Electronic Circular Dichroism of the Cas9 Protein and gRNA:Cas9 Ribonucleoprotein Complex
Abstract
1. Introduction
2. Results
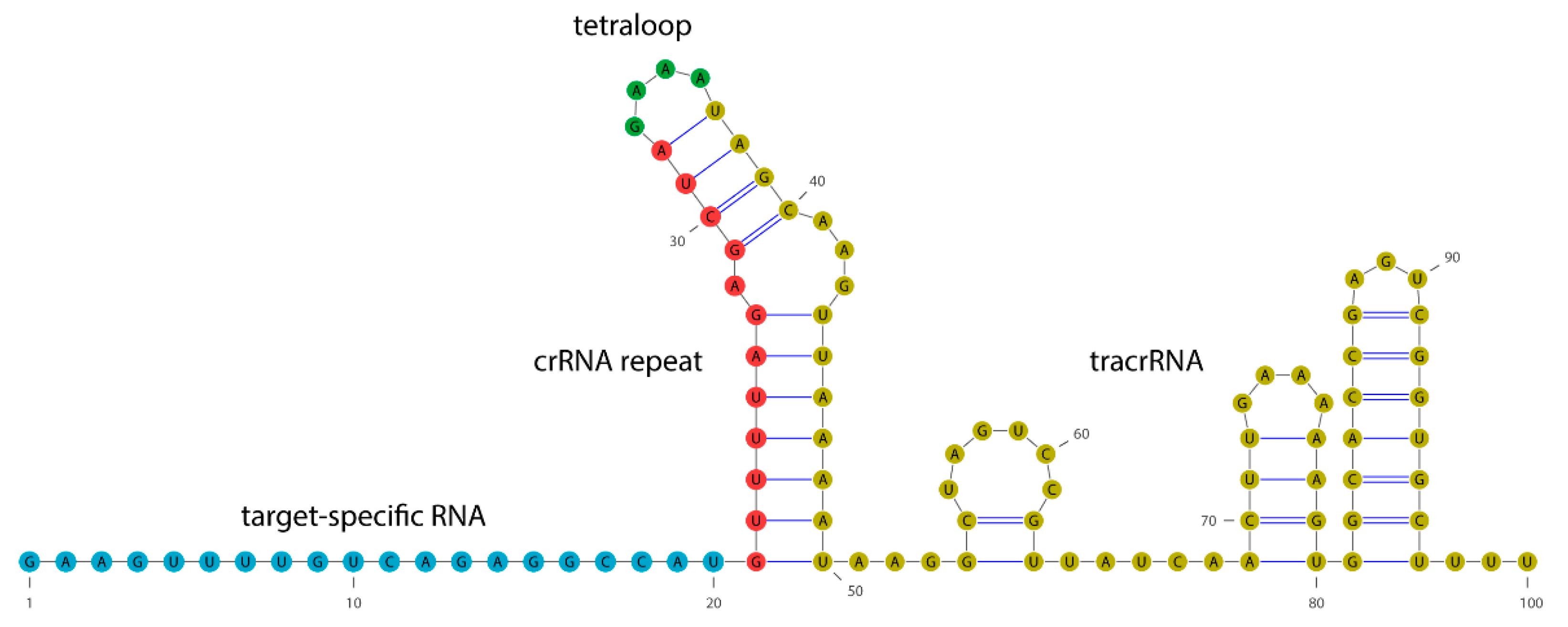
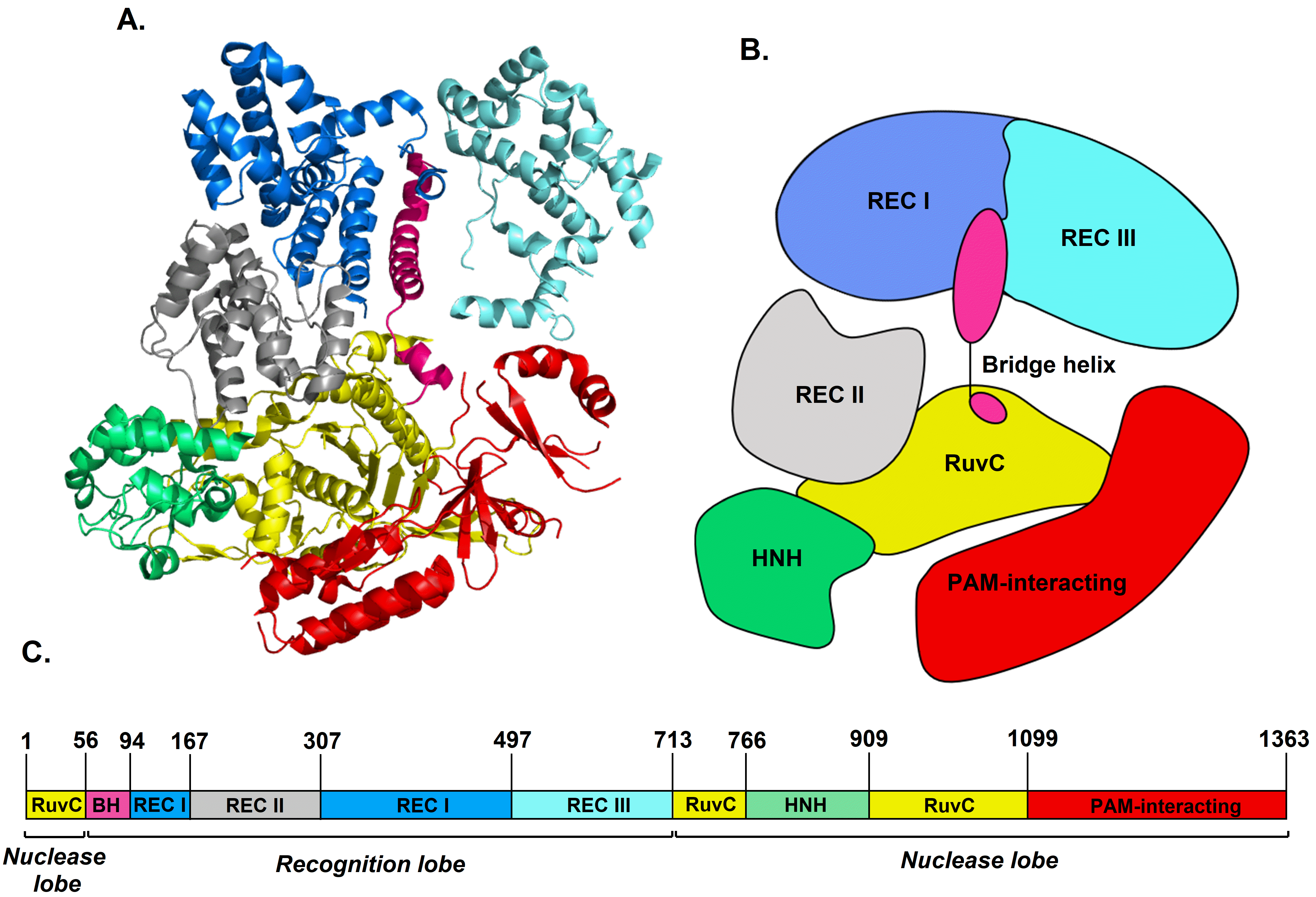
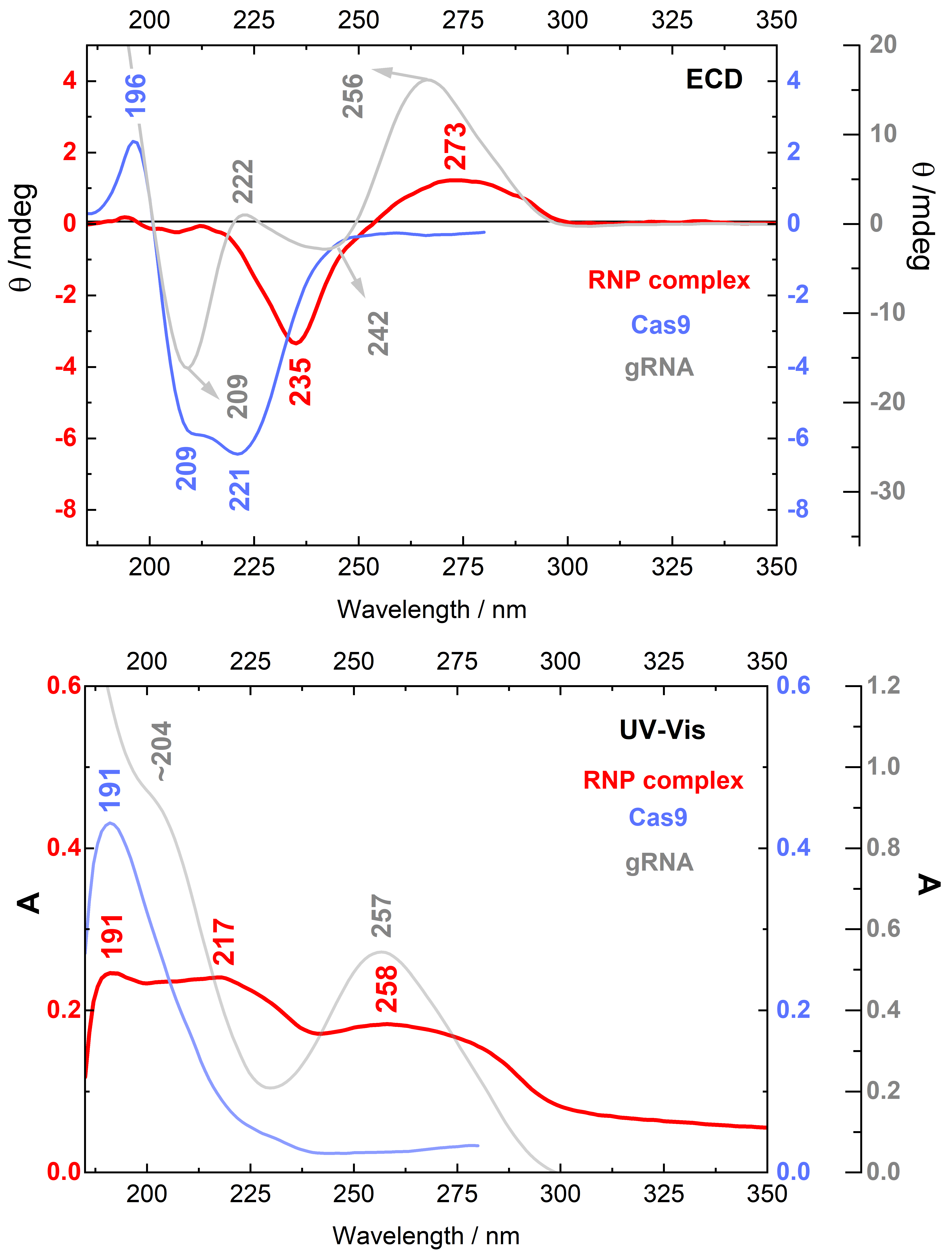
2.1. Circular Dichroism of Cas9 Protein and gRNA
2.2. Circular Dichroism of the RNP Complex
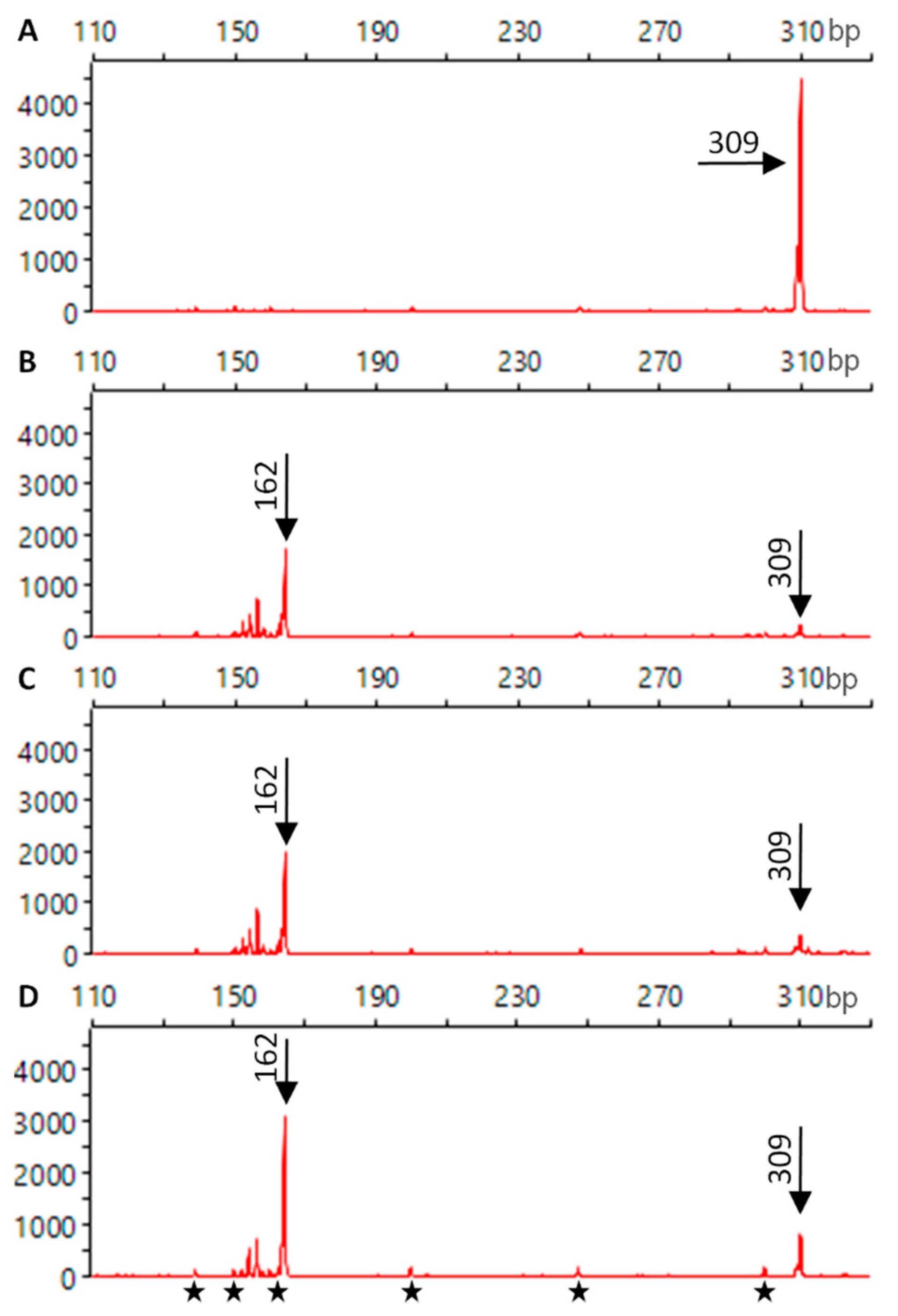
2.3. Biological Activity of the Cas9 Protein and RNP Complex
3. Discussion
3.1. Cas9 and gRNA:Cas9 Complex Structures Revealed by ECD Spectroscopy
3.2. Cas9 Binds gRNA and Retains Cleavage Activity after ECD Measurements
4. Materials and Methods
4.1. Cas9 Protein
4.2. gRNA Design and Transcription
4.3. Formation of the gRNA:Cas9 RNP Complex
4.4. UV-Vis/ECD Measurements
4.5. DNA Amplification
4.6. In Vitro DNA Cleavage
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Belhaj, K.; Chaparro-Garcia, A.; Kamoun, S.; Nekrasov, V. Plant genome editing made easy: Targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods 2013, 9, 39. [Google Scholar] [CrossRef] [PubMed]
- Travis, J. Making the cut. Science 2015, 350, 1456–1457. [Google Scholar] [CrossRef] [PubMed]
- Pickar-Oliver, A.; Gersbach, C.A. The next generation of CRISPR–Cas technologies and applications. Nat. Rev. Mol. Cell Biol. 2019, 20, 490–507. [Google Scholar] [CrossRef] [PubMed]
- Uddin, F.; Rudin, C.M.; Sen, T. CRISPR gene therapy: Applications, limitations, and implications for the future. Front. Oncol. 2020, 10, 1387. [Google Scholar] [CrossRef]
- Jacinto, F.V.; Link, W.; Ferreira, B.I. CRISPR/Cas9-mediated genome editing: From basic research to translational medicine. J. Cell. Mol. Med. 2020, 24, 3766–3778. [Google Scholar] [CrossRef]
- Zhang, M.; Eshraghian, E.A.; Jammal, O.A.; Zhang, Z.; Zhu, X. CRISPR technology: The engine that drives cancer therapy. Biomed. Pharm. 2021, 133, 111007. [Google Scholar] [CrossRef]
- Manghwar, H.; Lindsey, K.; Zhang, X.; Jin, S. CRISPR/Cas system: Recent advances and future prospects for genome editing. Trends Plant Sci. 2019, 24, 1102–1125. [Google Scholar] [CrossRef] [PubMed]
- Ku, H.K.; Ha, S.H. Improving nutritional and functional quality by genome editing of crops: Status and perspectives. Front. Plant Sci. 2020, 11, 577313. [Google Scholar] [CrossRef]
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR provides acquired resistance against viruses in Prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar] [CrossRef]
- Ishino, Y.; Shinagawa, H.; Makino, K.; Amemura, M.; Nakata, A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 1987, 169, 5429–5433. [Google Scholar] [CrossRef]
- Makarova, K.S.; Wolf, Y.I.; Alkhnbashi, O.S.; Costa, F.; Shah, S.A.; Saunders, S.J.; Barrangou, R.; Brouns, S.J.J.; Charpentier, E.; Haft, D.H.; et al. An updated evolutionary classification of CRISPR–Cas systems. Nat. Rev. Microbiol. 2015, 13, 722–736. [Google Scholar] [CrossRef] [PubMed]
- Mojica, F.; Díez-Villaseñor, C.; García-Martínez, J.; Soria, E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 2005, 60, 174–182. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Anders, C.; Niewoehner, O.; Duerst, A.; Jinek, M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 2014, 513, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Garneau, J.E.; Dupuis, M.-È.; Villion, M.; Romero, D.A.; Barrangou, R.; Boyaval, P.; Fremaux, C.; Horvath, P.; Magadán, A.H.; Moineau, S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 2010, 468, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Li, J. Review, analysis, and optimization of the CRISPR Streptococcus pyogenes Cas9 system. Med. Drug Discov. 2021, 9, 100080. [Google Scholar] [CrossRef]
- Babu, K.; Amrani, N.; Jiang, W.; Yogesha, S.D.; Nguyen, R.; Qin, P.Z.; Rajan, R. Bridge helix of Cas9 modulates target DNA cleavage and mismatch tolerance. Biochemistry 2019, 58, 1905–1917. [Google Scholar] [CrossRef]
- Horvath, P.; Romero, D.A.; Coûté-Monvoisin, A.-C.; Richards, M.; Deveau, H.; Moineau, S.; Boyaval, P.; Fremaux, C.; Barrangou, R. Diversity, activity, and evolution of CRISPR Loci in streptococcus thermophilus. J. Bacteriol. 2008, 190, 1401–1412. [Google Scholar] [CrossRef]
- Jinek, M.; Jiang, F.; Taylor, D.W.; Sternberg, S.H.; Kaya, E.; Ma, E.; Anders, C.; Hauer, M.; Zhou, K.; Lin, S.; et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science 2014, 343, 1247997. [Google Scholar] [CrossRef]
- Jiang, F.; Doudna, J.A. CRISPR–Cas9 Structures and Mechanisms. Annu. Rev. Biophys. 2017, 46, 505–529. [Google Scholar] [CrossRef]
- Berova, N.; Di Bari, L.; Pescitelli, G. Application of electronic circular dichroism in configurational and conformational analysis of organic compounds. Chem. Soc. Rev. 2007, 36, 914–931. [Google Scholar] [CrossRef]
- Pescitelli, G.; Di Bari, L.; Berova, N. Conformational aspects in the studies of organic compounds by electronic circular dichroism. Chem. Soc. Rev. 2011, 40, 4603–4625. [Google Scholar] [CrossRef]
- Brahms, J.; Mommaerts, W.F.H.M. A study of conformation of nucleic acids in solution by means of circular dichroism. J. Mol. Biol. 1964, 10, 73–88. [Google Scholar] [CrossRef]
- Padula, D.; Jurinovich, S.; Di Bari, L.; Mennucci, B. Simulation of electronic circular dichroism of nucleic acids: From the structure to the spectrum. Chem. A Eur. J. 2016, 22, 17011–17019. [Google Scholar] [CrossRef]
- Johnson, W.C. Secondary structure of proteins through circular dichroism spectroscopy. Annu. Rev. Biophys. Biophys. Chem. 1988, 17, 145–166. [Google Scholar] [CrossRef] [PubMed]
- Whitmore, L.; Wallace, B.A. Protein secondary structure analyses from circular dichroism spectroscopy: Methods and reference databases. Biopolymers 2008, 89, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Rogers, D.M.; Jasim, S.B.; Dyer, N.T.; Auvray, F.; Réfrégiers, M.; Hirst, J.D. Electronic circular dichroism spectroscopy of proteins. Chem 2019, 5, 2751–2774. [Google Scholar] [CrossRef]
- Kelly, S.M.; Jess, T.J.; Price, N.C. How to study proteins by circular dichroism. Biochim. Biophys. Acta Proteins Proteom. 2005, 1751, 119–139. [Google Scholar] [CrossRef]
- Sreerama, N.; Woody, R.W. Computation and analysis of protein circular dichroism spectra. Methods Enzymol. 2004, 383, 318–351. [Google Scholar] [CrossRef]
- Lim, Y.; Bak, S.; Sung, K.; Jeong, E.; Lee, S.H.; Kim, J.S.; Bae, S.; Kim, S. Structural roles of guide RNAs in the nuclease activity of Cas9 endonuclease. Nat. Commun. 2016, 7, 13350. [Google Scholar] [CrossRef]
- Wang, M.; Sintim, H.O. Discriminating cyclic from linear nucleotides—CRISPR/Cas-related cyclic hexaadenosine monophosphate as a case study. Anal. Biochem. 2019, 567, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Thavalingam, A.; Cheng, Z.; Garcia, B.; Huang, X.; Megha, S.; Sun, W.; Wang, M.; Harrington, L.; Hwang, S.; Reyes, Y.H.; et al. Inhibition of CRISPR-Cas9 ribonucleoprotein complex assembly by anti-CRISPR AcrIIC2. Nat. Commun. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Pawluk, A.; Shah, M.; Mejdani, M.; Calmettes, C.; Moraes, T.F.; Davidson, A.R.; Maxwell, K.L. Disabling a Type I-E CRISPR-Cas Nuclease with a Bacteriophage-Encoded Anti-CRISPR Protein. MBio 2017, 8, e01751-17. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, R.; Sheet, T. Ratio of ellipticities between 192 and 208 nm (R1): An effective electronic circular dichroism parameter for characterization of the helical components of proteins and peptides. Proteins Struct. Funct. Bioinform. 2017, 85, 1975–1982. [Google Scholar] [CrossRef]
- Hirst, J.D.; Colella, K.; Gilbert, A.T.B. Electronic circular dichroism of proteins from first-principles calculations. J. Phys. Chem. B 2003, 107, 11813–11819. [Google Scholar] [CrossRef]
- Nishimasu, H.; Ran, F.A.; Hsu, P.D.; Konermann, S.; Shehata, S.I.; Dohmae, N.; Ishitani, R.; Zhang, F.; Nureki, O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 2014, 156, 935–949. [Google Scholar] [CrossRef]
- Heinig, M.; Frishman, D. STRIDE: A web server for secondary structure assignment from known atomic coordinates of proteins. Nucleic Acids Res. 2004, 32, W500–W502. [Google Scholar] [CrossRef]
- Frishman, D.; Argos, P. Knowledge-based protein secondary structure assignment. Proteins Struct. Funct. Bioinform. 1995, 23, 566–579. [Google Scholar] [CrossRef]
- Zhang, Y.; Sagui, C. Secondary structure assignment for conformationally irregular peptides: Comparison between DSSP, STRIDE and KAKSI. J. Mol. Graph. Model. 2015, 55, 72–84. [Google Scholar] [CrossRef]
- Manning, M.C.; Woody, R.W. Theoretical CD studies of polypeptide helices: Examination of important electronic and geometric factors. Biopolymers 1991, 31, 569–586. [Google Scholar] [CrossRef]
- Enkhbayar, P.; Hikichi, K.; Osaki, M.; Kretsinger, R.H.; Matsushima, N. 310-helices in proteins are parahelices. Proteins Struct. Funct. Bioinform. 2006, 64, 691–699. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Hermans, J. 310 Helix Versus α-Helix: A molecular dynamics study of conformational preferences of aib and alanine. J. Am. Chem. Soc. 1994, 116, 11915–11921. [Google Scholar] [CrossRef]
- Topol, I.A.; Burt, S.K.; Deretey, E.; Tang, T.H.; Perczel, A.; Rashin, A.; Csizmadia, I.G. α- and 310-Helix Interconversion: A quantum-chemical study on polyalanine systems in the gas phase and in aqueous solvent. J. Am. Chem. Soc. 2001, 123, 6054–6060. [Google Scholar] [CrossRef] [PubMed]
- Toniolo, C.; Polese, A.; Formaggio, F.; Crisma, M.; Kamphuis, J. Circular dichroism spectrum of a peptide 310-helix. J. Am. Chem. Soc. 1996, 118, 2744–2745. [Google Scholar] [CrossRef]
- Woody, R.W. Electronic circular dichroism of proteins. In Comprehensive Chiroptical Spectroscopy; Chapter 14; Berova, N., Polvarapu, P.L., Nakanishi, K., Woody, R.W., Eds.; Wiley: Hoboken, NJ, USA, 2012; Volume 2, pp. 475–497. [Google Scholar]
- Migliore, M.; Bonvicini, A.; Tognetti, V.; Guilhaudis, L.; Baaden, M.; Oulyadi, H.; Joubert, L.; Ségalas-Milazzo, I. Characterization of β-turns by electronic circular dichroism spectroscopy: A coupled molecular dynamics and time-dependent density functional theory computational study. Phys. Chem. Chem. Phys. 2019, 22. [Google Scholar] [CrossRef]
- Bandekar, J.; Evans, D.J.; Krimm, S.; Leach, S.J.; Lee, S.; Mcquie, J.R.; Minasian, E.; Nemethy, G.; Pottle, M.S.; Scheraga, H.A.; et al. Conformations of cyclo(L-alanyl-L-alanyl-ε-aminocaproyl) and of cyclo(L-alany1-D-alanyl-ε-aminocaproyl); cyclized dipeptide models for specific types of β-bends. Int. J. Pept. Protein Res. 1982, 19, 187–205. [Google Scholar] [CrossRef]
- Rodger, A. UV absorbance spectroscopy of biological macromolecules. In Encyclopedia of Biophysics; Roberts, G.C.K., Ed.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 2714–2718. [Google Scholar] [CrossRef]
- Johnson, W.C. Determination of the Conformation of Nucleic Acids by Electronic CD. In Circular Dichroism and the Conformational Analysis of Biomolecules; Fasman, G.D., Ed.; Springer: Boston, MA, USA, 1996; pp. 433–468. [Google Scholar]
- Baranowski, D.S.; Kotkowiak, W.; Kierzek, R.; Pasternak, A. Hybridization Properties of RNA Containing 8-Methoxyguanosine and 8-Benzyloxyguanosine. PLoS ONE 2015, 10, e0137674. [Google Scholar] [CrossRef]
- Langkjær, N.; Pasternak, A.; Wengel, J. UNA (unlocked nucleic acid): A flexible RNA mimic that allows engineering of nucleic acid duplex stability. Bioorg. Med. Chem. 2009, 17, 5420–5425. [Google Scholar] [CrossRef]
- Yamada, M.; Watanabe, Y.; Gootenberg, J.S.; Hirano, H.; Ran, F.A.; Nakane, T.; Ishitani, R.; Zhang, F.; Nishimasu, H.; Nureki, O. Crystal structure of the minimal Cas9 from campylobacter jejuni reveals the molecular diversity in the CRISPR-Cas9 systems. Mol. Cell 2017, 65, 1109–1121. [Google Scholar] [CrossRef]
- Davis, A.R.; Kirkpatrick, C.C.; Znosko, B.M. Structural characterization of naturally occurring RNA single mismatches. Nucleic Acids Res. 2011, 39, 1081–1094. [Google Scholar] [CrossRef][Green Version]
- Johnson, W.C., Jr.; Tinoco, I., Jr. Circular dichroism of polynucleotides: A simple theory. Biopolymers 1969, 7, 727–749. [Google Scholar] [CrossRef]
- Gray, D.M. Circular Dichroism of Protein–Nucleic Acid Interactions. In Comprehensive Chiroptical Spectroscopy; Chapter 19; Berova, N., Polvarapu, P.L., Nakanishi, K., Woody, R.W., Eds.; Wiley: Hoboken, NJ, USA, 2012; Volume 2, pp. 615–633. [Google Scholar]
- Gray, D.M. Circular dichroism of protein-nucleic acid interactions. In Circular Dichroism and the Conformational Analysis of Biomolecules; Fasman, G.D., Ed.; Springer: Boston, MA, USA, 1996; pp. 469–500. [Google Scholar]
- Jiang, F.; Zhou, K.; Ma, L.; Gressel, S.; Doudna, J.A. A Cas9–guide RNA complex preorganized for target DNA recognition. Science 2015, 348, 1477–1481. [Google Scholar] [CrossRef] [PubMed]
- Ciubotaru, M.; Ptaszek, L.M.; Baker, G.A.; Baker, S.N.; Bright, F.V.; Schatz, D.G. RAG1-DNA Binding in V(D)J Recombination: Specificity and DNA-induced conformational changes revealed by fluorescence and CD spectroscopy. J. Biol. Chem. 2003, 278, 5584–5596. [Google Scholar] [CrossRef] [PubMed]
- Kaluzhny, D.; Laufman, O.; Timofeev, E.; Borisova, O.; Manor, H.; Shchyolkina, A. Conformational Changes Induced in the Human Protein Translin and in the Single-stranded Oligodeoxynucleotides d(GT) 12 and d(TTAGGG) 5 Upon Binding of These Oligodeoxynucleotides by Translin. J. Biomol. Struct. Dyn. 2006, 23, 257–265. [Google Scholar] [CrossRef]
- Hjelm, R.P.; Chih, R.; Huang, C. sheet of RNA and nonhistone proteins to the circular dichroism spectrum of chromatin. Biochemistry 1975, 14, 1682–1688. [Google Scholar] [CrossRef]
- Oda, Y.; Iwai, S.; Ohtsuka, E.; Ishikawa, M.; Ikehara, M.; Nakamura, H. Binding of nucleic acids to E. coli RNase HI observed by NMR and CD spectroscopy. Nucleic Acids Res. 1993, 21, 4690–4695. [Google Scholar] [CrossRef][Green Version]
- Klimek-Chodacka, M.; Oleszkiewicz, T.; Lowder, L.G.; Qi, Y.; Baranski, R. Efficient CRISPR/Cas9-based genome editing in carrot cells. Plant Cell Rep. 2018, 37, 575–586. [Google Scholar] [CrossRef]
- Mehravar, M.; Shirazi, A.; Mehrazar, M.M.; Nazari, M. In Vitro Pre-validation of Gene Editing by CRISPR/Cas9 Ribonucleoprotein. Avicenna J. Med. Biotechnol. 2019, 11, 259–263. [Google Scholar]
- Ramlee, M.K.; Yan, T.; Cheung, A.M.S.; Chuah, C.T.H.; Li, S. High-throughput genotyping of CRISPR/Cas9-mediated mutants using fluorescent PCR-capillary gel electrophoresis. Sci. Rep. 2015, 5, 15587. [Google Scholar] [CrossRef]
- Kocak, D.D.; Josephs, E.A.; Bhandarkar, V.; Adkar, S.S.; Kwon, J.B.; Gersbach, C.A. Increasing the specificity of CRISPR systems with engineered RNA secondary structures. Nat. Biotechnol. 2019, 37, 657–666. [Google Scholar] [CrossRef]
- Creutzburg, S.C.A.; Wu, W.Y.; Mohanraju, P.; Swartjes, T.; Alkan, F.; Gorodkin, J.; Staals, R.H.J.; van der Oost, J. Good guide, bad guide: Spacer sequence-dependent cleavage efficiency of Cas12a. Nucleic Acids Res. 2020, 48, 3228–3243. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.; Kim, H.K.; Lee, S.; Seo, J.H.; Choi, J.W.; Park, J.; Min, S.; Yoon, S.; Cho, S.R.; Kim, H.H. Prediction of the sequence-specific cleavage activity of Cas9 variants. Nat. Biotechnol. 2020, 38, 1328–1336. [Google Scholar] [CrossRef] [PubMed]
- Xiao, A.; Cheng, Z.; Kong, L.; Zhu, Z.; Lin, S.; Gao, G.; Zhang, B. CasOT: A genome-wide Cas9/gRNA off-target searching tool. Bioinformatics 2014, 30, 1180–1182. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Halat, M.; Klimek-Chodacka, M.; Orleanska, J.; Baranska, M.; Baranski, R. Electronic Circular Dichroism of the Cas9 Protein and gRNA:Cas9 Ribonucleoprotein Complex. Int. J. Mol. Sci. 2021, 22, 2937. https://doi.org/10.3390/ijms22062937
Halat M, Klimek-Chodacka M, Orleanska J, Baranska M, Baranski R. Electronic Circular Dichroism of the Cas9 Protein and gRNA:Cas9 Ribonucleoprotein Complex. International Journal of Molecular Sciences. 2021; 22(6):2937. https://doi.org/10.3390/ijms22062937
Chicago/Turabian StyleHalat, Monika, Magdalena Klimek-Chodacka, Jagoda Orleanska, Malgorzata Baranska, and Rafal Baranski. 2021. "Electronic Circular Dichroism of the Cas9 Protein and gRNA:Cas9 Ribonucleoprotein Complex" International Journal of Molecular Sciences 22, no. 6: 2937. https://doi.org/10.3390/ijms22062937
APA StyleHalat, M., Klimek-Chodacka, M., Orleanska, J., Baranska, M., & Baranski, R. (2021). Electronic Circular Dichroism of the Cas9 Protein and gRNA:Cas9 Ribonucleoprotein Complex. International Journal of Molecular Sciences, 22(6), 2937. https://doi.org/10.3390/ijms22062937






