Bioluminescence Measurement of Time-Dependent Dynamic Changes of CYP-Mediated Cytotoxicity in CYP-Expressing Luminescent HepG2 Cells
Abstract
1. Introduction
2. Results
2.1. Generation of CYP-Expressing Luminescent HepG2 Cells
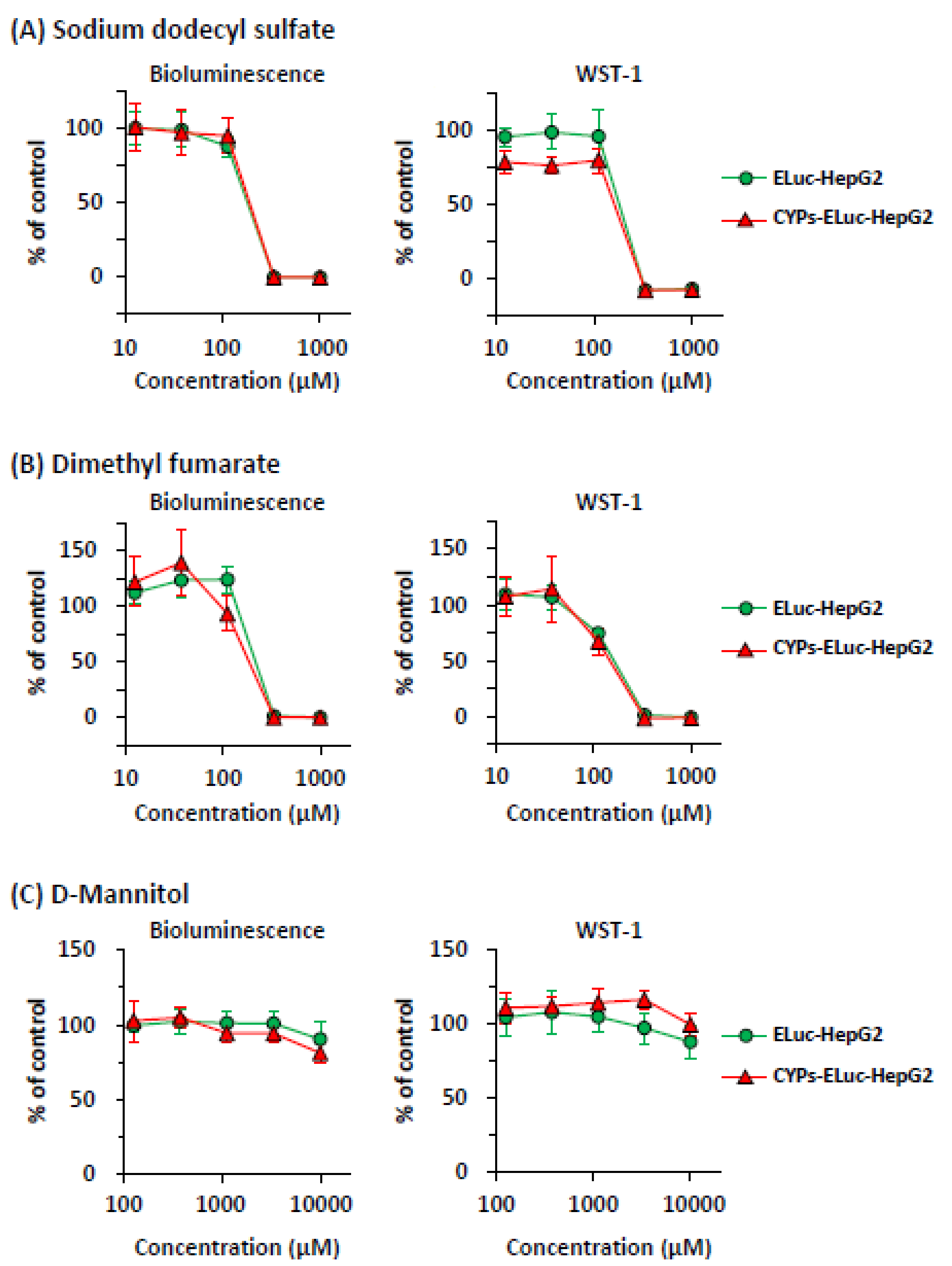
2.2. Comparison of Sensitivity of ELuc-HepG2 and CYPs-ELuc HepG2 Cells to CYP-Independent Toxicants
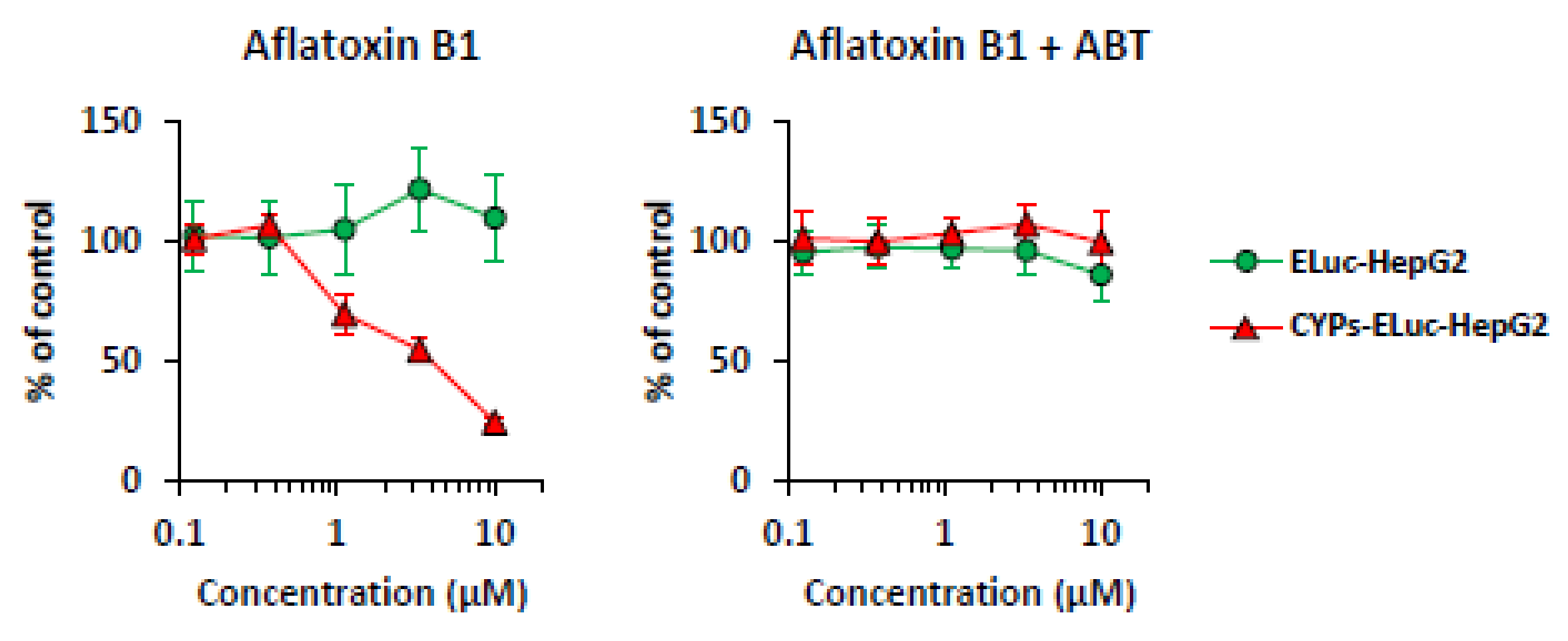
2.3. CYP-Mediated Cytotoxicity in CYPs-ELuc-HepG2 Cells
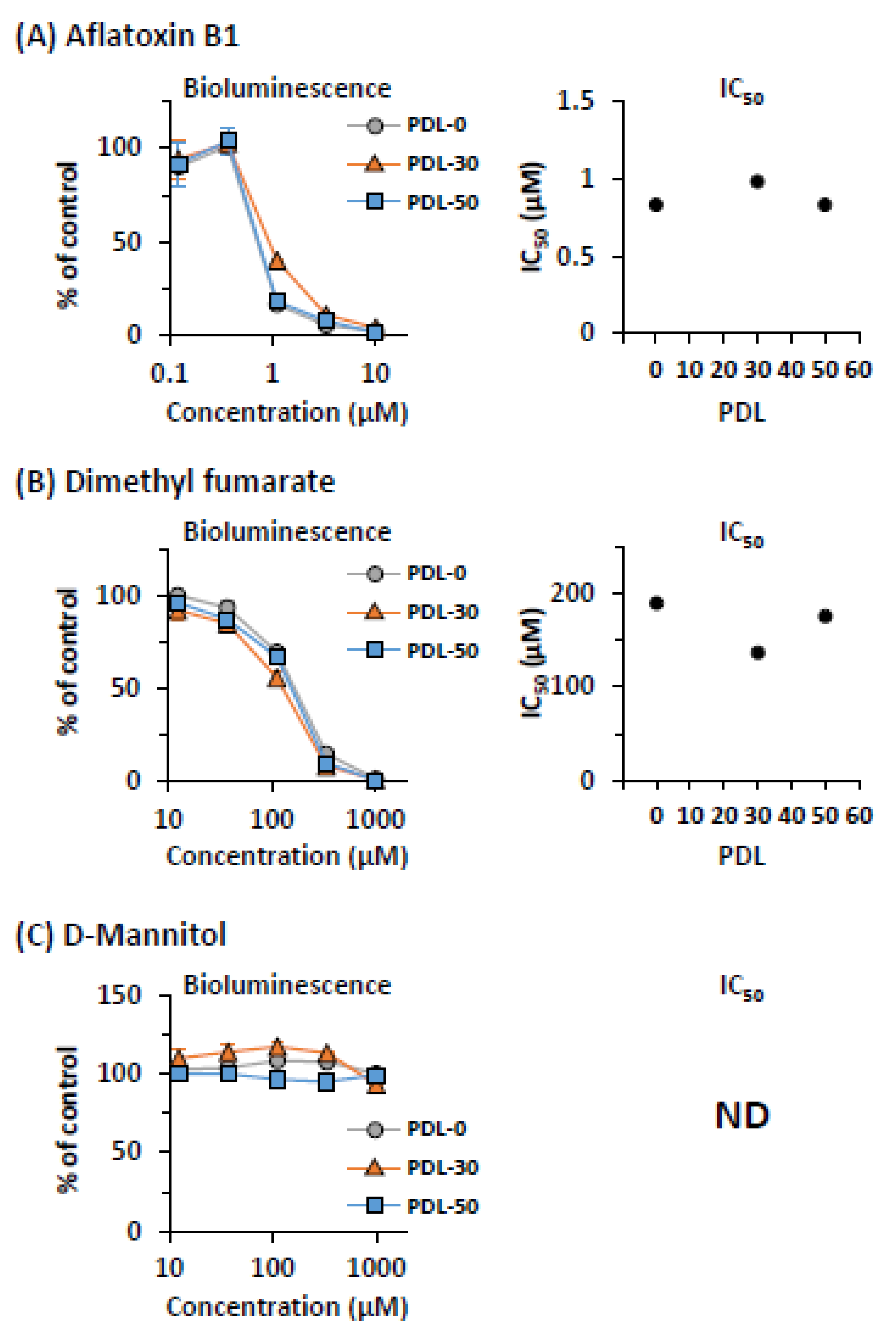
2.4. Stability of CYP-Mediated Cytotoxicity in CYPs-ELuc-HepG2 Cells during Long-Term Culture
2.5. Real-Time Bioluminescence Measurement of CYP-Mediated Cytotoxicity in CYPs-ELuc-HepG2 Cells
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Cell Culture
4.3. Microcell-Mediated Chromosome Transfer (MMCT)
4.4. FISH Analysis
4.5. Genomic PCR
4.6. Measurement of CYP Activities
4.7. Non-Destructive Bioluminescence Measurement and WST-1 Assay
4.8. Real-Time Bioluminescence Measurement
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Godoy, P.; Hewitt, N.J.; Albrecht, U.; Andersen, M.E.; Ansari, N.; Bhattacharya, S.; Bode, J.G.; Bolleyn, J.; Borner, C.; Böttger, J.; et al. Recent advances in 2D and 3D in vitro systems using primary hepatocytes, alternative hepatocyte sources and non-parenchymal liver cells and their use in investigating mechanisms of hepatotoxicity, cell signaling and ADME. Arch. Toxicol. 2013, 87, 1315–1530. [Google Scholar]
- Soldatow, V.Y.; Lecluyse, E.L.; Griffith, L.G.; Rusyn, I. In vitro models for liver toxicity testing. Toxicol. Res. 2013, 2, 23–39. [Google Scholar] [CrossRef]
- Afshari, A.; Shamdani, S.; Uzan, G.; Naserian, S.; Azarpira, N. Different approaches for transformation of mesenchymal stem cells into hepatocyte-like cells. Stem Cell Res. Ther. 2020, 11, 54. [Google Scholar] [CrossRef]
- Andersson, T.B.; Kanebratt, K.P.; Kenna, J.G. The HepaRG cell line: A unique in vitro tool for understanding drug metabolism and toxicology in human. Expert Opin. Drug Metab. Toxicol. 2012, 8, 909–920. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Matsuzaki, J.; Katsuda, T.; Saito, Y.; Saito, H.; Ochiya, T. Generation of functional human hepatocytes in vitro: Current status and future prospects. Inflamm. Regen. 2019, 39, 13. [Google Scholar] [CrossRef]
- Wilkening, S.; Stahl, F.; Bader, A. Comparison of primary human hepatocytes and hepatoma cell line Hepg2 with regard to their biotransformation properties. Drug Metab. Dispos. 2003, 31, 1035–1042. [Google Scholar] [CrossRef] [PubMed]
- Saito, J.; Okamura, A.; Takeuchi, K.; Hanioka, K.; Okada, A.; Ohata, T. High content analysis assay for prediction of human hepatotoxicity in HepaRG and HepG2 cells. Toxicol. Vitro 2016, 33, 63–70. [Google Scholar] [CrossRef]
- Satoh, D.; Iwado, S.; Abe, S.; Kazuki, K.; Wakuri, S.; Oshimura, M.; Kazuki, Y. Establishment of a novel hepatocyte model that expresses four cytochrome P450 genes stably via mammalian-derived artificial chromosome for pharmacokinetics and toxicity studies. PLoS ONE 2017, 12, e0187072. [Google Scholar] [CrossRef] [PubMed]
- Paine, M.F.; Hart, H.L.; Ludington, S.S.; Haining, R.L.; Rettie, A.E.; Zeldin, D.C. The human intestinal cytochrome P450 “pie”. Drug Metab. Dispos. 2006, 34, 880–886. [Google Scholar] [CrossRef] [PubMed]
- Miyawaki, A.; Niino, Y. Molecular spies for bioimaging--fluorescent protein-based probes. Mol. Cell 2015, 58, 632–643. [Google Scholar] [CrossRef] [PubMed]
- Specht, E.A.; Braselmann, E.; Palmer, A.E. A Critical and Comparative Review of Fluorescent Tools for Live-Cell Imaging. Annu. Rev. Physiol. 2017, 79, 93–117. [Google Scholar] [CrossRef]
- Roda, A.; Guardigli, M. Analytical chemiluminescence and bioluminescence: Latest achievements and new horizons. Anal. Bioanal. Chem. 2012, 402, 69–76. [Google Scholar] [CrossRef]
- Nakajima, Y.; Ohmiya, Y. Bioluminescence assays: Multicolor luciferase assay, secreted luciferase assay and imaging luciferase assay. Expert Opin. Drug Discov. 2010, 5, 835–849. [Google Scholar] [CrossRef] [PubMed]
- Gross, S.; Piwnica-Worms, D. Spying on cancer: Molecular imaging in vivo with genetically encoded reporters. Cancer Cell 2005, 7, 5–15. [Google Scholar]
- Class, B.; Thorne, N.; Aguisanda, F.; Southall, N.; McKew, J.C.; Zheng, W. High-throughput viability assay using an autonomously bioluminescent cell line with a bacterial Lux reporter. J. Lab. Autom. 2015, 20, 164–174. [Google Scholar] [CrossRef] [PubMed]
- Kimura, Y.; Fujimura, C.; Aiba, S. The modified IL-8 Luc assay, an in vitro skin sensitisation test, can significantly improve the false-negative judgment of lipophilic sensitizers with logKow values > 3.5. Arch. Toxicol. 2021, 95, 749–758. [Google Scholar] [CrossRef]
- Matta, H.; Gopalakrishnan, R.; Choi, S.; Prakash, R.; Natarajan, V.; Prins, R.; Gong, S.; Chitnis, S.D.; Kahn, M.; Han, X.; et al. Development and characterization of a novel luciferase based cytotoxicity assay. Sci. Rep. 2018, 8, 199. [Google Scholar] [CrossRef]
- Tsuji, S.; Ohbayashi, T.; Yamakage, K.; Oshimura, M.; Tada, M. A Cytoplasmic Form of Gaussia luciferase Provides a Highly Sensitive Test for Cytotoxicity. PLoS ONE 2016, 11, e0156202. [Google Scholar] [CrossRef]
- Wakuri, S.; Yamakage, K.; Kazuki, Y.; Kazuki, K.; Oshimura, M.; Aburatani, S.; Yasunaga, M.; Nakajima, Y. Correlation between luminescence intensity and cytotoxicity in cell-based cytotoxicity assay using luciferase. Anal. Biochem. 2017, 522, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Yasunaga, M.; Fujita, Y.; Saito, R.; Oshimura, M.; Nakajima, Y. Continuous long-term cytotoxicity monitoring in 3D spheroids of beetle luciferase-expressing hepatocytes by nondestructive bioluminescence measurement. BMC Biotechnol. 2017, 17, 54. [Google Scholar] [CrossRef]
- Uno, K.; Murotomi, K.; Kazuki, Y.; Oshimura, M.; Nakajima, Y. Bioluminescence-based cytotoxicity assay for simultaneous evaluation of cell viability and membrane damage in human hepatoma HepG2 cells. Luminescence 2018, 33, 616–624. [Google Scholar] [CrossRef]
- Gandelman, O.; Allue, I.; Bowers, K.; Cobbold, P. Cytoplasmic factors that affect the intensity and stability of bioluminescence from firefly luciferase in living mammalian cells. J. Biolumin. Chemilumin. 1994, 9, 363–371. [Google Scholar] [CrossRef]
- Ignowski, J.M.; Schaffer, D.V. Kinetic analysis and modeling of firefly luciferase as a quantitative reporter gene in live mammalian cells. Biotechnol. Bioeng. 2004, 86, 827–834. [Google Scholar] [CrossRef] [PubMed]
- Luker, K.E.; Luker, G.D. Applications of bioluminescence imaging to antiviral research and therapy: Multiple luciferase enzymes and quantitation. Antivir. Res. 2008, 78, 179–187. [Google Scholar] [CrossRef]
- Awais, M.; Ozawa, T. Illuminating intracellular signaling and molecules for single cell analysis. Mol. Biosyst. 2011, 7, 1376–1387. [Google Scholar] [CrossRef]
- Welsh, D.K.; Noguchi, T. Cellular bioluminescence imaging. Cold Spring Harb. Protoc. 2012, 2012, top070607. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, Y.; Yamazaki, T.; Nishii, S.; Noguchi, T.; Hoshino, H.; Niwa, K.; Viviani, V.R.; Ohmiya, Y. Enhanced beetle luciferase for high-resolution bioluminescence imaging. PLoS ONE 2010, 5, e10011. [Google Scholar] [CrossRef] [PubMed]
- Kazuki, K.; Takehara, S.; Uno, N.; Imaoka, N.; Abe, S.; Takiguchi, M.; Hiramatsu, K.; Oshimura, M.; Kazuki, Y. Highly stable maintenance of a mouse artificial chromosome in human cells and mice. Biochem. Biophys. Res. Commun. 2013, 442, 44–50. [Google Scholar] [CrossRef]
- Oshimura, M.; Uno, N.; Kazuki, Y.; Katoh, M.; Inoue, T. A pathway from chromosome transfer to engineering resulting in human and mouse artificial chromosomes for a variety of applications to bio-medical challenges. Chromosome Res. 2015, 23, 111–133. [Google Scholar] [CrossRef]
- Aubets, J.; Jansat, J.M.; Salva, M.; Birks, V.M.; Cole, R.J.; Lewis, J.; Pitcher, A.; Hall, M. No evidence for interactions of dimethylfumarate (DMF) and its main metabolite monomethylfumarate (MMF) with human cytochrome P450 (CYP) enzymes and the P-glycoprotein (P-gp) drug transporter. Pharmacol. Res. Perspect. 2019, 7, e00540. [Google Scholar] [CrossRef]
- Takahashi, T.; Kimura, Y.; Saito, R.; Nakajima, Y.; Ohmiya, Y.; Yamasaki, K.; Aiba, S. An in vitro test to screen skin sensitizers using a stable THP-1-derived IL-8 reporter cell line, THP-G8. Toxicol. Sci. 2011, 124, 359–369. [Google Scholar] [CrossRef]
- Dohnal, V.; Wu, Q.; Kuca, K. Metabolism of aflatoxins: Key enzymes and interindividual as well as interspecies differences. Arch. Toxicol. 2014, 88, 1635–1644. [Google Scholar] [CrossRef]
- Kamdem, L.K.; Meineke, I.; Godtel-Armbrust, U.; Brockmoller, J.; Wojnowski, L. Dominant contribution of P450 3A4 to the hepatic carcinogenic activation of aflatoxin B1. Chem. Res. Toxicol. 2006, 19, 577–586. [Google Scholar] [CrossRef] [PubMed]
- Orlando, R.; Piccoli, P.; De Martin, S.; Padrini, R.; Palatini, P. Effect of the CYP3A4 inhibitor erythromycin on the pharmacokinetics of lignocaine and its pharmacologically active metabolites in subjects with normal and impaired liver function. Br. J. Clin. Pharmacol. 2003, 55, 86–93. [Google Scholar] [CrossRef] [PubMed]
- Christensen, M.M.H.; Haastrup, M.B.; Øhlenschlaeger, T.; Esbech, P.; Pedersen, S.A.; Dunvald, A.B.; Stage, T.B.; Henriksen, D.P.; Pedersen, A.J.T. Interaction potential between clarithromycin and individual statins-A systematic review. Basic Clin. Pharmacol. Toxicol. 2020, 126, 307–317. [Google Scholar] [CrossRef] [PubMed]
- Marcsisin, S.R.; Reichard, G.; Pybus, B.S. Primaquine pharmacology in the context of CYP 2D6 pharmacogenomics: Current state of the art. Pharmacol. Ther. 2016, 161, 1–10. [Google Scholar] [CrossRef]
- Potter, B.M.J.; Xie, L.H.; Vuong, C.; Zhang, J.; Duan, D.; Luong, T.-L.T.; Herath, H.M.T.B.; Nanayakkara, N.P.D.; Tekwani, B.L.; Walker, L.A.; et al. Differential CYP 2D6 metabolism alters primaquine pharmacokinetics. Antimicrob. Agents Chemother. 2015, 59, 2380–2387. [Google Scholar] [CrossRef]
- Abdel-Rahman, S.M.; Gotschall, R.R.; Kauffman, R.E.; Leeder, J.S.; Kearns, G.L. Investigation of terbinafine as a CYP2D6 inhibitor in vivo. Clin. Pharmacol. Ther. 1999, 65, 465–472. [Google Scholar] [CrossRef]
- Akiyoshi, T.; Ishiuchi, M.; Imaoka, A.; Ohtani, H. Variation in the inhibitory potency of terbinafine among genetic variants of CYP2D6. Drug Metab. Pharmacokinet. 2015, 30, 321–324. [Google Scholar] [CrossRef]
- Kobayashi, M.; Yamauchi, Y.; Tanaka, A.; Shimamura, S. Improved dicistronic mRNA expression vectors for efficient selection of transfectants highly expressing foreign genes. BioTechniques 1996, 21, 398–402. [Google Scholar] [CrossRef]
- Niwa, K.; Ichino, Y.; Kumata, S.; Nakajima, Y.; Hiraishi, Y.; Kato, D.; Viviani, V.R.; Ohmiya, Y. Quantum yields and kinetics of the firefly bioluminescence reaction of beetle luciferases. Photochem. Photobiol. 2010, 86, 1046–1049. [Google Scholar] [CrossRef]
- Tanabe, M.; Niwa, K.; Kinoshita, K. Absolute optical responsivity down to the photon counting level with a photomultiplier tube. Rev. Sci. Instrum. 2017, 88, 043104. [Google Scholar] [CrossRef] [PubMed]
- Katoh, M.; Kazuki, Y.; Kazuki, K.; Kajitani, N.; Takiguchi, M.; Nakayama, Y.; Nakamura, T.; Oshimura, M. Exploitation of the interaction of measles virus fusogenic envelope proteins with the surface receptor CD46 on human cells for microcell-mediated chromosome transfer. BMC Biotechnol. 2010, 10, 37. [Google Scholar] [CrossRef] [PubMed]
- Katoh, M.; Ayabe, F.; Norikane, S.; Okada, T.; Masumoto, H.; Horike, S.; Shirayoshi, Y.; Oshimura, M. Construction of a novel human artificial chromosome vector for gene delivery. Biochem. Biophys. Res. Commun. 2004, 321, 280–290. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iwado, S.; Abe, S.; Oshimura, M.; Kazuki, Y.; Nakajima, Y. Bioluminescence Measurement of Time-Dependent Dynamic Changes of CYP-Mediated Cytotoxicity in CYP-Expressing Luminescent HepG2 Cells. Int. J. Mol. Sci. 2021, 22, 2843. https://doi.org/10.3390/ijms22062843
Iwado S, Abe S, Oshimura M, Kazuki Y, Nakajima Y. Bioluminescence Measurement of Time-Dependent Dynamic Changes of CYP-Mediated Cytotoxicity in CYP-Expressing Luminescent HepG2 Cells. International Journal of Molecular Sciences. 2021; 22(6):2843. https://doi.org/10.3390/ijms22062843
Chicago/Turabian StyleIwado, Satoru, Satoshi Abe, Mitsuo Oshimura, Yasuhiro Kazuki, and Yoshihiro Nakajima. 2021. "Bioluminescence Measurement of Time-Dependent Dynamic Changes of CYP-Mediated Cytotoxicity in CYP-Expressing Luminescent HepG2 Cells" International Journal of Molecular Sciences 22, no. 6: 2843. https://doi.org/10.3390/ijms22062843
APA StyleIwado, S., Abe, S., Oshimura, M., Kazuki, Y., & Nakajima, Y. (2021). Bioluminescence Measurement of Time-Dependent Dynamic Changes of CYP-Mediated Cytotoxicity in CYP-Expressing Luminescent HepG2 Cells. International Journal of Molecular Sciences, 22(6), 2843. https://doi.org/10.3390/ijms22062843





