PubMed]
- McDevitt, T.C.; Angello, J.C.; Whitney, M.L.; Reinecke, H.; Hauschka, S.D.; Murry, C.E.; Stayton, P.S. In vitro generation of differentiated cardiac myofibers on micropatterned laminin surfaces. J Biomed. Mater. Res. 2002, 60, 472–479. [Google Scholar] [CrossRef]
- Zhang, W.; Kong, C.W.; Tong, M.H.; Chooi, W.H.; Huang, N.; Li, R.A.; Chan, B.P. Maturation of human embryonic stem cell-derived cardiomyocytes (hESC-CMs) in 3D collagen matrix: Effects of niche cell supplementation and mechanical stimulation. Acta Biomater. 2017, 49, 204–217. [Google Scholar] [CrossRef] [PubMed]
- MacLennan, D.H.; Kranias, E.G. Phospholamban: A crucial regulator of cardiac contractility. Nat. Rev. Mol. Cell Biol. 2003, 4, 566–577. [Google Scholar] [CrossRef] [PubMed]
- Greiser, M.; Kerfant, B.-G.; Williams, G.S.B.; Voigt, N.; Harks, E.; Dibb, K.M.; Giese, A.; Meszaros, J.; Verheule, S.; Ravens, U.; et al. Tachycardia-induced silencing of subcellular Ca2+ signaling in atrial myocytes. J. Clin. Investig. 2014, 124, 4759–4772. [Google Scholar] [CrossRef] [PubMed]
- Györke, S.; Carnes, C. Dysregulated sarcoplasmic reticulum calcium release: Potential pharmacological target in cardiac disease. Pharmacol. Ther. 2008, 119, 340–354. [Google Scholar] [CrossRef] [PubMed]
- Lopaschuk, G.D.; Jaswal, J.S. Energy Metabolic Phenotype of the Cardiomyocyte During Development, Differentiation, and Postnatal Maturation. J. Cardiovasc. Pharmacol. 2010, 56, 130–140. [Google Scholar] [CrossRef] [PubMed]
- Stanley, W.C.; Recchia, F.A.; Lopaschuk, G.D. Myocardial Substrate Metabolism in the Normal and Failing Heart. Physiol. Rev. 2005, 85, 1093–1129. [Google Scholar] [CrossRef] [PubMed]
- Lopaschuk, G.D.; Spafford, M.A.; Marsh, D.R. Glycolysis is predominant source of myocardial ATP production immediately after birth. Am. J. Physiol. Heart Circ. Physiol. 1991, 261, H1698–H1705. [Google Scholar] [CrossRef] [PubMed]
- Karakikes, I.; Ameen, M.; Termglinchan, V.; Wu, J.C. Human induced pluripotent stem cell-derived cardiomyocytes: Insights into molecular, cellular, and functional phenotypes. Circ. Res. 2015, 117, 80–88. [Google Scholar] [CrossRef]
- Correia, C.; Koshkin, A.; Duarte, P.; Hu, D.; Carido, M.; Sebastião, M.J.; Gomes-Alves, P.; Elliott, D.A.; Domian, I.J.; Teixeira, A.P.; et al. 3D aggregate culture improves metabolic maturation of human pluripotent stem cell derived cardiomyocytes. Biotechnol. Bioeng. 2018, 115, 630–644. [Google Scholar] [CrossRef] [PubMed]
- DeLaughter, D.M.; Bick, A.G.; Wakimoto, H.; McKean, D.; Gorham, J.M.; Kathiriya, I.S.; Hinson, J.T.; Homsy, J.; Gray, J.; Pu, W. Single-cell resolution of temporal gene expression during heart development. Dev. Cell 2016, 39, 480–490. [Google Scholar] [CrossRef] [PubMed]
- Uosaki, H.; Taguchi, Y. Comparative gene expression analysis of mouse and human cardiac maturation. Genom. Proteom. Bioinform. 2016, 14, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wang, L.; Liu, Z.; Alimohamadi, S.; Yin, C.; Liu, J.; Qian, L. Comparative Gene Expression Analyses Reveal Distinct Molecular Signatures between Differentially Reprogrammed Cardiomyocytes. Cell Rep. 2017, 20, 3014–3024. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.B.; Moretti, A.; Mederos y Schnitzler, M.; Iop, L.; Storch, U.; Bellin, M.; Dorn, T.; Ruppenthal, S.; Pfeiffer, S.; Goedel, A.; et al. Dantrolene rescues arrhythmogenic RYR2 defect in a patient-specific stem cell model of catecholaminergic polymorphic ventricular tachycardia. EMBO Mol. Med. 2012, 4, 180–191. [Google Scholar] [CrossRef] [PubMed]
- Rao, C.; Prodromakis, T.; Kolker, L.; Chaudhry, U.A.R.; Trantidou, T.; Sridhar, A.; Weekes, C.; Camelliti, P.; Harding, S.E.; Darzi, A.; et al. The effect of microgrooved culture substrates on calcium cycling of cardiac myocytes derived from human induced pluripotent stem cells. Biomaterials 2013, 34, 2399–2411. [Google Scholar] [CrossRef] [PubMed]
- Itzhaki, I.; Rapoport, S.; Huber, I.; Mizrahi, I.; Zwi-Dantsis, L.; Arbel, G.; Schiller, J.; Gepstein, L. Calcium Handling in Human Induced Pluripotent Stem Cell Derived Cardiomyocytes. PLoS ONE 2011, 6, e18037. [Google Scholar] [CrossRef] [PubMed]
- Van den Berg, C.W.; Okawa, S.; Chuva de Sousa Lopes, S.M.; van Iperen, L.; Passier, R.; Braam, S.R.; Tertoolen, L.G.; del Sol, A.; Davis, R.P.; Mummery, C.L. Transcriptome of human foetal heart compared with cardiomyocytes from pluripotent stem cells. Development 2015, 142, 3231–3238. [Google Scholar] [CrossRef]
- Yin, Z.; Ren, J.; Guo, W. Sarcomeric protein isoform transitions in cardiac muscle: A journey to heart failure. Biochim. Biophys. Acta BBA Mol. Basis Dis. 2015, 1852, 47–52. [Google Scholar] [CrossRef]
- Guo, Y.; Pu William, T. Cardiomyocyte Maturation. Circ. Res. 2020, 126, 1086–1106. [Google Scholar] [CrossRef] [PubMed]
- Miklas, J.W.; Clark, E.; Levy, S.; Detraux, D.; Leonard, A.; Beussman, K.; Showalter, M.R.; Smith, A.T.; Hofsteen, P.; Yang, X.; et al. TFPa/HADHA is required for fatty acid beta-oxidation and cardiolipin re-modeling in human cardiomyocytes. Nat. Commun. 2019, 10, 4671. [Google Scholar] [CrossRef]
- Lewandowski, J.; Rozwadowska, N.; Kolanowski, T.J.; Malcher, A.; Zimna, A.; Rugowska, A.; Fiedorowicz, K.; Łabędź, W.; Kubaszewski, Ł.; Chojnacka, K.; et al. The impact of in vitro cell culture duration on the maturation of human cardiomyocytes derived from induced pluripotent stem cells of myogenic origin. Cell Transpl. 2018, 27, 1047–1067. [Google Scholar] [CrossRef]
- Kamakura, T.; Makiyama, T.; Sasaki, K.; Yoshida, Y.; Wuriyanghai, Y.; Chen, J.; Hattori, T.; Ohno, S.; Kita, T.; Horie, M.; et al. Ultrastructural maturation of human-induced pluripotent stem cell-derived cardiomyocytes in a long-term culture. Circ. J. 2013, 77, 1307–1314. [Google Scholar] [CrossRef]
- Piccini, I.; Rao, J.; Seebohm, G.; Greber, B. Human pluripotent stem cell-derived cardiomyocytes: Genome-wide expression profiling of long-term in vitro maturation in comparison to human heart tissue. Genom. Data 2015, 4, 69–72. [Google Scholar] [CrossRef] [PubMed]
- Macadangdang, J.; Guan, X.; Smith, A.S.; Lucero, R.; Czerniecki, S.; Childers, M.K.; Mack, D.L.; Kim, D.H. Nanopatterned Human iPSC-based Model of a Dystrophin-Null Cardiomyopathic Phenotype. Cell Mol. Bioeng. 2015, 8, 320–332. [Google Scholar] [CrossRef] [PubMed]
- Jacot, J.G.; Martin, J.C.; Hunt, D.L. Mechanobiology of cardiomyocyte development. J. Biomech. 2010, 43, 93–98. [Google Scholar] [CrossRef]
- Feaster, T.K.; Cadar, A.G.; Wang, L.; Williams, C.H.; Chun, Y.W.; Hempel, J.E.; Bloodworth, N.; Merryman, W.D.; Lim, C.C.; Wu, J.C.; et al. Matrigel Mattress: A Method for the Generation of Single Contracting Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes. Circ. Res. 2015, 117, 995–1000. [Google Scholar] [CrossRef] [PubMed]
- Martewicz, S.; Serena, E.; Zatti, S.; Keller, G.; Elvassore, N. Substrate and mechanotransduction influence SERCA2a localization in human pluripotent stem cell-derived cardiomyocytes affecting functional performance. Stem Cell Res. 2017, 25, 107–114. [Google Scholar] [CrossRef]
- MacQueen, L.A.; Sheehy, S.P.; Chantre, C.O.; Zimmerman, J.F.; Pasqualini, F.S.; Liu, X.; Goss, J.A.; Campbell, P.H.; Gonzalez, G.M.; Park, S.-J.; et al. A tissue-engineered scale model of the heart ventricle. Nat. Biomed. Eng. 2018, 2, 930–941. [Google Scholar] [CrossRef]
- Blazeski, A.; Lowenthal, J.; Zhu, R.; Ewoldt, J.; Boheler, K.R.; Tung, L. Functional Properties of Engineered Heart Slices Incorporating Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes. Stem Cell Rep. 2019, 12, 982–995. [Google Scholar] [CrossRef]
- Camelliti, P.; Borg, T.K.; Kohl, P. Structural and functional characterisation of cardiac fibroblasts. Cardiovasc. Res. 2005, 65, 40–51. [Google Scholar] [CrossRef]
- Brutsaert, D.L. Cardiac endothelial-myocardial signaling: Its role in cardiac growth, contractile performance, and rhythmicity. Physiol. Rev. 2003, 83, 59–115. [Google Scholar] [CrossRef] [PubMed]
- Pinto, A.R.; Ilinykh, A.; Ivey, M.J.; Kuwabara, J.T.; D’Antoni, M.L.; Debuque, R.; Chandran, A.; Wang, L.; Arora, K.; Rosenthal, N.A.; et al. Revisiting Cardiac Cellular Composition. Circ. Res. 2016, 118, 400–409. [Google Scholar] [CrossRef] [PubMed]
- Tirziu, D.; Giordano, F.J.; Simons, M. Cell communications in the heart. Circulation 2010, 122, 928–937. [Google Scholar] [CrossRef]
- Abecasis, B.; Gomes-Alves, P.; Rosa, S.; Gouveia, P.J.; Ferreira, L.; Serra, M.; Alves, P.M. Unveiling the molecular crosstalk in a human induced pluripotent stem cell-derived cardiac model. Biotechnol. Bioeng. 2019, 116, 1245–1252. [Google Scholar] [CrossRef] [PubMed]
- Dunn, K.K.; Reichardt, I.M.; Simmons, A.D.; Jin, G.; Floy, M.E.; Hoon, K.M.; Palecek, S.P. Coculture of Endothelial Cells with Human Pluripotent Stem Cell-Derived Cardiac Progenitors Reveals a Differentiation Stage-Specific Enhancement of Cardiomyocyte Maturation. Biotechnol. J. 2019, 14, e1800725. [Google Scholar] [CrossRef]
- Giacomelli, E.; Meraviglia, V.; Campostrini, G.; Cochrane, A.; Cao, X.; van Helden, R.W.J.; Krotenberg Garcia, A.; Mircea, M.; Kostidis, S.; Davis, R.P.; et al. Human-iPSC-Derived Cardiac Stromal Cells Enhance Maturation in 3D Cardiac Microtissues and Reveal Non-cardiomyocyte Contributions to Heart Disease. Cell Stem Cell 2020, 26, 862–879. [Google Scholar] [CrossRef] [PubMed]
- Horikoshi, Y.; Yan, Y.; Terashvili, M.; Wells, C.; Horikoshi, H.; Fujita, S.; Bosnjak, Z.J.; Bai, X. Fatty Acid-Treated Induced Pluripotent Stem Cell-Derived Human Cardiomyocytes Exhibit Adult Cardiomyocyte-Like Energy Metabolism Phenotypes. Cells 2019, 8, 1095. [Google Scholar] [CrossRef] [PubMed]
- Correia, C.; Koshkin, A.; Duarte, P.; Hu, D.; Teixeira, A.; Domian, I.; Serra, M.; Alves, P.M. Distinct carbon sources affect structural and functional maturation of cardiomyocytes derived from human pluripotent stem cells. Sci. Rep. 2017, 7, 8590. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Rodriguez, M.L.; Leonard, A.; Sun, L.; Fischer, K.A.; Wang, Y.; Ritterhoff, J.; Zhao, L.; Kolwicz, S.C.; Pabon, L.; et al. Fatty Acids Enhance the Maturation of Cardiomyocytes Derived from Human Pluripotent Stem Cells. Stem Cell Rep. 2019, 13, 657–668. [Google Scholar] [CrossRef]
- Nakano, H.; Minami, I.; Braas, D.; Pappoe, H.; Wu, X.; Sagadevan, A.; Vergnes, L.; Fu, K.; Morselli, M.; Dunham, C.; et al. Glucose inhibits cardiac muscle maturation through nucleotide biosynthesis. eLife 2017, 6, e29330. [Google Scholar] [CrossRef]
- Li, M.; Iismaa, S.E.; Naqvi, N.; Nicks, A.; Husain, A.; Graham, R.M. Thyroid hormone action in postnatal heart development. Stem Cell Res. 2014, 13, 582–591. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Rodriguez, M.; Pabon, L.; Fischer, K.A.; Reinecke, H.; Regnier, M.; Sniadecki, N.J.; Ruohola-Baker, H.; Murry, C.E. Tri-iodo-l-thyronine promotes the maturation of human cardiomyocytes-derived from induced pluripotent stem cells. J. Mol. Cell. Cardiol. 2014, 72, 296–304. [Google Scholar] [CrossRef]
- Rog-Zielinska, E.A.; Craig, M.A.; Manning, J.R.; Richardson, R.V.; Gowans, G.J.; Dunbar, D.R.; Gharbi, K.; Kenyon, C.J.; Holmes, M.C.; Hardie, D.G.; et al. Glucocorticoids promote structural and functional maturation of foetal cardiomyocytes: A role for PGC-1α. Cell Death Differ. 2015, 22, 1106–1116. [Google Scholar] [CrossRef] [PubMed]
- Rog-Zielinska, E.A.; Thomson, A.; Kenyon, C.J.; Brownstein, D.G.; Moran, C.M.; Szumska, D.; Michailidou, Z.; Richardson, J.; Owen, E.; Watt, A.; et al. Glucocorticoid receptor is required for foetal heart maturation. Hum. Mol. Genet. 2013, 22, 3269–3282. [Google Scholar] [CrossRef]
- Wu, L.; Jia, Z.; Yan, L.; Wang, W.; Wang, J.; Zhang, Y.; Zhou, C. Angiotensin II promotes cardiac differentiation of embryonic stem cells via angiotensin type 1 receptor. Differentiation 2013, 86, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Földes, G.; Mioulane, M.; Wright, J.S.; Liu, A.Q.; Novak, P.; Merkely, B.; Gorelik, J.; Schneider, M.D.; Ali, N.N.; Harding, S.E. Modulation of human embryonic stem cell-derived cardiomyocyte growth: A testbed for studying human cardiac hypertrophy? J. Mol. Cell. Cardiol. 2011, 50, 367–376. [Google Scholar] [CrossRef]
- Nakamura, M.; Sadoshima, J. Mechanisms of physiological and pathological cardiac hypertrophy. Nat. Rev. Cardiol. 2018, 15, 387–407. [Google Scholar] [CrossRef] [PubMed]
- Zuppinger, C. 3D culture for cardiac cells. Biochim. Biophys. Acta 2016, 1863, 1873–1881. [Google Scholar] [CrossRef] [PubMed]
- Mirbagheri, M.; Adibnia, V.; Hughes, B.R.; Waldman, S.D.; Banquy, X.; Hwang, D.K. Advanced cell culture platforms: A growing quest for emulating natural tissues. Mater. Horiz. 2019, 6, 45–71. [Google Scholar] [CrossRef]
- Tiburcy, M.; Hudson James, E.; Balfanz, P.; Schlick, S.; Meyer, T.; Chang Liao, M.-L.; Levent, E.; Raad, F.; Zeidler, S.; Wingender, E.; et al. Defined Engineered Human Myocardium with Advanced Maturation for Applications in Heart Failure Modeling and Repair. Circulation 2017, 135, 1832–1847. [Google Scholar] [CrossRef]
- Zhang, D.; Shadrin, I.Y.; Lam, J.; Xian, H.Q.; Snodgrass, H.R.; Bursac, N. Tissue-engineered cardiac patch for advanced functional maturation of human ESC-derived cardiomyocytes. Biomaterials 2013, 34, 5813–5820. [Google Scholar] [CrossRef] [PubMed]
- Hirt, M.N.; Hansen, A.; Eschenhagen, T. Cardiac tissue engineering: State of the art. Circ. Res. 2014, 114, 354–367. [Google Scholar] [CrossRef]
- Ronaldson-Bouchard, K.; Ma, S.P.; Yeager, K.; Chen, T.; Song, L.; Sirabella, D.; Morikawa, K.; Teles, D.; Yazawa, M.; Vunjak-Novakovic, G. Advanced maturation of human cardiac tissue grown from pluripotent stem cells. Nature 2018, 556, 239–243. [Google Scholar] [CrossRef]
- Ravenscroft, S.M.; Pointon, A.; Williams, A.W.; Cross, M.J.; Sidaway, J.E. Cardiac Non-myocyte Cells Show Enhanced Pharmacological Function Suggestive of Contractile Maturity in Stem Cell Derived Cardiomyocyte Microtissues. Toxicol. Sci. 2016, 152, 99–112. [Google Scholar] [CrossRef]
- Giacomelli, E.; Bellin, M.; Sala, L.; van Meer, B.J.; Tertoolen, L.G.; Orlova, V.V.; Mummery, C.L. Three-dimensional cardiac microtissues composed of cardiomyocytes and endothelial cells co-differentiated from human pluripotent stem cells. Development 2017, 144, 1008–1017. [Google Scholar] [CrossRef] [PubMed]
- Balistreri, M.; Davis, J.A.; Campbell, K.F.; Da Rocha, A.M.; Treadwell, M.C.; Herron, T.J. Effect of Glucose on 3D Cardiac Microtissues Derived from Human Induced Pluripotent Stem Cells. Pediatr. Cardiol. 2017, 38, 1575–1582. [Google Scholar] [CrossRef] [PubMed]
- Kadota, S.; Pabon, L.; Reinecke, H.; Murry, C.E. In Vivo Maturation of Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes in Neonatal and Adult Rat Hearts. Stem Cell Rep. 2017, 8, 278–289. [Google Scholar] [CrossRef]
- Cho, G.S.; Lee, D.I.; Tampakakis, E.; Murphy, S.; Andersen, P.; Uosaki, H.; Chelko, S.; Chakir, K.; Hong, I.; Seo, K.; et al. Neonatal Transplantation Confers Maturation of PSC-Derived Cardiomyocytes Conducive to Modeling Cardiomyopathy. Cell Rep. 2017, 18, 571–582. [Google Scholar] [CrossRef]
- Funakoshi, S.; Miki, K.; Takaki, T.; Okubo, C.; Hatani, T.; Chonabayashi, K.; Nishikawa, M.; Takei, I.; Oishi, A.; Narita, M.; et al. Enhanced engraftment, proliferation, and therapeutic potential in heart using optimized human iPSC-derived cardiomyocytes. Sci. Rep. 2016, 6, 19111. [Google Scholar] [CrossRef]
- Godier-Furnémont, A.F.; Tiburcy, M.; Wagner, E.; Dewenter, M.; Lämmle, S.; El-Armouche, A.; Lehnart, S.E.; Vunjak-Novakovic, G.; Zimmermann, W.H. Physiologic force-frequency response in engineered heart muscle by electromechanical stimulation. Biomaterials 2015, 60, 82–91. [Google Scholar] [CrossRef]
- Han, F.; Wang, J.; Ding, L.; Hu, Y.; Li, W.; Yuan, Z.; Guo, Q.; Zhu, C.; Yu, L.; Wang, H.; et al. Tissue Engineering and Regenerative Medicine: Achievements, Future, and Sustainability in Asia. Front. Bioeng. Biotechnol. 2020, 8. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Bai, X.; Ding, Y.; Lee, I.-S. Electrical stimulation as a novel tool for regulating cell behavior in tissue engineering. Biomater. Res. 2019, 23, 25. [Google Scholar] [CrossRef]
- Meier, E.; Lam, M. Role of Mechanical Stimulation in Stem Cell Differentiation. Biotechnol. Bioeng. 2016, 3, 1069. [Google Scholar]
- Schofield, Z.; Meloni, G.N.; Tran, P.; Zerfass, C.; Sena, G.; Hayashi, Y.; Grant, M.; Contera, S.A.; Minteer, S.D.; Kim, M.; et al. Bioelectrical understanding and engineering of cell biology. J. R. Soc. Interface 2020, 17, 20200013. [Google Scholar] [CrossRef] [PubMed]
- Levin, M. Molecular bioelectricity: How endogenous voltage potentials control cell behavior and instruct pattern regulation in vivo. Mol. Biol. Cell 2014, 25, 3835–3850. [Google Scholar] [CrossRef]
- Tyler, S.E.B. Nature’s Electric Potential: A Systematic Review of the Role of Bioelectricity in Wound Healing and Regenerative Processes in Animals, Humans, and Plants. Front. Physiol. 2017, 8. [Google Scholar] [CrossRef]
- Balint, R.; Cassidy, N.J.; Cartmell, S.H. Electrical Stimulation: A Novel Tool for Tissue Engineering. Tissue Eng. Part B Rev. 2012, 19, 48–57. [Google Scholar] [CrossRef]
- Aquilina, O. A brief history of cardiac pacing. Images Paediatr. Cardiol. 2006, 8, 17–81. [Google Scholar] [PubMed]
- Jeon, Y.H. Spinal cord stimulation in pain management: A review. Korean J. Pain 2012, 25, 143–150. [Google Scholar] [CrossRef]
- Eshraghi, A.A.; Nazarian, R.; Telischi, F.F.; Rajguru, S.M.; Truy, E.; Gupta, C. The cochlear implant: Historical aspects and future prospects. Anat. Rec. 2012, 295, 1967–1980. [Google Scholar] [CrossRef] [PubMed]
- Howland, R.H. Vagus Nerve Stimulation. Curr. Behav. Neurosci. Rep. 2014, 1, 64–73. [Google Scholar] [CrossRef] [PubMed]
- Lozano, A.M.; Lipsman, N.; Bergman, H.; Brown, P.; Chabardes, S.; Chang, J.W.; Matthews, K.; McIntyre, C.C.; Schlaepfer, T.E.; Schulder, M.; et al. Deep brain stimulation: Current challenges and future directions. Nat. Rev. Neurol. 2019, 15, 148–160. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S. Electroceuticals in medicine—The brave new future. Indian Heart J. 2017, 69, 685–686. [Google Scholar] [CrossRef] [PubMed]
- Zhu, R.; Sun, Z.; Li, C.; Ramakrishna, S.; Chiu, K.; He, L. Electrical stimulation affects neural stem cell fate and function in vitro. Exp. Neurol. 2019, 319, 112963. [Google Scholar] [CrossRef] [PubMed]
- Tomaskovic-Crook, E.; Zhang, P.; Ahtiainen, A.; Kaisvuo, H.; Lee, C.Y.; Beirne, S.; Aqrawe, Z.; Svirskis, D.; Hyttinen, J.; Wallace, G.G.; et al. Human Neural Tissues from Neural Stem Cells Using Conductive Biogel and Printed Polymer Microelectrode Arrays for 3D Electrical Stimulation. Adv. Healthc. Mater. 2019. [Google Scholar] [CrossRef]
- Korolj, A.; Wang, E.Y.; Civitarese, R.A.; Radisic, M. Biophysical stimulation for in vitro engineering of functional cardiac tissues. Clin. Sci. 2017, 131, 1393–1404. [Google Scholar] [CrossRef] [PubMed]
- Cameron, I.L.; Hardman, W.E.; Winters, W.D.; Zimmerman, S.; Zimmerman, A.M. Environmental magnetic fields: Influences on early embryogenesis. J. Cell. Biochem. 1993, 51, 417–425. [Google Scholar] [CrossRef]
- Chan, Y.C.; Ting, S.; Lee, Y.K.; Ng, K.M.; Zhang, J.; Chen, Z.; Siu, C.W.; Oh, S.K.; Tse, H.F. Electrical stimulation promotes maturation of cardiomyocytes derived from human embryonic stem cells. J. Cardiovasc. Transl. Res. 2013, 6, 989–999. [Google Scholar] [CrossRef] [PubMed]
- Tandon, N.; Cannizzaro, C.; Chao, P.H.; Maidhof, R.; Marsano, A.; Au, H.T.; Radisic, M.; Vunjak-Novakovic, G. Electrical stimulation systems for cardiac tissue engineering. Nat. Protoc. 2009, 4, 155–173. [Google Scholar] [CrossRef]
- Radisic, M.; Park, H.; Shing, H.; Consi, T.; Schoen, F.J.; Langer, R.; Freed, L.E.; Vunjak-Novakovic, G. Functional assembly of engineered myocardium by electrical stimulation of cardiac myocytes cultured on scaffolds. Proc. Natl. Acad. Sci. USA 2004, 101, 18129–18134. [Google Scholar] [CrossRef] [PubMed]
- Eng, G.; Lee, B.W.; Protas, L.; Gagliardi, M.; Brown, K.; Kass, R.S.; Keller, G.; Robinson, R.B.; Vunjak-Novakovic, G. Autonomous beating rate adaptation in human stem cell-derived cardiomyocytes. Nat. Commun. 2016, 7, 10312. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Rafatian, N.; Feric, N.T.; Cox, B.J.; Aschar-Sobbi, R.; Wang, E.Y.; Aggarwal, P.; Zhang, B.; Conant, G.; Ronaldson-Bouchard, K.; et al. A Platform for Generation of Chamber-Specific Cardiac Tissues and Disease Modeling. Cell 2019, 176, 913–927. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.; Liang, J.; Huang, W.; Guo, L.; Cai, W.; Wang, L.; Paul, C.; Yang, H.-T.; Kim, H.W.; Wang, Y. Electrical Stimulation Enhances Cardiac Differentiation of Human Induced Pluripotent Stem Cells for Myocardial Infarction Therapy. Antioxid. Redox Signal. 2018, 28, 371–384. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).