Mitochondrial Unfolded Protein Responses in White Adipose Tissue: Lipoatrophy, Whole-Body Metabolism and Lifespan
Abstract
1. Introduction
2. Mitochondrial Unfolded Protein Response
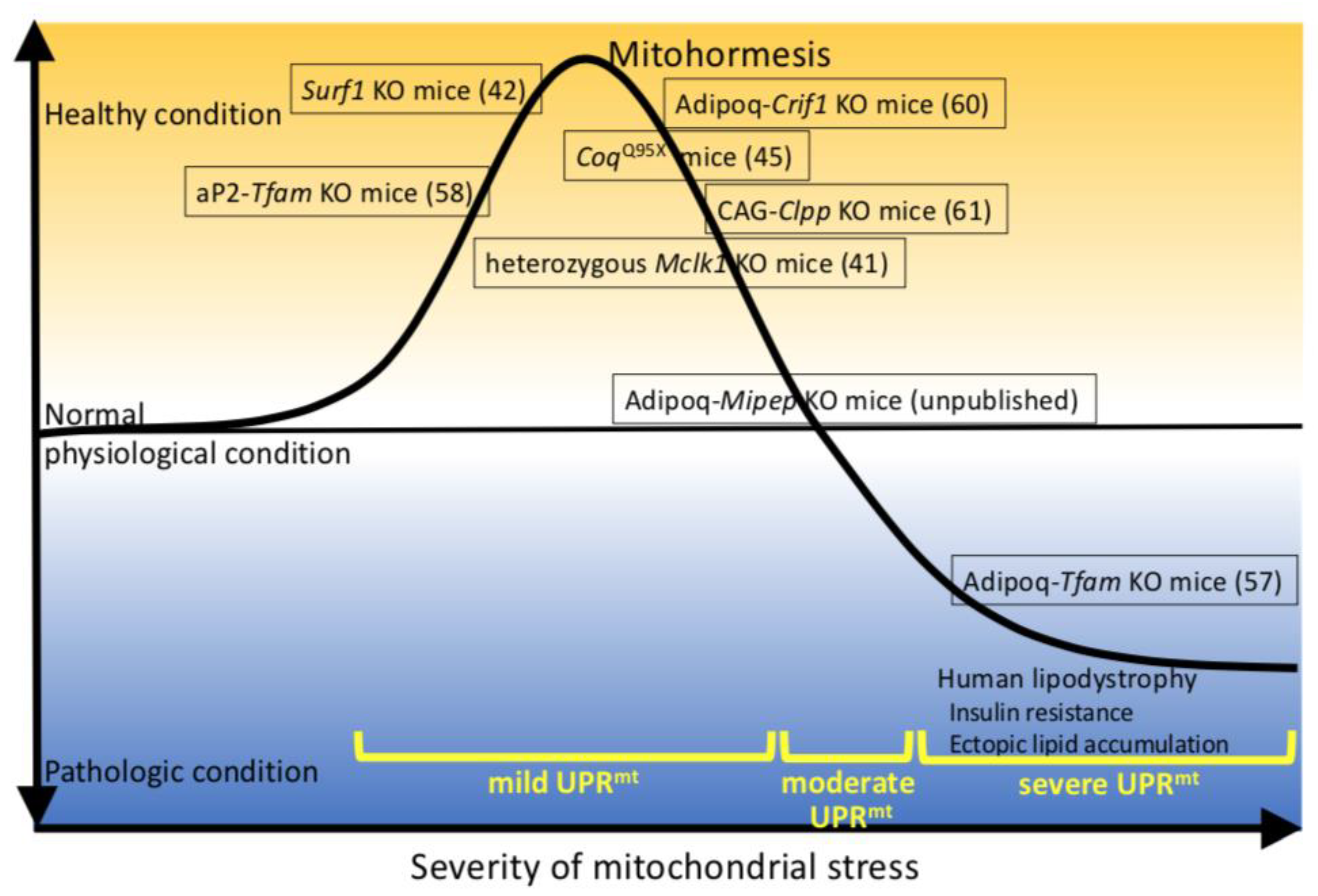
3. Genetically Modified Mice Live Longer Due to a Deficiency of Mitochondrial Proteins or Overexpression of Mitokines
4. Genetically Modified Mice Have Defective Mitochondrial Protein, Leading to Lipoatrophy, Which Regulates Whole-Body Metabolism
5. Discussion: Lessons Learned from Genetically Modified Mice that Develop Lipoatrophy Due to Defective Proteins Involved in Mitochondrial Function
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shpilka, T.; Haynes, C.M. The mitochondrial UPR: Mechanisms, physiological functions and implications in ageing. Nat. Rev. Mol. Cell Biol. 2018, 19, 109–120. [Google Scholar] [CrossRef]
- Yi, H.-S.; Chang, J.Y.; Shong, M. The mitochondrial unfolded protein response and mitohormesis: A perspective on metabolic diseases. J. Mol. Endocrinol. 2018, 61, R91–R105. [Google Scholar] [CrossRef]
- Quirós, P.M.; Langer, T.; López-Otín, C. New roles for mitochondrial proteases in health, ageing and disease. Nat. Rev. Mol. Cell Biol. 2015, 16, 345–359. [Google Scholar] [CrossRef] [PubMed]
- Ouchi, N.; Parker, J.L.; Lugus, J.J.; Walsh, K. Adipokines in inflammation and metabolic disease. Nat. Rev. Immunol. 2011, 11, 85–97. [Google Scholar] [CrossRef]
- Aguiar-Oliveira, M.H.; Bartke, A. Growth Hormone Deficiency: Health and Longevity. Endocr. Rev. 2019, 40, 575–601. [Google Scholar] [CrossRef] [PubMed]
- Accili, D.; Drago, J.; Lee, E.J.; Johnson, M.D.; Cool, M.H.; Salvatore, P.; Asico, L.D.; José, P.A.; Taylor, S.I.; Westphal, H. Early neonatal death in mice homozygous for a null allele of the insulin receptor gene. Nat. Genet. 1996, 12, 106–109. [Google Scholar] [CrossRef] [PubMed]
- Ueki, K.; Okada, T.; Hu, J.; Liew, C.W.; Assmann, A.; Dahlgren, G.M.; Peters, J.L.; Shackman, J.G.; Zhang, M.; Artner, I.; et al. Total insulin and IGF-I resistance in pancreatic beta cells causes overt diabetes. Nat. Genet. 2006, 38, 583–588. [Google Scholar] [CrossRef]
- Laustsen, P.G.; Russell, S.J.; Cui, L.; Entingh-Pearsall, A.; Holzenberger, M.; Liao, R.; Kahn, C.R. Essential role of insulin and insulin-like growth factor 1 receptor signaling in cardiac development and function. Mol. Cell Biol. 2007, 27, 1649–1664. [Google Scholar] [CrossRef]
- Michael, M.D.; Kulkarni, R.N.; Postic, C.; Previs, S.F.; Shulman, G.I.; Magnuson, M.A.; Kahn, C.R. Loss of insulin signaling in hepatocytes leads to severe insulin resistance and progressive hepatic dysfunction. Mol. Cell 2000, 6, 87–97. [Google Scholar] [CrossRef]
- Blüher, M.; Kahn, B.B.; Kahn, C.R. Extended longevity in mice lacking the insulin receptor in adipose tissue. Science 2003, 299, 572–574. [Google Scholar] [CrossRef]
- Otabe, S.; Yuan, X.; Fukutani, T.; Wada, N.; Hashinaga, T.; Nakayama, H.; Hirota, N.; Kojima, M.; Yamada, K. Overexpression of human adiponectin in transgenic mice results in suppression of fat accumulation and prevention of premature death by high-calorie diet. Am. J. Physiol. Endocrinol. Metab. 2007, 293, E210–E218. [Google Scholar] [CrossRef]
- Chen, S.S.; Chen, J.F.; Johnson, P.F.; Muppala, V.; Lee, Y.H. C/EBPβ, when expressed from the C/ebpα gene locus, can functionally replace C/EBPα in liver but not in adipose tissue. Mol. Cell Biol. 2000, 20, 7292–7299. [Google Scholar] [CrossRef]
- Chiu, C.H.; Lin, W.D.; Huang, S.Y.; Lee, Y.H. Effect of a C/EBP gene replacement on mitochondrial biogenesis in fat cells. Genes Dev. 2004, 18, 1970–1975. [Google Scholar] [CrossRef]
- Argmann, C.; Dobrin, R.; Heikkinen, S.; Auburtin, A.; Pouilly, L.; Cock, T.A.; Koutnikova, H.; Zhu, J.; Schadt, E.E.; Auwerx, J. Ppargamma2 is a key driver of longevity in the mouse. PLoS Genet. 2009, 5, 1000752. [Google Scholar] [CrossRef]
- Fujii, N.; Narita, T.; Okita, N.; Kobayashi, M.; Furuta, Y.; Chujo, Y.; Sakai, M.; Yamada, A.; Takeda, K.; Konishi, T.; et al. Sterol regulatory element-binding protein-1c orchestrates metabolic remodeling of white adipose tissue by caloric restriction. Aging Cell 2017, 16, 508–517. [Google Scholar] [CrossRef]
- Roufayel, R.; Kadry, S. Molecular Chaperone HSP70 and key regulators of apoptosis–A review. Curr. Mol. Med. 2019, 19, 315–325. [Google Scholar] [CrossRef] [PubMed]
- Böttinger, L.; Oeljeklaus, S.; Guiard, B.; Rospert, S.; Warscheid, B.; Becker, T. Mitochondrial heat shock protein (Hsp) 70 and Hsp10 cooperate in the formation of Hsp60 complexes. J. Biol. Chem. 2015, 290, 11611–11622. [Google Scholar] [CrossRef]
- D’Silva, P.; Liu, Q.; Walter, W.; Craig, E.A. Regulated interactions of mtHsp70 with Tim44 at the translocon in the mitochondrial inner membrane. Nat. Struct. Mol. Biol. 2004, 11, 1084–1091. [Google Scholar] [CrossRef]
- Konovalova, S.; Liu, X.; Manjunath, P.; Baral, S.; Neupane, N.; Hilander, T.; Yang, Y.; Balboa, D.; Terzioglu, M.; Euro, L.; et al. Redox regulation of GRPEL2 nucleotide exchange factor for mitochondrial HSP70 chaperone. Redox Biol. 2018, 19, 37–45. [Google Scholar] [CrossRef]
- Ghosh, J.C.; Dohi, T.; Kang, B.H.; Altieri, D.C. Hsp60 regulation of tumor cell apoptosis. J. Biol. Chem. 2008, 283, 5188–5194. [Google Scholar] [CrossRef] [PubMed]
- Ngo, J.K.; Pomatto, L.C.D.; Bota, D.A.; Koop, A.L.; Davies, K.J.A. Impairment of lon-induced protection against the accumulation of oxidized proteins in senescent wi-38 fibroblasts. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2011, 66, 1178–1185. [Google Scholar] [CrossRef] [PubMed]
- Gatsogiannis, C.; Balogh, D.; Merino, F.; Sieber, S.A.; Raunser, S. Cryo-EM structure of the ClpXP protein degradation machinery. Nat. Struct. Mol. Biol. 2019, 26, 946–954. [Google Scholar] [CrossRef]
- Restelli, L.M.; Oettinghaus, B.; Halliday, M.; Agca, C.; Licci, M.; Sironi, L.; Savoia, C.; Hench, J.; Tolnay, M.; Neutzner, A.; et al. Neuronal mitochondrial dysfunction activates the integrated stress response to induce fibroblast growth factor 21. Cell Rep. 2018, 24, 1407–1414. [Google Scholar] [CrossRef] [PubMed]
- Fujita, Y.; Taniguchi, Y.; Shinkai, S.; Tanaka, M.; Ito, M. Secreted growth differentiation factor 15 as a potential biomarker for mitochondrial dysfunctions in aging and age-related disorders. Geriatr. Gerontol. Int. 2016, 16, 17–29. [Google Scholar] [CrossRef]
- Koga, Y.; Povalko, N.; Inoue, E.; Nashiki, K.; Tanaka, M. Biomarkers and clinical rating scales for sodium pyruvate therapy in patients with mitochondrial disease. Mitochondrion 2019, 48, 11–15. [Google Scholar] [CrossRef]
- Tsai, V.W.W.; Husaini, Y.; Sainsbury, A.; Brown, D.A.; Breit, S.N. The MIC-1/GDF15-GFRAL pathway in energy homeostasis: Implications for obesity, cachexia, and other associated diseases. Cell Metab. 2018, 28, 353–368. [Google Scholar] [CrossRef] [PubMed]
- Bootcov, M.R.; Bauskin, A.R.; Valenzuela, S.M.; Moore, A.G.; Bansal, M.; He, X.Y.; Zhang, H.P.; Donnellan, M.; Mahler, S.; Pryor, K.; et al. MIC-1, a novel macrophage inhibitory cytokine, is a divergent member of the TGF-beta superfamily. Proc. Natl. Acad. Sci. USA 1997, 94, 11514–11519. [Google Scholar] [CrossRef] [PubMed]
- Böttner, M.; Suter-Crazzolara, C.; Schober, A.; Unsicker, K. Expression of a novel member of the TGF-beta superfamily, growth/differentiation factor-15/macrophage-inhibiting cytokine-1 (GDF-15/MIC-1) in adult rat tissues. Cell Tissue Res. 1999, 297, 103–110. [Google Scholar] [CrossRef]
- Ding, Q.; Mracek, T.; Gonzalez-Muniesa, P.; Kos, K.; Wilding, J.; Trayhurn, P.; Bing, C. Identification of macrophage inhibitory cytokine-1 in adipose tissue and its secretion as an adipokine by human adipocytes. Endocrinology 2009, 150, 1688–1696. [Google Scholar] [CrossRef]
- Chung, H.K.; Ryu, D.; Kim, K.S.; Chang, J.Y.; Kim, Y.K.; Yi, H.S.; Kang, S.G.; Choi, M.J.; Lee, S.E.; Jung, S.B.; et al. Growth differentiation factor 15 is a myomitokine governing systemic energy homeostasis. J. Cell Biol. 2017, 216, 149–165. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.K.; Kim, J.T.; Kim, H.W.; Kwon, M.; Kim, S.Y.; Shong, M.; Kim, K.S.; Yi, H.S. GDF15 deficiency exacerbates chronic alcohol- and carbon tetrachloride-induced liver injury. Sci. Rep. 2017, 7, 17238. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.; Alvarez-Guaita, A.; Melvin, A.; Rimmington, D.; Dattilo, A.; Miedzybrodzka, E.L.; Cimino, I.; Maurin, A.C.; Roberts, G.P.; Meek, C.L.; et al. GDF15 provides an endocrine signal of nutritional stress in mice and humans. Cell Metab. 2019, 29, 707–718. [Google Scholar] [CrossRef] [PubMed]
- Hsu, J.Y.; Crawley, S.; Chen, M.; Ayupova, D.A.; Lindhout, D.A.; Higbee, J.; Kutach, A.; Joo, W.; Gao, Z.; Fu, D.; et al. Non-homeostatic body weight regulation through a brainstem-restricted receptor for GDF15. Nature 2017, 550, 255–259. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, T.; Nakatake, Y.; Konishi, M.; Itoh, N. Identification of a novel FGF, FGF-21, preferentially expressed in the liver. Biochim. Biophys. Acta 2000, 1492, 203–206. [Google Scholar] [CrossRef]
- Kharitonenkov, A.; Shiyanova, T.L.; Koester, A.; Ford, A.M.; Micanovic, R.; Galbreath, E.J.; Sandusky, G.E.; Hammond, L.J.; Moyers, J.S.; Owens, R.A.; et al. FGF-21 as a novel metabolic regulator. J. Clin. Invest. 2005, 115, 1627–1635. [Google Scholar] [CrossRef]
- Kurosu, H.; Choi, M.; Ogawa, Y.; Dickson, A.S.; Goetz, R.; Eliseenkova, A.V.; Mohammadi, M.; Rosenblatt, K.P.; Kliewer, S.A.; Kuro-o, M. Tissue-specific expression of betaKlotho and fibroblast growth factor (FGF) receptor isoforms determines metabolic activity of FGF19 and FGF21. J. Biol. Chem. 2007, 282, 26687–26695. [Google Scholar] [CrossRef]
- Ding, X.; Boney-Montoya, J.; Owen, B.M.; Bookout, A.L.; Coate, K.C.; Mangelsdorf, D.J.; Kliewer, S.A. betaKlotho is required for fibroblast growth factor 21 effects on growth and metabolism. Cell Metab. 2012, 16, 387–393. [Google Scholar] [CrossRef]
- Fon Tacer, K.; Bookout, A.L.; Ding, X.; Kurosu, H.; John, G.B.; Wang, L.; Goetz, R.; Mohammadi, M.; Kuro-o, M.; Mangelsdorf, D.J.; et al. Research resource: Comprehensive expression atlas of the fibroblast growth factor system in adult mouse. Mol. Endocrinol. 2010, 24, 2050–2064. [Google Scholar] [CrossRef]
- Fujii, N.; Uta, S.; Kobayashi, M.; Sato, T.; Okita, N.; Higami, Y. Impact of aging and caloric restriction on fibroblast growth factor 21 signaling in rat white adipose tissue. Exp. Gerontol. 2019, 118, 55–64. [Google Scholar] [CrossRef]
- Bárcena, C.; Mayoral, P.; Quirós, P.M. Mitohormesis, an antiaging paradigm. Int. Rev. Cell Mol. Biol. 2018, 340, 35–77. [Google Scholar] [CrossRef]
- Liu, X.; Jiang, N.; Hughes, B.; Bigras, E.; Shoubridge, E.; Hekimi, S. Evolutionary conservation of the clk-1-dependent mechanism of longevity: Loss of mclk1 increases cellular fitness and lifespan in mice. Genes Dev. 2005, 19, 2424–2434. [Google Scholar] [CrossRef]
- Dell’agnello, C.; Leo, S.; Agostino, A.; Szabadkai, G.; Tiveron, C.; Zulian, A.; Prelle, A.; Roubertoux, P.; Rizzuto, R.; Zeviani, M. Increased longevity and refractoriness to Ca(2+)-dependent neurodegeneration in Surf1 knockout mice. Hum. Mol. Genet. 2007, 16, 431–444. [Google Scholar] [CrossRef]
- Lapointe, J.; Hekimi, S. Early mitochondrial dysfunction in long-lived Mclk1+/− mice. J. Biol. Chem. 2008, 283, 26217–26227. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Wang, Y.; Argyriou, C.; Carrière, A.; Malo, D.; Hekimi, S. An enhanced immune response of Mclk1+/− mutant mice is associated with partial protection from fibrosis, cancer and the development of biomarkers of aging. PLoS ONE 2012, 7, 49606. [Google Scholar] [CrossRef][Green Version]
- Luna-Sánchez, M.; Díaz-Casado, E.; Barca, E.; Tejada, M.Á.; Montilla-García, Á.; Cobos, E.J.; Escames, G.; Acuña-Castroviejo, D.; Quinzii, C.M.; López, L.C. The clinical heterogeneity of coenzyme Q10 deficiency results from genotypic differences in the Coq9 gene. EMBO Mol. Med. 2015, 7, 670–687. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Hidalgo, M.; Luna-Sánchez, M.; Hidalgo-Gutiérrez, A.; Barriocanal-Casado, E.; Mascaraque, C.; Acuña-Castroviejo, D.; Rivera, M.; Escames, G.; López, L.C. Reduction in the levels of CoQ biosynthetic proteins is related to an increase in lifespan without evidence of hepatic mitohormesis. Sci. Rep. 2018, 8, 14013. [Google Scholar] [CrossRef]
- Monaghan, R.M.; Barnes, R.G.; Fisher, K.; Andreou, T.; Rooney, N.; Poulin, G.B.; Whitmarsh, A.J. A nuclear role for the respiratory enzyme CLK-1 in regulating mitochondrial stress responses and longevity. Nat. Cell Biol. 2015, 17, 782–792. [Google Scholar] [CrossRef] [PubMed]
- Deepa, S.S.; Pulliam, D.; Hill, S.; Shi, Y.; Walsh, M.E.; Salmon, A.; Sloane, L.; Zhang, N.; Zeviani, M.; Viscomi, C.; et al. Improved insulin sensitivity associated with reduced mitochondrial complex IV assembly and activity. FASEB J. 2013, 27, 1371–1380. [Google Scholar] [CrossRef]
- Pulliam, D.A.; Deepa, S.S.; Liu, Y.; Hill, S.; Lin, A.L.; Bhattacharya, A.; Shi, Y.; Sloane, L.; Viscomi, C.; Zeviani, M.; et al. Complex IV-deficient Surf1(−/−) mice initiate mitochondrial stress responses. Biochem. J. 2014, 462, 359–371. [Google Scholar] [CrossRef] [PubMed]
- Deepa, S.S.; Pharaoh, G.; Kinter, M.; Diaz, V.; Fok, W.C.; Riddle, K.; Pulliam, D.; Hill, S.; Fischer, K.E.; Soto, V.; et al. Lifelong reduction in complex IV induces tissue-specific metabolic effects but does not reduce lifespan or healthspan in mice. Aging Cell 2018, 17, 12769. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xie, Y.; Berglund, E.D.; Coate, K.C.; He, T.T.; Katafuchi, T.; Xiao, G.; Potthoff, M.J.; Wei, W.; Wan, Y.; et al. The starvation hormone, fibroblast growth factor-21, extends lifespan in mice. Elife 2012, 1, 00065. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chrysovergis, K.; Kosak, J.; Kissling, G.; Streicker, M.; Moser, G.; Li, R.; Eling, T.E. HNAG-1 increases lifespan by regulating energy metabolism and insulin/IGF-1/mTOR signaling. Aging (Albany NY) 2014, 6, 690–704. [Google Scholar] [CrossRef] [PubMed]
- Chrysovergis, K.; Wang, X.; Kosak, J.; Lee, S.H.; Kim, J.S.; Foley, J.F.; Travlos, G.; Singh, S.; Baek, S.J.; Eling, T.E. NAG-1/GDF-15 prevents obesity by increasing thermogenesis, lipolysis and oxidative metabolism. Int. J. Obes. (Lond.) 2014, 38, 1555–1564. [Google Scholar] [CrossRef]
- Luan, H.H.; Wang, A.; Hilliard, B.K.; Carvalho, F.; Rosen, C.E.; Ahasic, A.M.; Herzog, E.L.; Kang, I.; Pisani, M.A.; Yu, S.; et al. GDF15 is an inflammation-induced central mediator of tissue tolerance. Cell 2019, 178, 1231–1244.e11. [Google Scholar] [CrossRef]
- Jung, S.B.; Choi, M.J.; Ryu, D.; Yi, H.S.; Lee, S.E.; Chang, J.Y.; Chung, H.K.; Kim, Y.K.; Kang, S.G.; Lee, J.H.; et al. Reduced oxidative capacity in macrophages results in systemic insulin resistance. Nat. Commun. 2018, 9, 1551. [Google Scholar] [CrossRef]
- Gaspari, M.; Larsson, N.G.; Gustafsson, C.M. The transcription machinery in mammalian mitochondria. Biochim. Biophys. Acta 2004, 1659, 148–152. [Google Scholar] [CrossRef] [PubMed]
- Vernochet, C.; Damilano, F.; Mourier, A.; Bezy, O.; Mori, M.A.; Smyth, G.; Rosenzweig, A.; Larsson, N.G.; Kahn, C.R. Adipose tissue mitochondrial dysfunction triggers a lipodystrophic syndrome with insulin resistance, hepatosteatosis, and cardiovascular complications. FASEB J. 2014, 28, 4408–4419. [Google Scholar] [CrossRef]
- Vernochet, C.; Mourier, A.; Bezy, O.; Macotela, Y.; Boucher, J.; Rardin, M.J.; An, D.; Lee, K.Y.; Ilkayeva, O.R.; Zingaretti, C.M.; et al. Adipose-specific deletion of TFAM increases mitochondrial oxidation and protects mice against obesity and insulin resistance. Cell Metab. 2012, 16, 765–776. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Kwon, M.C.; Ryu, M.J.; Chung, H.K.; Tadi, S.; Kim, Y.K.; Kim, J.M.; Lee, S.H.; Park, J.H.; Kweon, G.R.; et al. CRIF1 is essential for the synthesis and insertion of oxidative phosphorylation polypeptides in the mammalian mitochondrial membrane. Cell Metab. 2012, 16, 274–283. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.J.; Jung, S.B.; Lee, S.E.; Kang, S.G.; Lee, J.H.; Ryu, M.J.; Chung, H.K.; Chang, J.Y.; Kim, Y.K.; Hong, H.J.; et al. An adipocyte-specific defect in oxidative phosphorylation increases systemic energy expenditure and protects against diet-induced obesity in mouse models. Diabetologia 2020, 63, 837–852. [Google Scholar] [CrossRef] [PubMed]
- Bhaskaran, S.; Pharaoh, G.; Ranjit, R.; Murphy, A.; Matsuzaki, S.; Nair, B.C.; Forbes, B.; Gispert, S.; Auburger, G.; Humphries, K.M.; et al. Loss of mitochondrial protease ClpP protects mice from diet-induced obesity and insulin resistance. EMBO Rep. 2018, 19, 45009. [Google Scholar] [CrossRef]
- Kobayashi, M.; Takeda, K.; Narita, T.; Nagai, K.; Okita, N.; Sudo, Y.; Miura, Y.; Tsumoto, H.; Nakagawa, Y.; Shimano, H.; et al. Mitochondrial intermediate peptidase is a novel regulator of sirtuin-3 activation by caloric restriction. FEBS Lett. 2017, 591, 4067–4073. [Google Scholar] [CrossRef]
- Mann, J.P.; Savage, D.B. What lipodystrophies teach us about the metabolic syndrome. J. Clin. Investig. 2019, 129, 4009–4021. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Barak, Y.; Hevener, A.; Olson, P.; Liao, D.; Le, J.; Nelson, M.; Ong, E.; Olefsky, J.M.; Evans, R.M. Adipose-specific peroxisome proliferator-activated receptor gamma knockout causes insulin resistance in fat and liver but not in muscle. Proc. Natl. Acad. Sci. USA 2003, 100, 15712–15717. [Google Scholar] [CrossRef]
- Wang, F.; Mullican, S.E.; DiSpirito, J.R.; Peed, L.C.; Lazar, M.A. Lipoatrophy and severe metabolic disturbance in mice with fat-specific deletion of PPARγ. Proc. Natl. Acad. Sci. USA 2013, 110, 18656–18661. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.V.; Deng, Y.; Wang, Q.A.; Sun, K.; Scherer, P.E. Identification and characterization of a promoter cassette conferring adipocyte-specific gene expression. Endocrinology 2010, 151, 2933–2939. [Google Scholar] [CrossRef]
- Mullican, S.E.; Tomaru, T.; Gaddi, C.A.; Peed, L.C.; Sundaram, A.; Lazar, M.A. A novel adipose-specific gene deletion model demonstrates potential pitfalls of existing methods. Mol. Endocrinol. 2013, 27, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Molenaars, M.; Janssens, G.E.; Williams, E.G.; Jongejan, A.; Lan, J.; Rabot, S.; Joly, F.; Moerland, P.D.; Schomakers, B.V.; Lezzerini, M.; et al. A conserved mito-cytosolic translational balance links two longevity pathways. Cell Metab. 2020, 31, 549–563. [Google Scholar] [CrossRef] [PubMed]
| Mice Genotype | Function of Target Protein | Body Weigh | Alteration of WAT and Adipocytes | Alteration Of Mitochondria Function | Glucose Metabolism | Uprmt | Adipocyte Differ. | Others | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Genetically modified mice, which live longer due to deficiency of mitochondrial proteins | |||||||||
| Heterozygous Mclk1 KO | Ubiquinone biosynthesis | → | n.r. | Complex II activity in ES cells ↓ Complex II, Complex I-III, Complex II-III activity in liver → | n.r. | n.r. | n.r. | ・Resistant to oxidative stress and infection ・Enhanced immune response | [41,43,44] |
| CoqQ95X | Ubiquinone biosynthesis | ↓ | n.r. | Proteins involved in ubiquinone biosynthesis in brain, heart, kidney and SKM ↓, in liver → Complex I-III, Complex II-III activity in female kidney, SKM ↓, in brain → | n.r. | Factors involved in chaperone and protease in liver → | n.r. | ・oxidative stress → ・Voluntary running distance ↓ | [45,46] |
| Surf1 KO | Complex IV assembly | ↓ | WAT weight: ↓ Adipocyte size: ↓ | Complex IV activity in liver, heart, SKM and WAT ↓ Complex I, II, III activity in heart, SKM → Mitochondria biogenesis in WAT ↑ Amount of Complex II, V in WAT ↑ | Improved | Factors involved in chaperone and protease in liver, heart and/or SKM↑ | ↑/→ | ・Plasma insulin ↓ ・Resistant to Ca2+- dependent neurodegeneration | [42,48,49,50] |
| Genetically modified mice, which defect mitochondrial protein, induce lipoatrophy | |||||||||
| Tfam KO (aP2-Cre) | Stabilization and transcription of mtDNA | ↓ | WAT weight: ↓ Adipocyte size: ↓ | mtDNA derived factors in WAT ↓ CS in WAT ↑ Complex I, IV activity in WAT ↓ | ND: improved HFD: improved | n.r. | n.r. | ・Plasma insulin ↓ | [58] |
| Tfam KO (Adipoq-Cre) | Stabilization and transcription of mtDNA | ↓ | WAT weight: ↓ Adipocyte size: → | Factors encoded mtDNA in WAT ↓ CS activity in WAT ↑ OXPHOS subunits encoded nuclear DNA in WAT ↓ Complex I, II-III, IV activity in WAT ↓ | ND: worsen HFD: worsen | n.r. | n.r. | ・Fatty liver ・Inflammation ・Hypertension ・Cardiac dysfunction ・Plasma insulin ↑ | [57] |
| Crif1 KO (Adipoq-Cre) | Translation in mitochondria | ↓ | WAT weight: ↓ Adipocyte size: ↓ | OXPHOS formation in WAT ↓ | ND: → HFD: improved | GDF15, FGF21 ↑ Factors involved in chaperone and protease ↑ | → | ・Macrophagein WAT (predominantly M2) ↑ ・Fatty liver ↓ | [60] |
| Clpp KO (Cag-Cre) | Mitochondrial protease | ↓ | WAT weight: ↓ Adipocyte size: ↓ | Mitochondria biogenesis in WAT ↑ βoxidation in WAT ↓ OXPHOS Complex Ⅱ in WAT ↑ | ND: improved HFD: markedly improved | Factors involved in chaperone and protease ↑ | → | ・Plasma insulin ↓ ・Thermogenesis ↑ | [61] |
| Mipep KO (Adipoq-Cre) | Mitochondrial signal peptidase | ↓ | WAT weight: ↓ Adipocyte size: → | Proteins encoded nuclear DNA (COX4, SIRT3) in WAT ↓ Proteins encoded nuclear DNA (MDH2) in WAT ↑ CS activity in WAT → | ND: worsen HFD: slightly worsen | GDF15, FGF21 ↑ Factors involved in chaperone and protease ↓ | ↓ | ・Plasma insulin ↑ | unpublished |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kobayashi, M.; Nezu, Y.; Tagawa, R.; Higami, Y. Mitochondrial Unfolded Protein Responses in White Adipose Tissue: Lipoatrophy, Whole-Body Metabolism and Lifespan. Int. J. Mol. Sci. 2021, 22, 2854. https://doi.org/10.3390/ijms22062854
Kobayashi M, Nezu Y, Tagawa R, Higami Y. Mitochondrial Unfolded Protein Responses in White Adipose Tissue: Lipoatrophy, Whole-Body Metabolism and Lifespan. International Journal of Molecular Sciences. 2021; 22(6):2854. https://doi.org/10.3390/ijms22062854
Chicago/Turabian StyleKobayashi, Masaki, Yuichiro Nezu, Ryoma Tagawa, and Yoshikazu Higami. 2021. "Mitochondrial Unfolded Protein Responses in White Adipose Tissue: Lipoatrophy, Whole-Body Metabolism and Lifespan" International Journal of Molecular Sciences 22, no. 6: 2854. https://doi.org/10.3390/ijms22062854
APA StyleKobayashi, M., Nezu, Y., Tagawa, R., & Higami, Y. (2021). Mitochondrial Unfolded Protein Responses in White Adipose Tissue: Lipoatrophy, Whole-Body Metabolism and Lifespan. International Journal of Molecular Sciences, 22(6), 2854. https://doi.org/10.3390/ijms22062854






