Unusually Fast bis-Histidyl Coordination in a Plant Hemoglobin
Abstract
1. Introduction
2. Results and Discussion
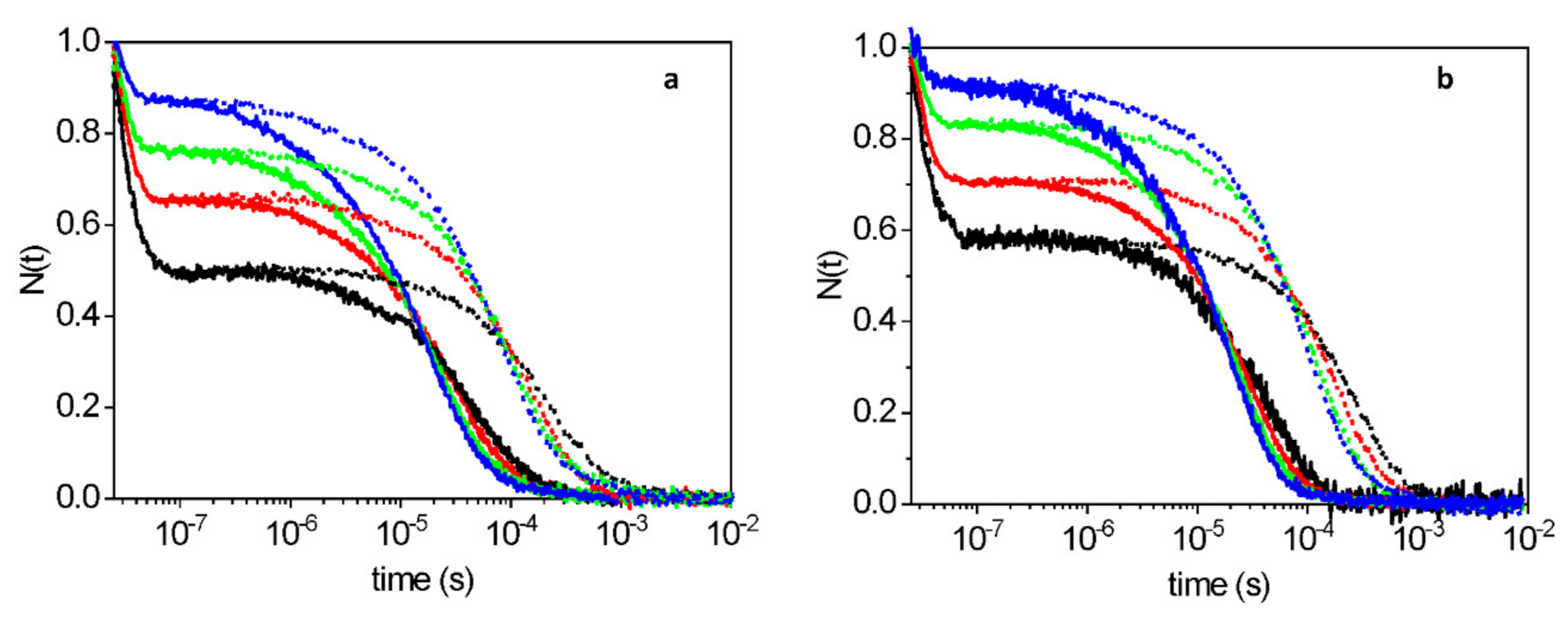
2.1. Unusually Fast CO Rebinding Kinetics to WT1 and WT4 after Nanosecond Laser Photolysis
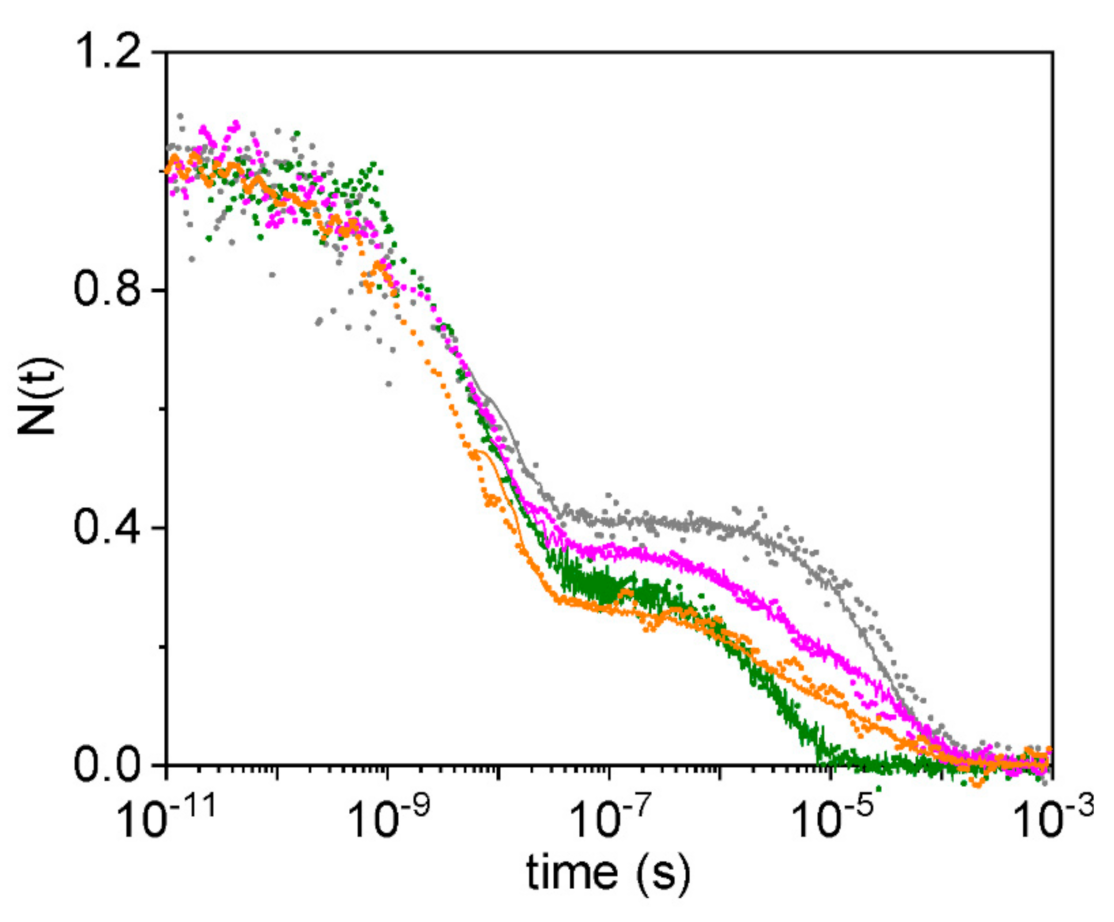
2.2. Experiments with Subnanosecond Resolution Reveal a Large Geminate Rebinding
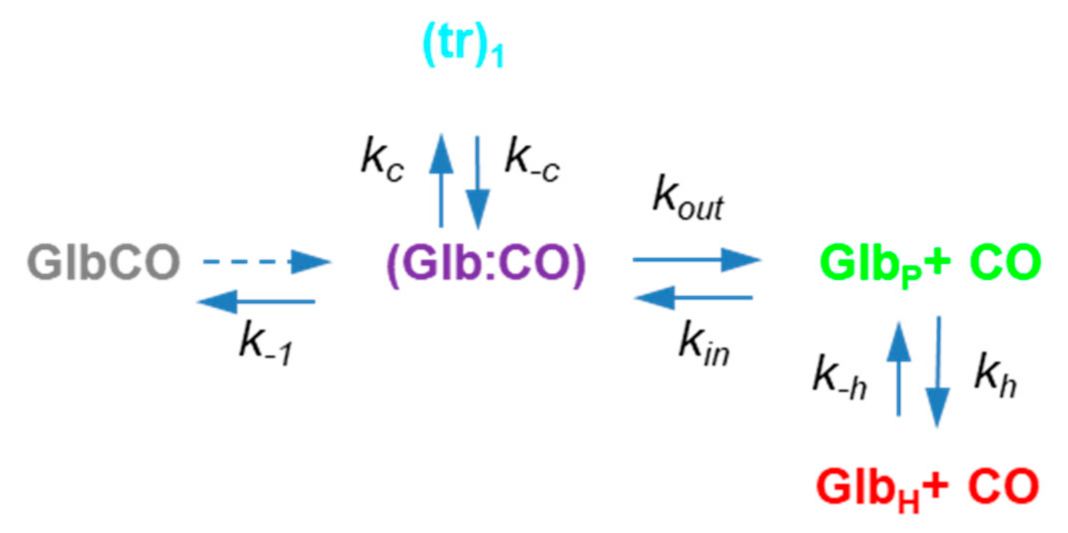
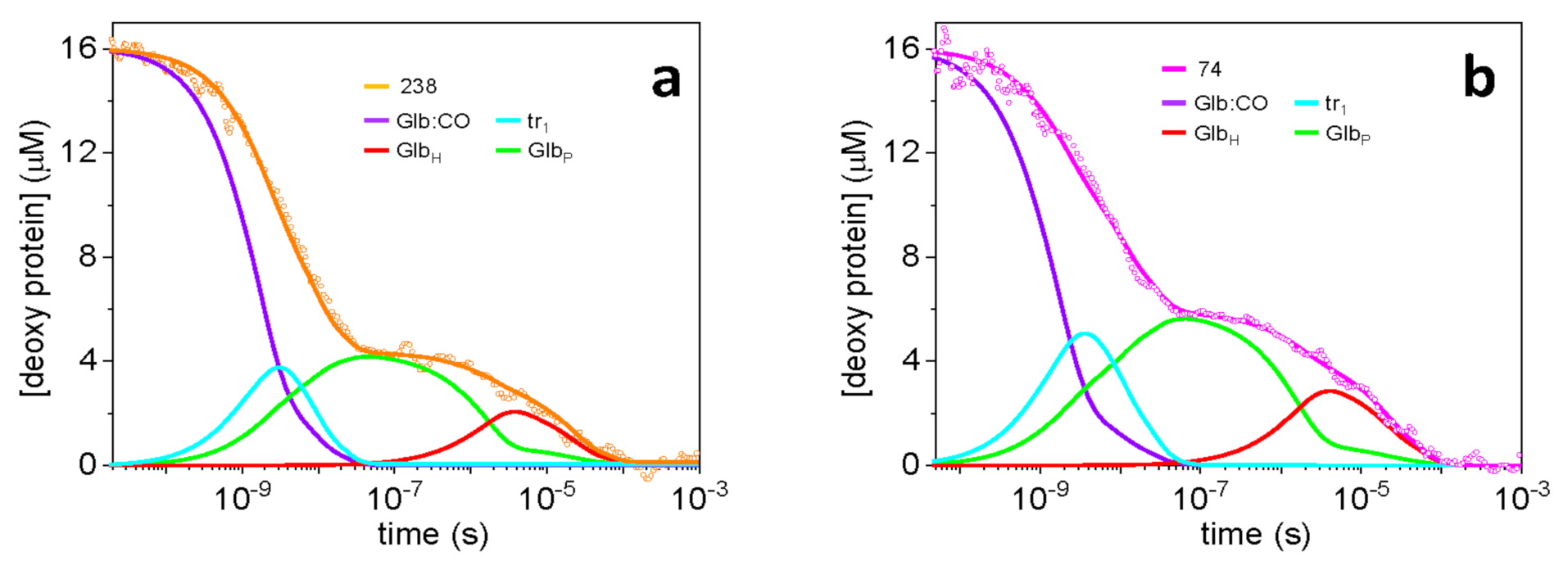
2.3. Description of the Rebinding Kinetics with a Microscopic Model
2.4. Rate Constants for Hisd Binding and Dissociation Are Extraordinarily High
2.5. Dissecting the Contribution of the Hisd in Each Domain of MtGlb1-2.1
2.6. Thermal Activation of Kinetic Processes
3. Conclusions
4. Materials and Methods
4.1. Protein Expression and Purification
4.2. Nanosecond Laser Flash Photolysis of CO Rebinding Kinetics
4.3. Ultrafast Spectroscopy of CO Rebinding Kinetics
4.4. Data Analyses
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| Glb | Phytoglobin (or nonsymbiotic hemoglobin) |
| Hisd | Distal histidine |
| LFP | Laser flash photolysis |
| NO | Nitric oxide |
| O2 | Molecular oxygen |
| CO | Carbon monoxide |
| WT | Wild type |
References
- Appleby, C.A. The origin and functions of hemoglobin in plants. Sci. Prog. 1992, 76, 365–398. [Google Scholar]
- Garrocho-Villegas, V.; Gopalasubramaniam, S.K.; Arredondo-Peter, R. Plant hemoglobins: What we know six decades after their discovery. Gene 2007, 398, 78–85. [Google Scholar] [CrossRef]
- Dordas, C. Nonsymbiotic hemoglobins and stress tolerance in plants. Plant Sci. 2009, 176, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Jokipii-Lukkari, S.; Frey, A.D.; Kallio, P.T.; Häggman, H. Intrinsic non-symbiotic and truncated haemoglobins and heterologous Vitreoscilla haemoglobin expression in plants. J. Exp. Bot. 2009, 60, 409–422. [Google Scholar] [CrossRef] [PubMed]
- Kakar, S.; Hoffman, F.G.; Storz, J.F.; Fabian, M.; Hargrove, M.S. Structure and reactivity of hexacoordinate hemoglobins. Biophys. Chem. 2010, 152, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Hill, R.D. Non-symbiotic haemoglobins—What’s happening beyond nitric oxide scavenging? AoB Plants 2012, 2012, pls004. [Google Scholar] [CrossRef] [PubMed]
- Becana, M.; Yruela, I.; Sarath, G.; Catalán, P.; Hargrove, M.S. Plant hemoglobins: A journey from unicellular green algae to vascular plants. New Phytol. 2020, 227, 1618–1635. [Google Scholar] [CrossRef] [PubMed]
- Wittenberg, J.B.; Bolognesi, M.; Wittenberg, B.A.; Guertin, M. Truncated hemoglobins: A new family of hemoglobins widely distributed in bacteria, unicellular eukaryotes, and plants. J. Biol. Chem. 2002, 277, 871–874. [Google Scholar] [CrossRef]
- Vinogradov, S.N.; Hoogewijs, D.; Bailly, X.; Arredondo-Peter, R.; Guertin, M.; Gough, J.; Dewilde, S.; Moens, L.; Vanfleteren, J.R. Three globin lineages belonging to two structural classes in genomes from the three kingdoms of life. Proc. Natl. Acad. Sci. USA 2005, 102, 11385–11389. [Google Scholar] [CrossRef] [PubMed]
- Smagghe, B.J.; Hoy, J.A.; Percifield, R.; Kundu, S.; Hargrove, M.S.; Sarath, G.; Hilbert, J.L.; Watts, R.A.; Dennis, E.S.; Peacock, W.J.; et al. Correlations between oxygen affinity and sequence classifications of plant hemoglobins. Biopolymers 2009, 91, 1083–1096. [Google Scholar] [CrossRef]
- Bruno, S.; Faggiano, S.; Spyrakis, F.; Mozzarelli, A.; Cacciatori, E.; Dominici, P.; Grandi, E.; Abbruzzetti, S.; Viappiani, C. Different roles of protein dynamics and ligand migration in non-symbiotic hemoglobins AHb1 and AHb2 from Arabidopsis thaliana. Gene 2007, 398, 224–233. [Google Scholar] [CrossRef]
- Bruno, S.; Faggiano, S.; Spyrakis, F.; Mozzarelli, A.; Abbruzzetti, S.; Grandi, E.; Viappiani, C.; Feis, A.; Mackowiak, S.; Smulevich, G.; et al. The reactivity with CO of AHb1 and AHb2 from Arabidopsis thaliana is controlled by the distal His E7 and internal hydrophobic cavities. J. Am. Chem. Soc. 2007, 129, 2880–2889. [Google Scholar] [CrossRef]
- Spyrakis, F.; Luque, F.J.; Viappiani, C. Structural analysis in nonsymbiotic hemoglobins: What can we learn from inner cavities? Plant Sci. 2011, 181, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Abbruzzetti, S.; Spyrakis, F.; Bidon-Chanal, A.; Luque, F.J.; Viappiani, C. Ligand migration through hemeprotein cavities: Insights from laser flash photolysis and molecular dynamics simulations. Phys. Chem. Chem. Phys. 2013, 15, 10686–10701. [Google Scholar] [CrossRef] [PubMed]
- Watts, R.A.; Hunt, P.W.; Hvitved, A.N.; Hargrove, M.S.; Peacock, W.J.; Dennis, E.S. A hemoglobin from plants homologous to truncated hemoglobins of microorganisms. Proc. Natl. Acad. Sci. USA 2001, 98, 10119–10124. [Google Scholar] [CrossRef]
- Wang, Y.H.; Kochian, L.V.; Doyle, J.J.; Garvin, D.F. Two tomato non-symbiotic haemoglobin genes are differentially expressed in response to diverse changes in mineral nutrient status. Plant Cell Environ. 2003, 26, 673–680. [Google Scholar] [CrossRef]
- Shimoda, Y.; Nagata, M.; Suzuki, A.; Abe, M.; Sato, S.; Kato, T.; Tabata, S.; Higashi, S.; Uchiumi, T. Symbiotic rhizobium and nitric oxide induce gene expression of non-symbiotic hemoglobin in Lotus japonicus. Plant Cell Physiol. 2005, 46, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Bustos-Sanmamed, P.; Tovar-Méndez, A.; Crespi, M.; Sato, S.; Tabata, S.; Becana, M. Regulation of nonsymbiotic and truncated hemoglobin genes of Lotus japonicus in plant organs and in response to nitric oxide and hormones. New Phytol. 2011, 189, 765–776. [Google Scholar] [CrossRef]
- Mira, M.M.; Hill, R.D.; Stasolla, C. Phytoglobins improve hypoxic root growth by alleviating apical meristem cell death. Plant Physiol. 2016, 172, 2044–2056. [Google Scholar] [CrossRef]
- Berger, A.; Guinand, S.; Boscari, A.; Puppo, A.; Brouquisse, R. Medicago truncatula phytoglobin 1.1 controls symbiotic nodulation and nitrogen fixation via the regulation of nitric oxide concentration. New Phytol. 2020, 227, 84–98. [Google Scholar] [CrossRef]
- Smagghe, B.J.; Trent, J.T., 3rd; Hargrove, M.S. NO dioxygenase activity in hemoglobins is ubiquitous in vitro, but limited by reduction in vivo. PLoS ONE 2008, 3, e2039. [Google Scholar] [CrossRef] [PubMed]
- Rubio, M.C.; Calvo-Begueria, L.; Díaz-Mendoza, M.; Elhiti, M.; Moore, M.; Matamoros, M.A.; James, E.K.; Díaz, I.; Pérez-Rontomé, C.; Villar, I.; et al. Phytoglobins in the nuclei, cytoplasm and chloroplasts modulate nitric oxide signaling and interact with abscisic acid. Plant J. 2019, 100, 38–54. [Google Scholar] [CrossRef] [PubMed]
- Trevaskis, B.; Watts, R.A.; Andersson, C.R.; Llewellyn, D.J.; Hargrove, M.S.; Olson, J.S.; Dennis, E.S.; Peacock, W.J. Two hemoglobin genes in Arabidopsis thaliana: The evolutionary origins of leghemoglobins. Proc. Natl. Acad. Sci. USA 1997, 94, 12230–12234. [Google Scholar] [CrossRef]
- Hunt, P.W.; Watts, R.A.; Trevaskis, B.; Llewelyn, D.J.; Burnell, J.; Dennis, E.S.; Peacock, W.J. Expression and evolution of functionally distinct haemoglobin genes in plants. Plant Mol. Biol. 2001, 47, 677–692. [Google Scholar] [CrossRef]
- Hebelstrup, K.H.; Shah, J.K.; Igamberdiev, A.U. The role of nitric oxide and hemoglobin in plant development and morphogenesis. Physiol. Plant. 2013, 148, 457–469. [Google Scholar] [CrossRef] [PubMed]
- Vieweg, M.F.; Hohnjec, N.; Küster, H. Two genes encoding different truncated hemoglobins are regulated during root nodule and arbuscular mycorrhiza symbioses of Medicago truncatula. Planta 2005, 220, 757–766. [Google Scholar] [CrossRef] [PubMed]
- Larrainzar, E.; Villar, I.; Rubio, M.C.; Pérez-Rontomé, C.; Huertas, R.; Sato, S.; Mun, J.-H.; Becana, M. Hemoglobins in the legume–Rhizobium symbiosis. New Phytol. 2020, 228, 472–484. [Google Scholar] [CrossRef]
- Villar, I.; Larrainzar, E.; Milazzo, L.; Pérez-Rontomé, C.; Rubio, M.C.; Smulevich, G.; Martínez, J.I.; Wilson, M.T.; Reeder, B.; Huertas, R.; et al. A plant gene encoding one-heme and two-heme hemoglobins with extreme reactivities toward diatomic gases and nitrite. Front. Plant Sci. 2020, 11, 1743. [Google Scholar] [CrossRef]
- Kriegl, J.M.; Bhattacharyya, A.J.; Nienhaus, K.; Deng, P.; Minkow, O.; Nienhaus, G.U. Ligand binding and protein dynamics in neuroglobin. Proc. Natl. Acad. Sci. USA 2002, 99, 7992–7997. [Google Scholar] [CrossRef]
- Abbruzzetti, S.; Bruno, S.; Faggiano, S.; Grandi, E.; Mozzarelli, A.; Viappiani, C. Time-resolved methods in Biophysics. 2. Monitoring haem proteins at work with nanosecond laser flash photolysis. Photochem. Photobiol. Sci. 2006, 5, 1109–1120. [Google Scholar] [CrossRef] [PubMed]
- Bisht, N.K.; Abbruzzetti, S.; Uppal, S.; Bruno, S.; Spyrakis, F.; Mozzarelli, A.; Viappiani, C.; Kundu, S. Ligand migration and hexacoordination in type 1 non-symbiotic rice hemoglobin. Biochim. Biophys. Acta-Proteins Proteom. 2011, 1814, 1042–1053. [Google Scholar] [CrossRef] [PubMed]
- Marcelli, A.; Abbruzzetti, S.; Bustamante, J.P.; Feis, A.; Bonamore, A.; Boffi, A.; Gellini, C.; Salvi, P.R.; Estrin, D.A.; Bruno, S.; et al. Following ligand migration pathways from picoseconds to milliseconds in type II truncated hemoglobin from Thermobifida fusca. PLoS ONE 2012, 7, e39884. [Google Scholar]
- Bustamante, J.P.; Abbruzzetti, S.; Marcelli, A.; Gauto, D.; Boechi, L.; Bonamore, A.; Boffi, A.; Bruno, S.; Feis, A.; Foggi, P.; et al. Ligand uptake modulation by internal water molecules and hydrophobic cavities in hemoglobins. J. Phys. Chem. B 2014, 118, 1234–1245. [Google Scholar] [CrossRef]
- Knipp, M.; Ogata, H.; Soavi, G.; Cerullo, G.; Allegri, A.; Abbruzzetti, S.; Bruno, S.; Viappiani, C.; Bidon-Chanal, A.; Luque, F.J. Structure and dynamics of the membrane attaching nitric oxide transporter nitrophorin 7. F100 Research 2015, 4, 45. [Google Scholar] [CrossRef]
- Benabbas, A.; Ye, X.; Kubo, M.; Zhang, Z.; Maes, E.M.; Montfort, W.R.; Champion, P.M. Ultrafast dynamics of diatomic ligand binding to nitrophorin 4. J. Am. Chem. Soc. 2010, 132, 2811–2820. [Google Scholar] [CrossRef]
- Yu, A.; Ye, X.; Ionascu, D.; Cao, W.; Champion, P.M. Two-color pump-probe laser spectroscopy instrument with picosecond time-resolved electronic delay and extended scan range. Rev. Sci. Instrum. 2005, 76, 114301. [Google Scholar] [CrossRef]
- Boubeta, F.M.; Boechi, L.; Estrin, D.; Patrizi, B.; Di Donato, M.; Iagatti, A.; Giordano, D.; Verde, C.; Bruno, S.; Abbruzzetti, S.; et al. Cold-adaptation signatures in the ligand rebinding kinetics to the truncated hemoglobin of the antarctic bacterium Pseudoalteromonas haloplanktis TAC125. J. Phys. Chem. B 2018, 122, 11649–11661. [Google Scholar] [CrossRef]
- Giordano, D.; Boubeta, F.; Prisco, G.D.; Estrin, D.; Smulevich, G.; Viappiani, C.; Verde, C. Conformational flexibility drives cold adaptation in Pseudoalteromonas haloplanktis TAC125 globins. Antiox. Redox Signal. 2020, 32, 396–411. [Google Scholar] [CrossRef] [PubMed]
- Abbruzzetti, S.; Allegri, A.; Bidon-Chanal, A.; Ogata, H.; Soavi, G.; Cerullo, G.; Bruno, S.; Montali, C.; Luque, F.J.; Viappiani, C. Electrostatic tuning of the ligand binding mechanism by Glu27 in nitrophorin 7. Sci. Rep. 2018, 8, 10855. [Google Scholar] [CrossRef]
- Sottini, S.; Abbruzzetti, S.; Viappiani, C.; Ronda, L.; Mozzarelli, A. Determination of microscopic rate constants for CO binding and migration in myoglobin encapsulated in silica gels. J. Phys. Chem. B 2005, 109, 19523–19528. [Google Scholar] [CrossRef]
- Abbruzzetti, S.; Faggiano, S.; Bruno, S.; Spyrakis, F.; Mozzarelli, A.; Dewilde, S.; Moens, L.; Viappiani, C. Ligand migration through the internal hydrophobic cavities in human neuroglobin. Proc. Natl. Acad. Sci. USA 2009, 106, 18984–18989. [Google Scholar] [CrossRef]
- Uzan, J.; Dewilde, S.; Burmester, T.; Hankeln, T.; Moens, L.; Hamdane, D.; Marden, M.C.; Kiger, L. Neuroglobin and other hexacoordinated hemoglobins show a weak temperature dependence of oxygen binding. Biophys. J. 2004, 87, 1196–1204. [Google Scholar] [CrossRef]
- Jasaitis, A.; Ouellet, H.; Lambry, J.C.; Martin, J.L.; Friedman, J.M.; Guertin, M.; Vos, M.H. Ultrafast heme-ligand recombination in truncated hemoglobin HbO from Mycobacterium tuberculosis: A ligand cage. Chem. Phys. 2012, 396, 10–16. [Google Scholar] [CrossRef]
- Faggiano, S.; Abbruzzetti, S.; Spyrakis, F.; Grandi, E.; Viappiani, C.; Bruno, S.; Mozzarelli, A.; Cozzini, P.; Astegno, A.; Dominici, P.; et al. Structural plasticity and functional implications of internal cavities in distal mutants of type 1 non-symbiotic hemoglobin AHb1 from Arabidopsis thaliana. J. Phys. Chem. B 2009, 113, 16028–16038. [Google Scholar] [CrossRef] [PubMed]
- Anselmi, M.; DiNola, A.; Amadei, A. Kinetics of carbon monoxide migration and binding in solvated neuroglobin as revealed by molecular dynamics simulations and quantum mechanical calculations. J. Phys. Chem. B 2011, 115, 2436–2446. [Google Scholar] [CrossRef] [PubMed]
- Gabba, M.; Abbruzzetti, S.; Bruno, F.S.S.; Mozzarelli, A.; Luque, F.J.; Viappiani, C.; Cozzini, P.; Nardini, M.; Germani, F.; Bolognesi, M.; et al. CO rebinding kinetics and molecular dynamics simulations highlight dynamic regulation of internal cavities in human cytoglobin. PLoS ONE 2013, 8, e49770. [Google Scholar] [CrossRef] [PubMed]
- Spyrakis, F.; Lucas, F.; Bidon-Chanal, A.; Viappiani, C.; Guallar, V.; Luque, F.J. Comparative analysis of inner cavities and ligand migration in non-symbiotic AHb1 and AHb2. Biochim. Biophys. Acta-Proteins Proteom. 2013, 1834, 1957–1967. [Google Scholar] [CrossRef]
- Brunori, M.; Giuffrè, A.; Nienhaus, K.; Nienhaus, G.U.; Scandurra, F.M.; Vallone, B. Neuroglobin, nitric oxide, and oxygen: Functional pathways and conformational changes. Proc. Natl. Acad. Sci. USA 2005, 102, 8483–8488. [Google Scholar] [CrossRef]
- Dewilde, S.; Ebner, B.; Vinck, E.; Gilany, K.; Hankeln, T.; Burmester, T.; Kreiling, J.; Reinisch, C.; Vanfleteren, J.R.; Kiger, L.; et al. The nerve hemoglobin of the bivalve mollusc Spisula solidissima: Molecular cloning, ligand binding studies, and phylogenetic analysis. J. Biol. Chem. 2006, 281, 5364–5372. [Google Scholar] [CrossRef]
- Manzoni, C.; Polli, D.; Cerullo, G. Two-color pump-probe system broadly tunable over the visible and the near infrared with sub-30 fs temporal resolution. Rev. Sci. Instrum. 2006, 77, 023103. [Google Scholar] [CrossRef]
- Barker, A.J.; Chen, K.; Hodgkiss, J.M. Distance distributions of photogenerated charge pairs in organic photovoltaic cells. J. Am. Chem. Soc. 2014, 136, 12018–12026. [Google Scholar] [CrossRef]

| Rate Constants | Units | MtGlb1-2.1 | MtGlb1-2.4 | ||||
|---|---|---|---|---|---|---|---|
| WT1 | 74 | 238 | 74/238 | WT4 | 109 | ||
| k−1 | ×108 s−1 | 2.0 ± 0.4 | 1.9 ± 0.4 | 2.6 ± 0.5 | 1.6 ± 0.3 | 14 ± 3 | 23 ± 4 |
| kh | ×105 s−1 | 8 ± 1 | 3.8 ± 0.7 | 4.2 ± 0.8 | - | 7 ± 1 | - |
| k−h | ×105 s−1 | 3.8 ± 0.7 | 1.3 ± 0.3 | 1.6 ± 0.3 | - | 3.4 ± 0.7 | - |
| kout | ×108 s−1 | 1.4 ± 0.3 | 1.1 ± 0.2 | 0.9 ± 0.2 | 0.7 ± 0.1 | 5 ± 1 | 2.1 ± 0.4 |
| kin | ×108 M−1s−1 | 1.4 ± 0.3 | 3.7 ± 0.7 | 3.4 ± 0.7 | 4.8 ± 0.9 | 1.0 ± 0.2 | 6.5 ± 0.9 |
| kc | ×108 s−1 | 2.3 ± 0.4 | 2.9 ± 0.6 | 2.5 ± 0.5 | 2.3 ± 0.4 | 6.8 ± 0.9 | 3.3 ± 0.7 |
| k−c | ×108 s−1 | 1.2 ± 0.2 | 1.8 ± 0.4 | 2.7 ± 0.5 | 2.5 ± 0.5 | 0.8 ± 0.2 | 1.7 ± 0.3 |
| kon | ×108 M−1s−1 | 1.0 ± 0.2 | 1.5 ± 0.3 | 2.0 ± 0.4 | 1.4 ± 0.3 | 2.4 ± 0.4 | 17 ± 3 |
| KH | 2.2 ± 0.4 | 2.9 ± 0.6 | 2.5 ± 0.5 | - | 2.0 ± 0.4 | - | |
| kobs | ×104 s−1 | 2.6 ± 0.5 | 2.7 ± 0.5 | 3.7 ± 0.7 | - | 6 ± 1 | - |
| Rate Constants | MtGlb1-2.1 | MtGlb1-2.4 | ||||
|---|---|---|---|---|---|---|
| WT1 | 74 | 238 | 74/238 | WT4 | 109 | |
| kout | 10.7 ± 0.4 | 8.2 ± 0.2 | 7.0 ± 0.5 | 6.1 ± 0.2 | 11 ± 1.0 | 4.1 ± 10.7 |
| kin | 13.6 ± 0.9 | 11.7 ± 0.1 | 11 ± 1.0 | 7.8 ± 0.3 | 14 ± 2.0 | - |
| kh | 1.1 ± 0.2 | 2.7 ± 1.4 | 4.8 ± 0.5 | - | 1.4 ± 0.4 | - |
| k−h | 5.1 ± 0.2 | 4.3 ± 0.6 | 3.4 ± 0.8 | - | 2.6 ± 0.2 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abbruzzetti, S.; Barker, A.J.; Villar, I.; Pérez-Rontomé, C.; Bruno, S.; Cerullo, G.; Viappiani, C.; Becana, M. Unusually Fast bis-Histidyl Coordination in a Plant Hemoglobin. Int. J. Mol. Sci. 2021, 22, 2740. https://doi.org/10.3390/ijms22052740
Abbruzzetti S, Barker AJ, Villar I, Pérez-Rontomé C, Bruno S, Cerullo G, Viappiani C, Becana M. Unusually Fast bis-Histidyl Coordination in a Plant Hemoglobin. International Journal of Molecular Sciences. 2021; 22(5):2740. https://doi.org/10.3390/ijms22052740
Chicago/Turabian StyleAbbruzzetti, Stefania, Alex J. Barker, Irene Villar, Carmen Pérez-Rontomé, Stefano Bruno, Giulio Cerullo, Cristiano Viappiani, and Manuel Becana. 2021. "Unusually Fast bis-Histidyl Coordination in a Plant Hemoglobin" International Journal of Molecular Sciences 22, no. 5: 2740. https://doi.org/10.3390/ijms22052740
APA StyleAbbruzzetti, S., Barker, A. J., Villar, I., Pérez-Rontomé, C., Bruno, S., Cerullo, G., Viappiani, C., & Becana, M. (2021). Unusually Fast bis-Histidyl Coordination in a Plant Hemoglobin. International Journal of Molecular Sciences, 22(5), 2740. https://doi.org/10.3390/ijms22052740










