Multiple Sclerosis-Associated hnRNPA1 Mutations Alter hnRNPA1 Dynamics and Influence Stress Granule Formation
Abstract
1. Introduction
2. Results
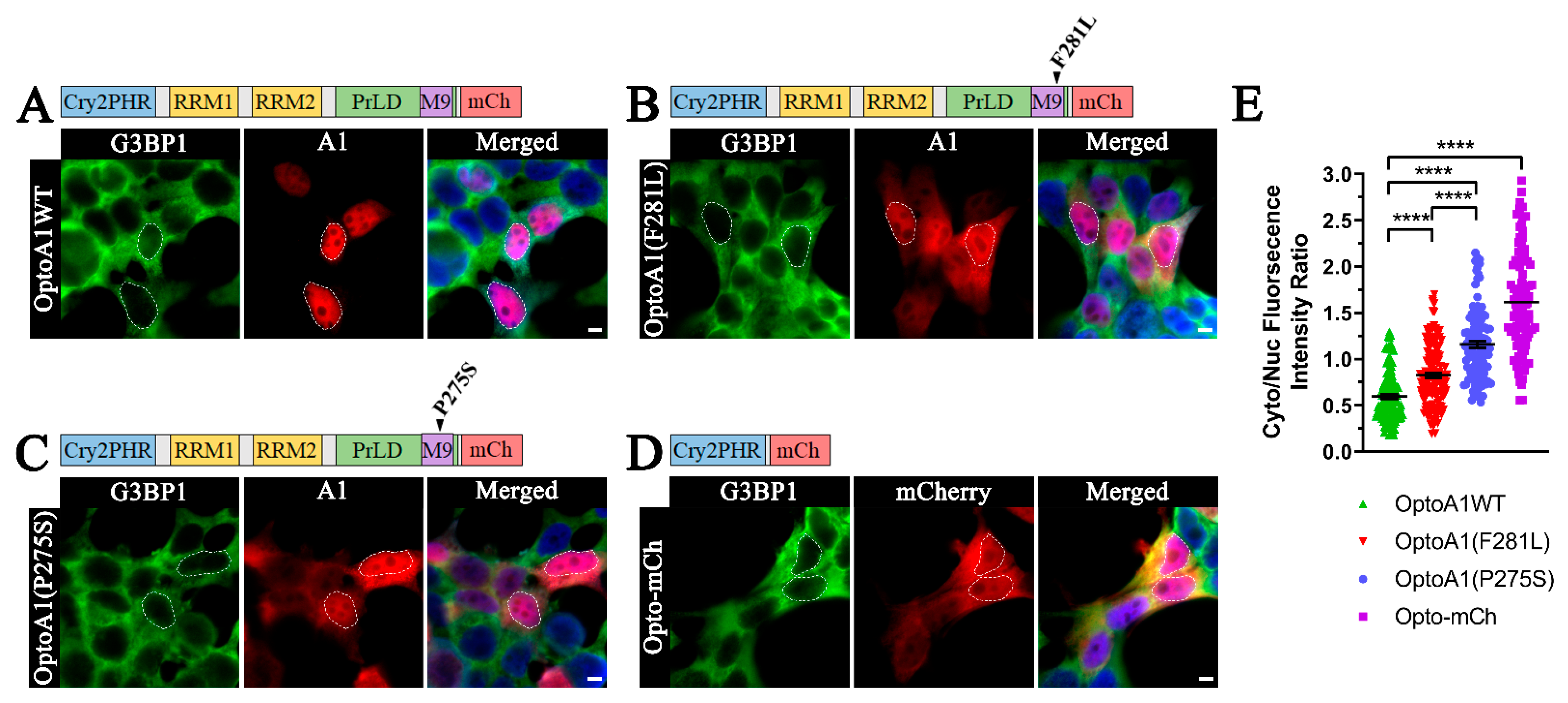
2.1. A1 PrLD Mutations Exacerbate A1 Cytoplasmic Mislocalization
2.2. Blue Light Stimulation Facilitates Reversible A1 Self-Association without Inducing Cell Stress
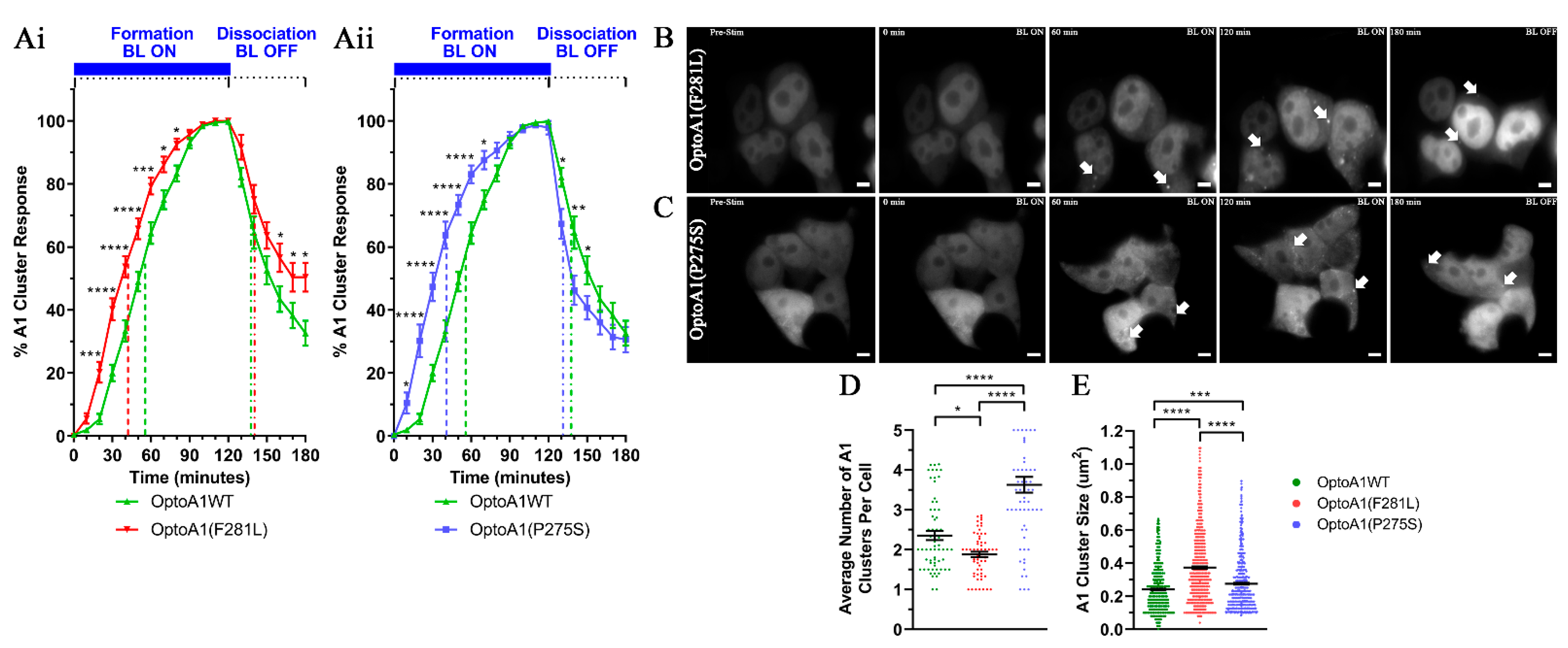
2.3. A1 Protein Self-Association Kinetics Are Altered by PrLD Mutations
2.4. A1 Protein Cluster Characteristics Are Altered by PrLD Mutations
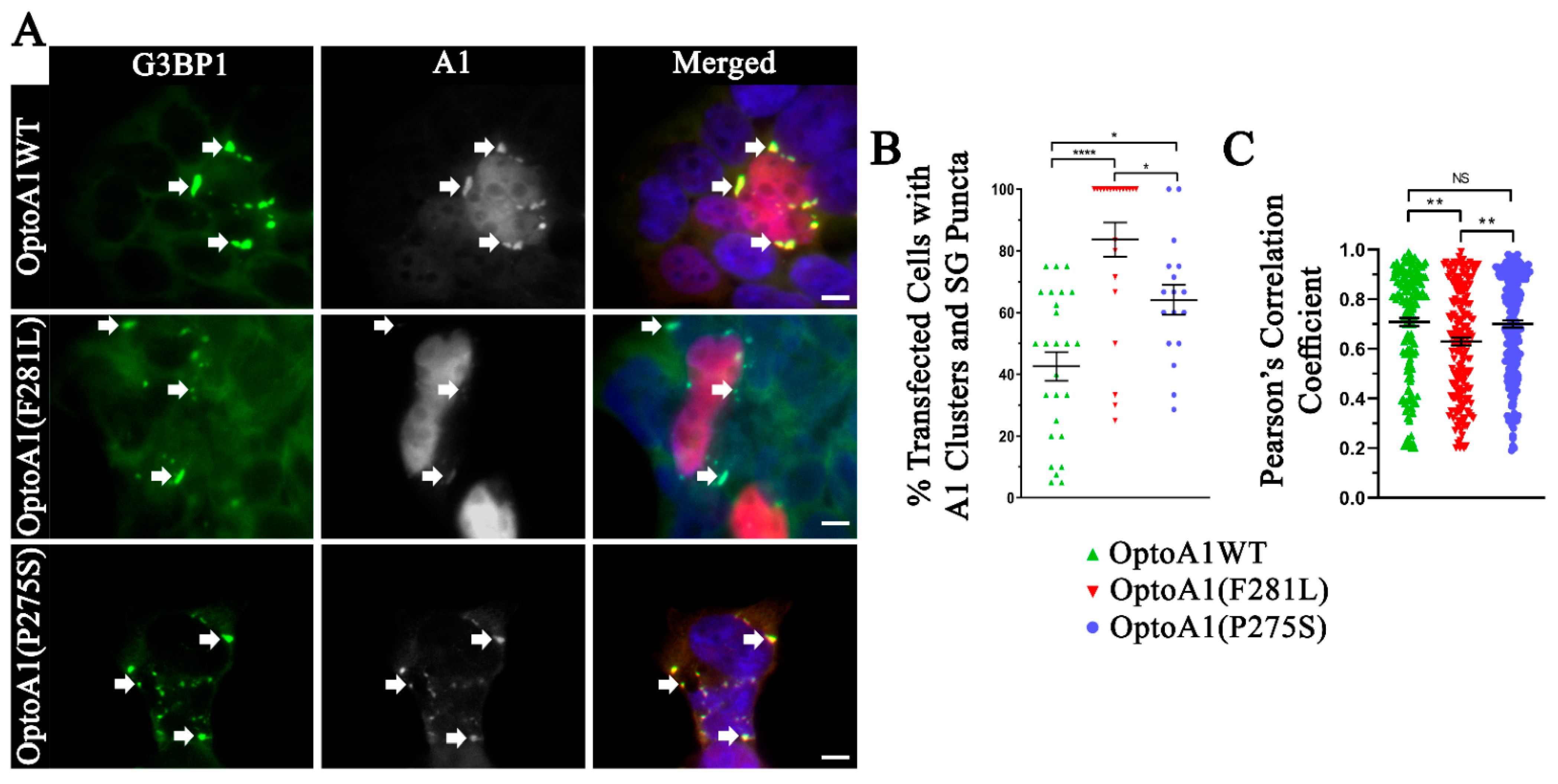
2.5. A1 Protein Clusters Co-Localize with Stress Granules
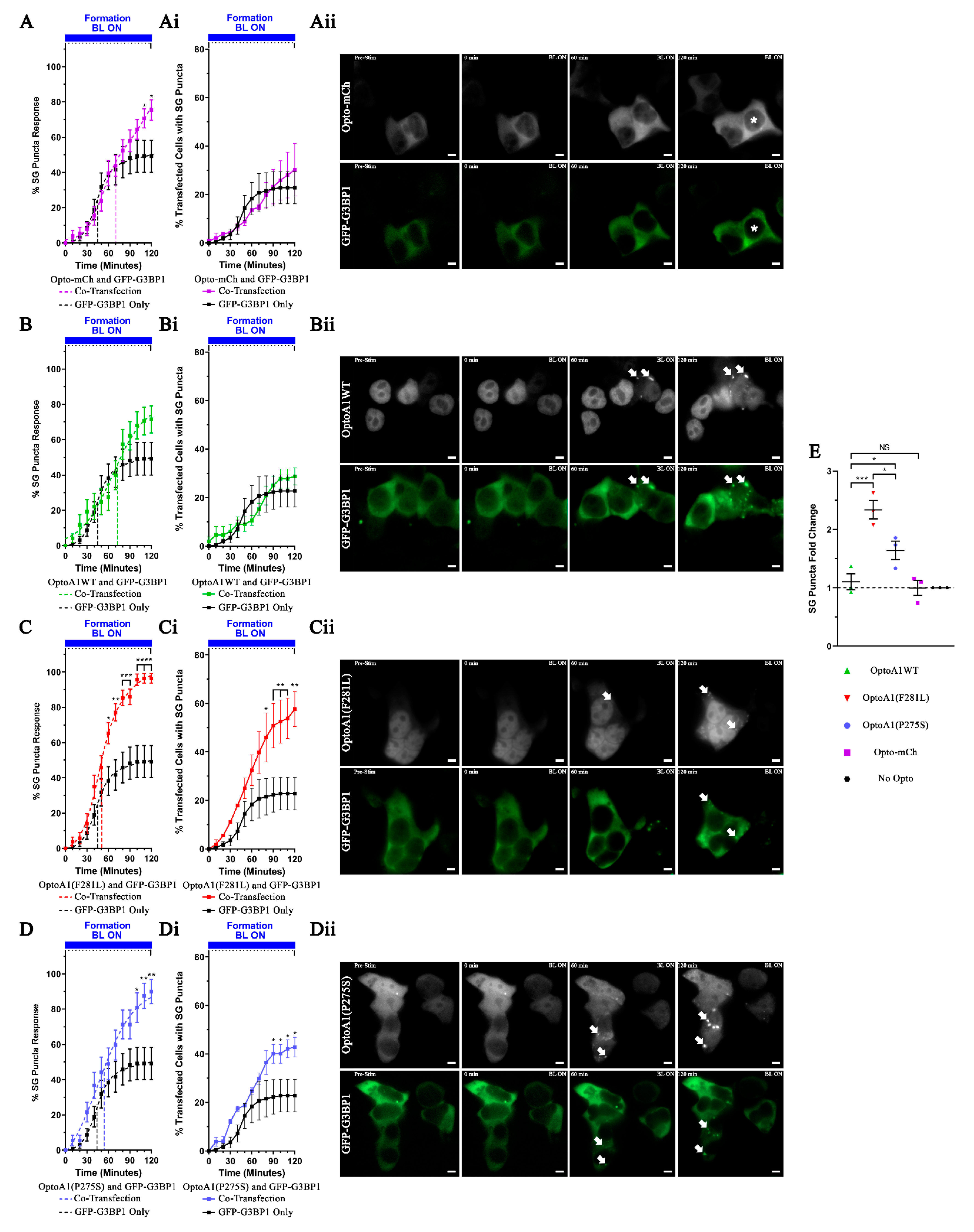
2.6. A1 Protein Clustering Promotes Stress Granule Formation
3. Discussion
4. Materials and Methods
4.1. Patient Material
4.2. Cell Culture
4.3. Cloning
4.4. Blue Light Treatments
4.5. Stress Treatments
4.6. Nuclear/Cytoplasmic Fractionation and SDS-PAGE/Western Immunoblotting
4.7. Immunocytochemistry
4.8. Live-Cell Imaging
4.9. Quantification and Statistical Analysis
4.9.1. Immunocytochemistry
4.9.2. Live-Cell Imaging
4.9.3. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lassmann, H.; van Horssen, J. The molecular basis of neurodegeneration in multiple sclerosis. FEBS Lett. 2011, 585, 3715–3723. [Google Scholar] [CrossRef] [PubMed]
- Dutta, R.; McDonough, J.; Yin, X.; Peterson, J.; Chang, A.; Torres, T.; Gudz, T.; Macklin, W.B.; Lewis, D.A.; Fox, R.J. Mitochondrial dysfunction as a cause of axonal degenerationin multiple sclerosis patients. Ann. Neurol. 2006, 59, 478–489. [Google Scholar] [CrossRef]
- De Vos, K.J.; Grierson, A.J.; Ackerley, S.; Miller, C.C.J. Role of axonal transport in neurodegenerative diseases. Annu. Rev. Neurosci. 2008, 31, 151–173. [Google Scholar] [CrossRef]
- Campbell, G.R.; Ziabreva, I.; Reeve, A.K.; Krishnan, K.J.; Reynolds, R.; Howell, O.; Lassmann, H.; Turnbull, D.M.; Mahad, D.J. Mitochondrial DNA deletions and neurodegeneration in multiple sclerosis. Ann. Neurol. 2010, 69, 481–492. [Google Scholar] [CrossRef] [PubMed]
- Haider, L.; Fischer, M.T.; Frischer, J.M.; Bauer, J.; Höftberger, R.; Botond, G.; Esterbauer, H.; Binder, C.J.; Witztum, J.L.; Lassmann, H. Oxidative damage in multiple sclerosis lesions. Brain 2011, 134, 1914–1924. [Google Scholar] [CrossRef] [PubMed]
- Fischer, M.T.; Sharma, R.; Lim, J.L.; Haider, L.; Frischer, J.M.; Drexhage, J.; Mahad, D.; Bradl, M.; Van Horssen, J.; Lassmann, H. NADPH oxidase expression in active multiple sclerosis lesions in relation to oxidative tissue damage and mitochondrial injury. Brain 2012, 135, 886–899. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.-W.; Zhang, X.; Huang, W.-J. Role of neuroinflammation in neurodegenerative diseases (Review). Mol. Med. Rep. 2016, 13, 3391–3396. [Google Scholar] [CrossRef]
- Ransohoff, R.M. How neuroinflammation contributes to neurodegeneration. Science 2016, 353, 777–783. [Google Scholar] [CrossRef]
- Douglas, J.N.; Gardner, L.A.; Levin, M.C. Antibodies to an Intracellular Antigen Penetrate Neuronal Cells and Cause Deleterious Effects. J. Clin. Cell. Immunol. 2013, 4, 1–7. [Google Scholar] [CrossRef]
- Kozin, M.S.; Kulakova, O.G.; Favorova, O.O. Involvement of Mitochondria in Neurodegeneration in Multiple Sclerosis. Biochemstry 2018, 83, 813–830. [Google Scholar] [CrossRef]
- Trapp, B.D.; Stys, P.K. Virtual hypoxia and chronic necrosis of demyelinated axons in multiple sclerosis. Lancet Neurol. 2009, 8, 280–291. [Google Scholar] [CrossRef]
- Trapp, B.D.; Ransohoff, R.; Rudick, R. Axonal pathology in multiple sclerosis: Relationship to neurologic disability. Curr. Opin. Neurol. 1999, 12, 295–302. [Google Scholar] [CrossRef]
- Levin, M.C.; Lee, S.M.; Kalume, F.; Morcos, Y.; Dohan, F.C., Jr.; Hasty, K.A.; Callaway, J.C.; Zunt, J.; Desiderio, D.M.; Stuart, J.M. Autoimmunity due to molecular mimicry as a cause of neurological disease. Nat. Med. 2002, 8, 509–513. [Google Scholar] [CrossRef]
- De March, A.K.; De Bouwerie, M.; Kolopp-Sarda, M.N.; Faure, G.C.; Bene, M.C.; Bernard, C.C.A. Anti-myelin oligodendrocyte glycoprotein B-cell responses in multiple sclerosis. J. Neuroimmunol. 2003, 135, 117–125. [Google Scholar] [CrossRef]
- Craner, M.J.; Newcombe, J.; Black, J.A.; Hartle, C.; Cuzner, M.L.; Waxman, S.G. Molecular changes in neurons in multiple sclerosis: Altered axonal expression of Nav1.2 and Nav1.6 sodium channels and Na+/Ca2+ exchanger. Proc. Natl. Acad. Sci. USA 2004, 101, 8168–8173. [Google Scholar] [CrossRef] [PubMed]
- Khalil, M.; Reindl, M.; Lutterotti, A.; Kuenz, B.; Ehling, R.; Gneiss, C.; Lackner, P.; Deisenhammer, F.; Berger, T. Epitope specificity of serum antibodies directed against the extracellular domain of myelin oligodendrocyte glycoprotein: Influence of relapses and immunomodulatory treatments. J. Neuroimmunol. 2006, 174, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Black, J.A.; Newcombe, J.; Trapp, B.D.; Waxman, S.G. Sodium Channel Expression Within Chronic Multiple Sclerosis Plaques. J. Neuropathol. Exp. Neurol. 2007, 66, 828–837. [Google Scholar] [CrossRef]
- Dutta, R.; Trapp, B.D. Pathogenesis of axonal and neuronal damage in multiple sclerosis. Neurological 2007, 68, S22–S31. [Google Scholar] [CrossRef]
- Bar-Or, A.; Fawaz, L.; Fan, B.; Darlington, P.J.; Rieger, A.; Msc, C.G.; Calabresi, P.A.; Waubant, E.; Hauser, S.L.; Zhang, J.; et al. Abnormal B-cell cytokine responses a trigger of T-cell-mediated disease in MS? Ann. Neurol. 2009, 67, 452–461. [Google Scholar] [CrossRef]
- Dutta, R.; Trapp, B.D. Mechanisms of neuronal dysfunction and degeneration in multiple sclerosis. Prog. Neurobiol. 2011, 93, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Dendrou, C.A.; Fugger, L.; Friese, M.A. Immunopathology of multiple sclerosis. Nat. Rev. Immunol. 2015, 15, 545–558. [Google Scholar] [CrossRef]
- Li, R.; Patterson, K.R.; Bar-Or, A. Reassessing B cell contributions in multiple sclerosis. Nat. Immunol. 2018, 19, 696–707. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.M.; Dunnavant, F.D.; Jang, H.; Zunt, J.; Levin, M.C. Autoantibodies that recognize functional domains of hnRNPA1 implicate molecular mimicry in the pathogenesis of neurological disease. Neurosci. Lett. 2006, 401, 188–193. [Google Scholar] [CrossRef] [PubMed]
- Levin, M.C.; Lee, S.; Gardner, L.A.; Shin, Y.; Douglas, J.N.; Cooper, C. Autoantibodies to Non-myelin Antigens as Contributors to the Pathogenesis of Multiple Sclerosis. J. Clin. Cell Immunol. 2013, 4. [Google Scholar] [CrossRef]
- Lee, S.; Levin, M. Novel somatic single nucleotide variants within the RNA binding protein hnRNP A1 in multiple sclerosis patients. F1000Research 2014, 3, 132. [Google Scholar] [CrossRef]
- Douglas, J.N.; Gardner, L.A.; Salapa, H.E.; Levin, M.C. Antibodies to the RNA Binding Protein Heterogeneous Nuclear Ribonucleoprotein A1 Colocalize to Stress Granules Resulting in Altered RNA and Protein Levels in a Model of Neurodegeneration in Multiple Sclerosis. J. Clin. Cell. Immunol. 2016, 7, 1–7. [Google Scholar] [CrossRef]
- Salapa, H.E.; Johnson, C.; Hutchinson, C.; Popescu, B.F.; Levin, M.C. Dysfunctional RNA binding proteins and stress granules in multiple sclerosis. J. Neuroimmunol. 2018, 324, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Salapa, H.E.; Levin, M.C. Localization of near-infrared labeled antibodies to the central nervous system in experimental autoimmune encephalomyelitis. PLoS ONE 2019, 14, e0212357. [Google Scholar] [CrossRef] [PubMed]
- Libner, C.D.; Salapa, H.E.; Hutchinson, C.; Lee, S.; Levin, M.C. Antibodies to the RNA binding protein heterogeneous nuclear ribonucleoprotein A1 contribute to neuronal cell loss in an animal model of multiple sclerosis. J. Comp. Neurol. 2019, 528, 1704–1724. [Google Scholar] [CrossRef] [PubMed]
- Salapa, H.E.; Libner, C.D.; Levin, M.C. Dysfunctional RNA-binding protein biology and neurodegeneration in experimental autoimmune encephalomyelitis in female mice. J. Neurosci. Res. 2019, 98, 704–717. [Google Scholar] [CrossRef] [PubMed]
- Salapa, H.E.; Hutchinson, C.; Popescu, B.F.; Levin, M.C. Neuronal RNA-binding protein dysfunction in multiple sclerosis cortex. Ann. Clin. Transl. Neurol. 2020, 7, 1214–1224. [Google Scholar] [CrossRef]
- Singh, R. RNA-protein interactions that regulate pre-mRNA splicing. Gene Expr. 2002, 10, 79–92. [Google Scholar]
- Kishore, S.; Luber, S.; Zavolan, M. Deciphering the role of RNA-binding proteins in the post-transcriptional control of gene expression. Brief. Funct. Genom. 2010, 9, 391–404. [Google Scholar] [CrossRef] [PubMed]
- Licatalosi, D.D.; Darnell, R.B. RNA processing and its regulation: Global insights into biological networks. Nat. Rev. Genet. 2010, 11, 75–87. [Google Scholar] [CrossRef]
- Han, K.; Yeo, G.; An, P.; Burge, C.B.; Grabowski, P.J. A Combinatorial Code for Splicing Silencing: UAGG and GGGG Motifs. PLoS Biol. 2005, 3, e158. [Google Scholar] [CrossRef] [PubMed]
- Roy, R.; Durie, D.; Lin, G.; Liu, B.-Q.; Skehel, J.M.; Mauri, F.; Cuorvo, L.V.; Barbareschi, M.; Guo, L.; Holcik, M.; et al. hnRNPA1 couples nuclear export and translation of specific mRNAs downstream of FGF-2/S6K2 signalling. Nucleic Acids Res. 2014, 42, 12483–12497. [Google Scholar] [CrossRef] [PubMed]
- Rebane, A.; Aab, A.; Steitz, J.A. Transportins 1 and 2 are redundant nuclear import factors for hnRNP A1 and HuR. RNA 2004, 10, 590–599. [Google Scholar] [CrossRef] [PubMed]
- Allemand, E.; Guil, S.; Myers, M.; Moscat, J.; Cáceres, J.F.; Krainer, A.R. Regulation of heterogenous nuclear ribonucleoprotein A1 transport by phosphorylation in cells stressed by osmotic shock. Proc. Natl. Acad. Sci. USA 2005, 102, 3605–3610. [Google Scholar] [CrossRef]
- He, Y.; Smith, R. Nuclear functions of heterogeneous nuclear ribonucleoproteins A/B. Cell. Mol. Life Sci. 2008, 66, 1239–1256. [Google Scholar] [CrossRef]
- Cartegni, L.; Maconi, M.; Morandi, E.; Cobianchi, F.; Riva, S.; Biamonti, G. hnRNP A1 Selectively Interacts Through its Gly-rich Domain with Different RNA-binding Proteins. J. Mol. Biol. 1996, 259, 337–348. [Google Scholar] [CrossRef]
- Fisette, J.-F.; Toutant, J.; Dugré-Brisson, S.; DesGroseillers, L.; Chabot, B. hnRNP A1 and hnRNP H can collaborate to modulate 5’ splice site selection. RNA 2009, 16, 228–238. [Google Scholar] [CrossRef]
- Kim, H.J.; Kim, N.C.; Wang, Y.-D.; Scarborough, E.A.; Moore, J.; Diaz, Z.; MacLea, K.S.; Freibaum, B.; Li, S.; Molliex, A.; et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature 2013, 495, 467–473. [Google Scholar] [CrossRef]
- Molliex, A.; Temirov, J.; Lee, J.; Coughlin, M.; Kanagaraj, A.P.; Kim, H.J.; Mittag, T.; Taylor, J.P. Phase Separation by Low Complexity Domains Promotes Stress Granule Assembly and Drives Pathological Fibrillization. Cell 2015, 163, 123–133. [Google Scholar] [CrossRef]
- Franzmann, T.M.; Alberti, S. Prion-like low-complexity sequences: Key regulators of protein solubility and phase behavior. J. Biol. Chem. 2019, 294, 7128–7136. [Google Scholar] [CrossRef]
- Lee, B.J.; Cansizoglu, A.E.; Suel, K.E.; Louis, T.H.; Zhang, Z.; Chook, Y.M. Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell 2006, 126, 543–558. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Xu, L.; Shin, Y.; Gardner, L.; Hartzes, A.; Dohan, F.C.; Raine, C.; Homayouni, R.; Levin, M.C. A potential link between autoimmunity and neurodegeneration in immune-mediated neurological disease. J. Neuroimmunol. 2011, 235, 56–69. [Google Scholar] [CrossRef]
- De Conti, L.; Baralle, M.; Buratti, E. Neurodegeneration and RNA-binding proteins. Wiley Interdiscip. Rev. RNA 2017, 8, e1394. [Google Scholar] [CrossRef] [PubMed]
- Harrison, A.F.; Shorter, J. RNA-binding proteins with prion-like domains in health and disease. Biochem. J. 2017, 474, 1417–1438. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Kim, H.J.; Wang, H.; Monaghan, J.; Freyermuth, F.; Sung, J.C.; O’Donovan, K.; Fare, C.M.; Diaz, Z.; Singh, N.; et al. Nuclear-Import Receptors Reverse Aberrant Phase Transitions of RNA-Binding Proteins with Prion-like Domains. Cell 2018, 173, 677–692. [Google Scholar] [CrossRef]
- Mann, J.R.; Gleixner, A.M.; Mauna, J.C.; Gomes, E.; DeChellis-Marks, M.R.; Needham, P.G.; Copley, K.E.; Hurtle, B.; Portz, B.; Pyles, N.J.; et al. RNA Binding Antagonizes Neurotoxic Phase Transitions of TDP-43. Neuron 2019, 102, 321–338.e8. [Google Scholar] [CrossRef]
- Chung, C.G.; Lee, H.; Lee, S.B. Mechanisms of protein toxicity in neurodegenerative diseases. Cell. Mol. Life Sci. 2018, 75, 3159–3180. [Google Scholar] [CrossRef]
- Protter, D.S.; Rao, B.S.; Van Treeck, B.; Lin, Y.; Mizoue, L.; Rosen, M.K.; Parker, R. Intrinsically Disordered Regions Can Contribute Promiscuous Interactions to RNP Granule Assembly. Cell Rep. 2018, 22, 1401–1412. [Google Scholar] [CrossRef]
- Mittag, T.; Parker, R. Multiple Modes of Protein–Protein Interactions Promote RNP Granule Assembly. J. Mol. Biol. 2018, 430, 4636–4649. [Google Scholar] [CrossRef]
- Protter, D.S.; Parker, R. Principles and Properties of Stress Granules. Trends Cell Biol. 2016, 26, 668–679. [Google Scholar] [CrossRef]
- Wheeler, J.R.; Matheny, T.; Jain, S.; Abrisch, R.; Parker, R. Distinct stages in stress granule assembly and disassembly. eLife 2016, 5, e18413. [Google Scholar] [CrossRef]
- Hyman, A.A.; Weber, C.A.; Jülicher, F. Liquid-Liquid Phase Separation in Biology. Annu. Rev. Cell Dev. Biol. 2014, 30, 39–58. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zuo, Z.; Wang, X.; Gu, L.; Yoshizumi, T.; Yang, Z.; Yang, L.; Liu, Q.; Liu, W.; Han, Y.-J.; et al. Photoactivation and inactivation of Arabidopsis cryptochrome 2. Science 2016, 354, 343–347. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Fan, B.; Yang, P.; Temirov, J.; Messing, J.; Kim, H.J.; Taylor, J.P. Chronic optogenetic induction of stress granules is cytotoxic and reveals the evolution of ALS-FTD pathology. eLife 2019, 8. [Google Scholar] [CrossRef]
- Shin, Y.; Berry, J.; Pannucci, N.; Haataja, M.P.; Toettcher, J.E.; Brangwynne, C.P. Spatiotemporal Control of Intracellular Phase Transitions Using Light-Activated optoDroplets. Cell 2017, 168, 159–171. [Google Scholar] [CrossRef]
- Tourriere, H.; Chebli, K.; Zekri, L.; Courselaud, B.; Blanchard, J.M.; Bertrand, E.; Tazi, J. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 2003, 160, 823–831. [Google Scholar] [CrossRef]
- Colombrita, C.; Zennaro, E.; Fallini, C.; Weber, M.; Sommacal, A.; Buratti, E.; Silani, V.; Ratti, A. TDP-43 is recruited to stress granules in conditions of oxidative insult. J. Neurochem. 2009, 111, 1051–1061. [Google Scholar] [CrossRef] [PubMed]
- Kedersha, N.; Chen, S.; Gilks, N.; Li, W.; Miller, I.J.; Stahl, J.; Anderson, P. Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol. Biol. Cell 2002, 13, 195–210. [Google Scholar] [CrossRef]
- Takahashi, M.; Higuchi, M.; Matsuki, H.; Yoshita, M.; Ohsawa, T.; Oie, M.; Fujii, M. Stress Granules Inhibit Apoptosis by Reducing Reactive Oxygen Species Production. Mol. Cell. Biol. 2013, 33, 815–829. [Google Scholar] [CrossRef]
- Reineke, L.C.; Dougherty, J.D.; Pierre, P.; Lloyd, R.E. Large G3BP-induced granules trigger eIF2α phosphorylation. Mol. Biol. Cell 2012, 23, 3499–3510. [Google Scholar] [CrossRef]
- Bracha, D.; Walls, M.T.; Wei, M.-T.; Zhu, L.; Kurian, M.; Avalos, J.L.; Toettcher, J.E.; Brangwynne, C.P. Mapping Local and Global Liquid Phase Behavior in Living Cells Using Photo-Oligomerizable Seeds. Cell 2018, 175, 1467–1480. [Google Scholar] [CrossRef] [PubMed]
- Hutten, S.; Usluer, S.; Bourgeois, B.; Simonetti, F.; Odeh, H.M.; Fare, C.M.; Czuppa, M.; Hruska-Plochan, M.; Hofweber, M.; Polymenidou, M.; et al. Nuclear Import Receptors Directly Bind to Arginine-Rich Dipeptide Repeat Proteins and Suppress Their Pathological Interactions. Cell Rep. 2020, 33, 108538. [Google Scholar] [CrossRef]
- Shorter, J.; Taylor, J.P. Disease mutations in the prion-like domains of hnRNPA1 and hnRNPA2/B1 introduce potent steric zippers that drive excess RNP granule assembly. Rare Dis. 2013, 1, e25200. [Google Scholar] [CrossRef]
- Guil, S.; Long, J.C.; Cáceres, J.F. hnRNP A1 Relocalization to the Stress Granules Reflects a Role in the Stress Response. Mol. Cell. Biol. 2006, 26, 5744–5758. [Google Scholar] [CrossRef] [PubMed]
- Markmiller, S.; Soltanieh, S.; Server, K.L.; Mak, R.; Jin, W.; Fang, M.Y.; Luo, E.-C.; Krach, F.; Yang, D.; Sen, A.; et al. Context-Dependent and Disease-Specific Diversity in Protein Interactions within Stress Granules. Cell 2018, 172, 590–604. [Google Scholar] [CrossRef] [PubMed]
- Youn, J.-Y.; Dyakov, B.J.; Zhang, J.; Knight, J.D.; Vernon, R.M.; Forman-Kay, J.D.; Gingras, A.-C. Properties of Stress Granule and P-Body Proteomes. Mol. Cell 2019, 76, 286–294. [Google Scholar] [CrossRef] [PubMed]
- Gui, X.; Luo, F.; Li, Y.; Zhou, H.; Qin, Z.; Liu, Z.; Gu, J.; Xie, M.; Zhao, K.; Dai, B.; et al. Structural basis for reversible amyloids of hnRNPA1 elucidates their role in stress granule assembly. Nat. Commun. 2019, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Iervolino, A.; Santilli, G.; Trotta, R.; Guerzoni, C.; Cesi, V.; Bergamaschi, A.; Passerini, C.G.; Calabretta, B.; Perrotti, D. hnRNP A1 Nucleocytoplasmic Shuttling Activity Is Required for Normal Myelopoiesis and BCR/ABL Leukemogenesis. Mol. Cell. Biol. 2002, 22, 2255–2266. [Google Scholar] [CrossRef]
- Hamilton, B.J.; Burns, C.M.; Nichols, R.C.; Rigby, W.F.C. Modulation of AUUUA Response Element Binding by Heterogeneous Nuclear Ribonucleoprotein A1 in Human T Lymphocytes. The roles of cytoplasmic location, transcription, and phosphorylation. J. Biol. Chem. 1997, 272, 28732–28741. [Google Scholar] [CrossRef]
- Rajani, D.K.; Walch, M.; Martinvalet, D.; Thomas, M.P.; Lieberman, J. Alterations in RNA processing during immune-mediated programmed cell death. Proc. Natl. Acad. Sci. USA 2012, 109, 8688–8693. [Google Scholar] [CrossRef]
- Allegretta, M.; Nicklas, J.A.; Sriram, S.; Albertini, R.J. T cells responsive to myelin basic protein in patients with multiple sclerosis. Science 1990, 247, 718–721. [Google Scholar] [CrossRef]
- Lodge, P.A.; Allegretta, M.; Steinman, L.; Sriram, S. Myelin basic protein peptide specificity and T-cell receptor gene usage of HPRT mutant T-cell clones in patients with multiple sclerosis. Ann. Neurol. 1994, 36, 734–740. [Google Scholar] [CrossRef] [PubMed]
- Valori, M.; Jansson, L.; Kiviharju, A.; Ellonen, P.; Rajala, H.; Awad, S.A.; Mustjoki, S.; Tienari, P.J. A novel class of somatic mutations in blood detected preferentially in CD8 + cells. Clin. Immunol. 2017, 175, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Van Horebeek, L.; Hilven, K.; Mallants, K.; Van Nieuwenhuijze, A.; Kelkka, T.; Savola, P.; Mustjoki, S.; Schlenner, S.M.; Liston, A.; Dubois, B.; et al. A robust pipeline with high replication rate for detection of somatic variants in the adaptive immune system as a source of common genetic variation in autoimmune disease. Hum. Mol. Genet. 2019, 28, 1369–1380. [Google Scholar] [CrossRef]
- Liu, Q.; Shu, S.; Wang, R.R.; Liu, F.; Cui, B.; Guo, X.N.; Lu, C.X.; Li, X.G.; Liu, M.S.; Peng, B.; et al. Whole-exome sequencing identifies a missense mutation in hnRNPA1 in a family with flail arm ALS. Neurological 2016, 87, 1763–1769. [Google Scholar] [CrossRef]
- Naruse, H.; Ishiura, H.; Mitsui, J.; Date, H.; Takahashi, Y.; Matsukawa, T.; Tanaka, M.; Ishii, A.; Tamaoka, A.; Hokkoku, K.; et al. Molecular epidemiological study of familial amyotrophic lateral sclerosis in Japanese population by whole-exome sequencing and identification of novel HNRNPA1 mutation. Neurobiol. Aging 2018, 61, 255.e9–255.e16. [Google Scholar] [CrossRef]
- Taylor, J.P.; Brown, R.H., Jr.; Cleveland, D.W. Decoding ALS: From genes to mechanism. Nat. Cell Biol. 2016, 539, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Deeb, O.; Nabulsi, M. Exploring Multiple Sclerosis (MS) and Amyotrophic Lateral Scler osis (ALS) as Neurodegenerative Diseases and their Treatments: A Review Study. Curr. Top. Med. Chem. 2020, 20, 2391–2403. [Google Scholar] [CrossRef] [PubMed]
- Bahi-Buisson, N.; Souville, I.; Fourniol, F.J.; Toussaint, A.; Moores, C.A.; Houdusse, A.; Lemaitre, J.Y.; Poirier, K.; Khalaf-Nazzal, R.; Hully, M.; et al. New insights into genotype–phenotype correlations for the doublecortin-related lissencephaly spectrum. Brain 2013, 136, 223–244. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Ramos, A.; Picher, A.J.; García, E.; Garrido, P.; Hernandez, F.; Soriano, E.; Avila, J. Validation of Suspected Somatic Single Nucleotide Variations in the Brain of Alzheimer’s Disease Patients. J. Alzheimer’s Dis. 2017, 56, 977–990. [Google Scholar] [CrossRef]
- McConnell, M.J.; Moran, J.V.; Abyzov, A.; Akbarian, S.; Bae, T.; Cortes-Ciriano, I.; Erwin, J.A.; Fasching, L.; Flasch, D.A.; Freed, D.; et al. Intersection of diverse neuronal genomes and neuropsychiatric disease: The Brain Somatic Mosaicism Network. Science 2017, 356, eaal1641. [Google Scholar] [CrossRef]

| Reagent or Resource | Source | Identifier |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-β-Actin (8H10D10) | New England BioLabs | Cat# 3700S |
| Rabbit monoclonal anti-EIF2S1 (EPR23098-50) | Abcam | Cat# ab242148 |
| Rabbit monoclonal anti-EIF2S1 (phospho S51) (E90) | Abcam | Cat# ab32157 |
| Rabbit polyclonal anti-G3BP (EPR13986(B)) | Abcam | Cat# ab217225 |
| Mouse monoclonal anti-hnRNPA1 (4B10) | Millipore Sigma | Cat# 05-1521 |
| Rabbit polyclonal anti-TDP-43 | Proteintech | Cat# 10782-2-AP |
| Alexa Fluor 488 Donkey anti-rabbit IgG (H + L) | Jackson ImmunoResearch | Cat# 711-546-152 |
| Alexa Fluor 647 Goat anti-rabbit IgG (H + L) | Jackson ImmunoResearch | Cat# 111-605-047 |
| Goat anti-mouse IgG (H + L) HRP Conjugate | Bio-Rad | Cat# 1706516 |
| Goat anti-rabbit IgG (H + L) HRP Conjugate | Bio-Rad | Cat# 1706515 |
| Chemicals | ||
| Dulbecco′s Modified Eagle Media (DMEM) | Fisher Scientific | Cat# MT-10-013-CV |
| FluoroBrite DMEM Media | Fisher Scientific | Cat# A1896701 |
| Formaldehyde, 37% by weight (with preservative/certified ACS) | Fisher Scientific | Cat# F79-500 |
| Gibco Fetal Bovine Growth Serum | Thermo Fisher Scientific | Cat# 12483020 |
| Invitrogen ProLong Gold Antifade Mountant with DAPI | Fisher Scientific | Cat# P36935 |
| Immobilon-FL PVDF Membrane | Millipore Sigma | Cat# IPVH00010 |
| Lipofectamine 2000 | Fisher Scientific | Cat# 11-668-019 |
| Opti-MEM Reduced Serum Media | Fisher Scientific | Cat# 31985062 |
| Sodium Arsenite Solution | Millipore Sigma | Cat# 106277 |
| Sodium Dodecyl Sulfate (SDS) | Fisher Scientific | Cat# BP166 |
| Triton X-100 | Fisher Scientific | Cat# AC215682500 |
| Tween 20 | Fisher Scientific | Cat# BP337 |
| Commercial Assays | ||
| Clarity Western ECL Substrate | Bio-Rad | Cat# 1705060 |
| NE-PER Nuclear Cytoplasmic Extraction Reagents Kit | Thermo Fisher Scientific | Cat# 78833 |
| Cell Lines | ||
| HEK293T | ATCC | Cat# CRL-11268 |
| Recombinant DNA Plasmids | ||
| pmCry2PHR-mCherry | Addgene | Cat# 26866 |
| pmTriEx-A1WT | [25] | N/A |
| pmTriEx-A1(F281L) | [25] | N/A |
| pmTriEx-A1(P275S) | [25] | N/A |
| pmA1WT-mCherry | This paper | N/A |
| pmA1(F281L)-mCherry | This paper | N/A |
| pmA1(P275S)-mCherry | This paper | N/A |
| pmCry2PHR-A1WT-mCherry | This paper | N/A |
| pmCry2PHR-A1(F281L)-mCherry | This paper | N/A |
| pmCry2PHR-A1(P275S)-mCherry | This paper | N/A |
| pmCry2PHR-A1(PrLD)-mCherry | This paper | N/A |
| pcDNA3.1/NT-GFP-TOPO | Thermo Fisher Scientific | Cat# K481001 |
| pDONR221-G3BP1 | DNASU | Cat# HsCD00042033 |
| pmGFP-G3BP1 | This paper | N/A |
| Primers | ||
| Gibson Assembly Primers (See Supplemental Table S1) | IDT | N/A |
| Software | ||
| Adobe Photoshop CS6 | Adobe | https://www.adobe.com |
| GraphPad Prism 9.0.0 | GraphPad Software Inc. | https://www.graphpad.com |
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| ZEN 3.1 Blue Edition | Carl Zeiss Microscopy, LLC | https://www.zeiss.com |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Clarke, J.-P.W.E.; Thibault, P.A.; Salapa, H.E.; Kim, D.E.; Hutchinson, C.; Levin, M.C. Multiple Sclerosis-Associated hnRNPA1 Mutations Alter hnRNPA1 Dynamics and Influence Stress Granule Formation. Int. J. Mol. Sci. 2021, 22, 2909. https://doi.org/10.3390/ijms22062909
Clarke J-PWE, Thibault PA, Salapa HE, Kim DE, Hutchinson C, Levin MC. Multiple Sclerosis-Associated hnRNPA1 Mutations Alter hnRNPA1 Dynamics and Influence Stress Granule Formation. International Journal of Molecular Sciences. 2021; 22(6):2909. https://doi.org/10.3390/ijms22062909
Chicago/Turabian StyleClarke, Joseph-Patrick W. E., Patricia A. Thibault, Hannah E. Salapa, David E. Kim, Catherine Hutchinson, and Michael C. Levin. 2021. "Multiple Sclerosis-Associated hnRNPA1 Mutations Alter hnRNPA1 Dynamics and Influence Stress Granule Formation" International Journal of Molecular Sciences 22, no. 6: 2909. https://doi.org/10.3390/ijms22062909
APA StyleClarke, J.-P. W. E., Thibault, P. A., Salapa, H. E., Kim, D. E., Hutchinson, C., & Levin, M. C. (2021). Multiple Sclerosis-Associated hnRNPA1 Mutations Alter hnRNPA1 Dynamics and Influence Stress Granule Formation. International Journal of Molecular Sciences, 22(6), 2909. https://doi.org/10.3390/ijms22062909







