Drug Discovery in Liver Disease Using Kinome Profiling
Abstract
1. Introduction
2. Viral Infection
3. Steatosis-Related Hepatitis
4. Liver Cancer
5. Need for Kinome Profiling Approaches in Liver Disease
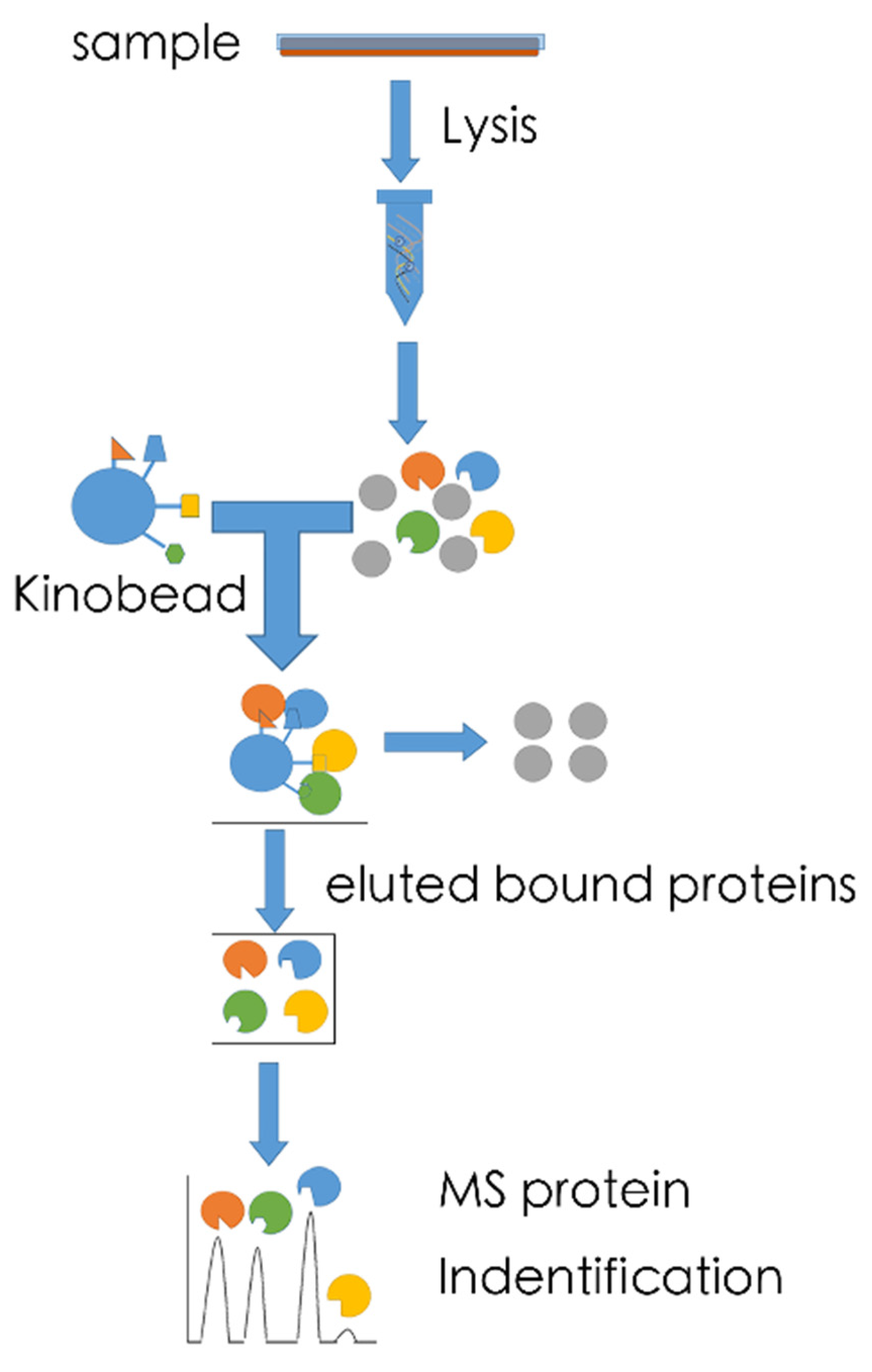
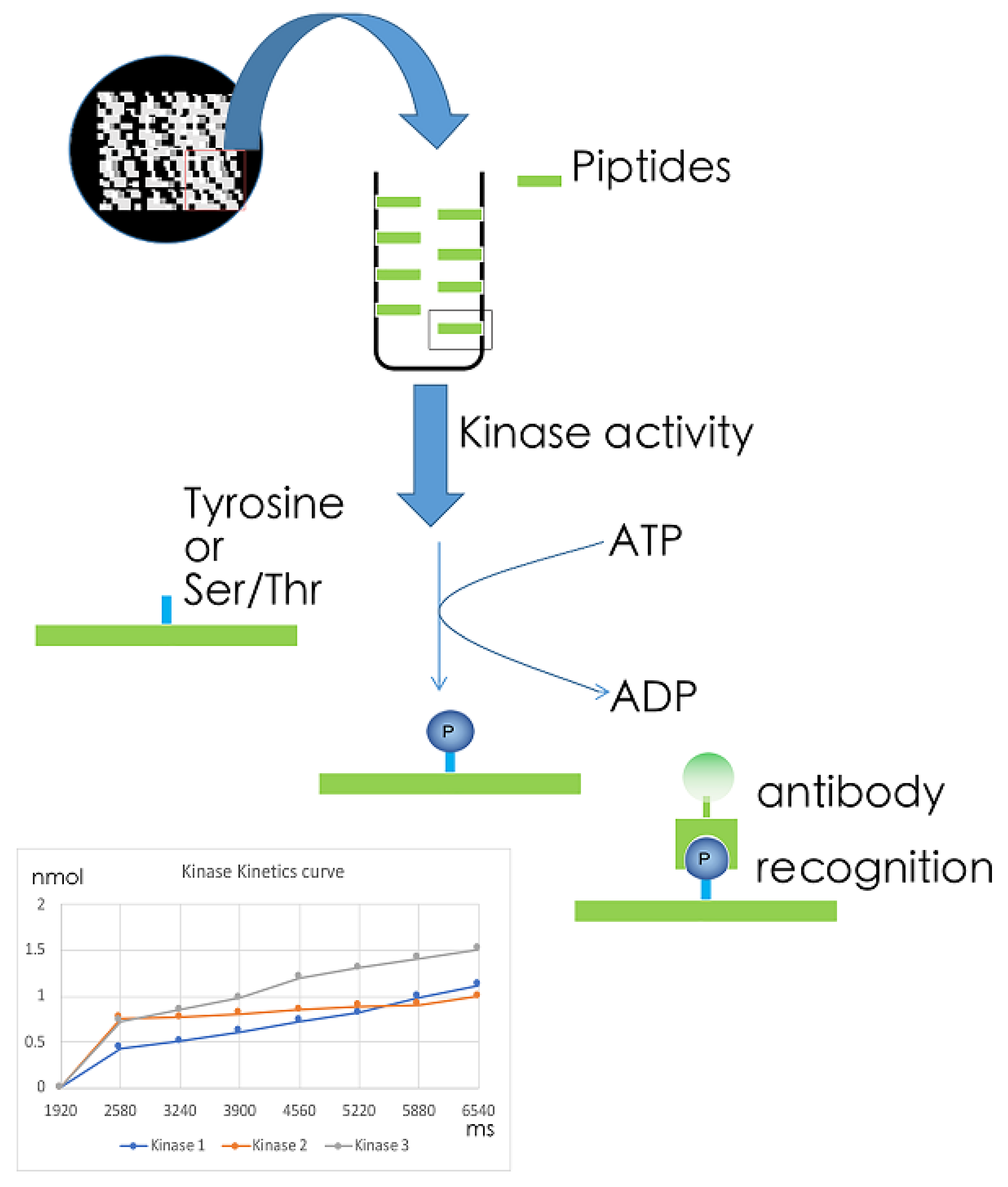
5.1. MS-Based Kinome Profiling
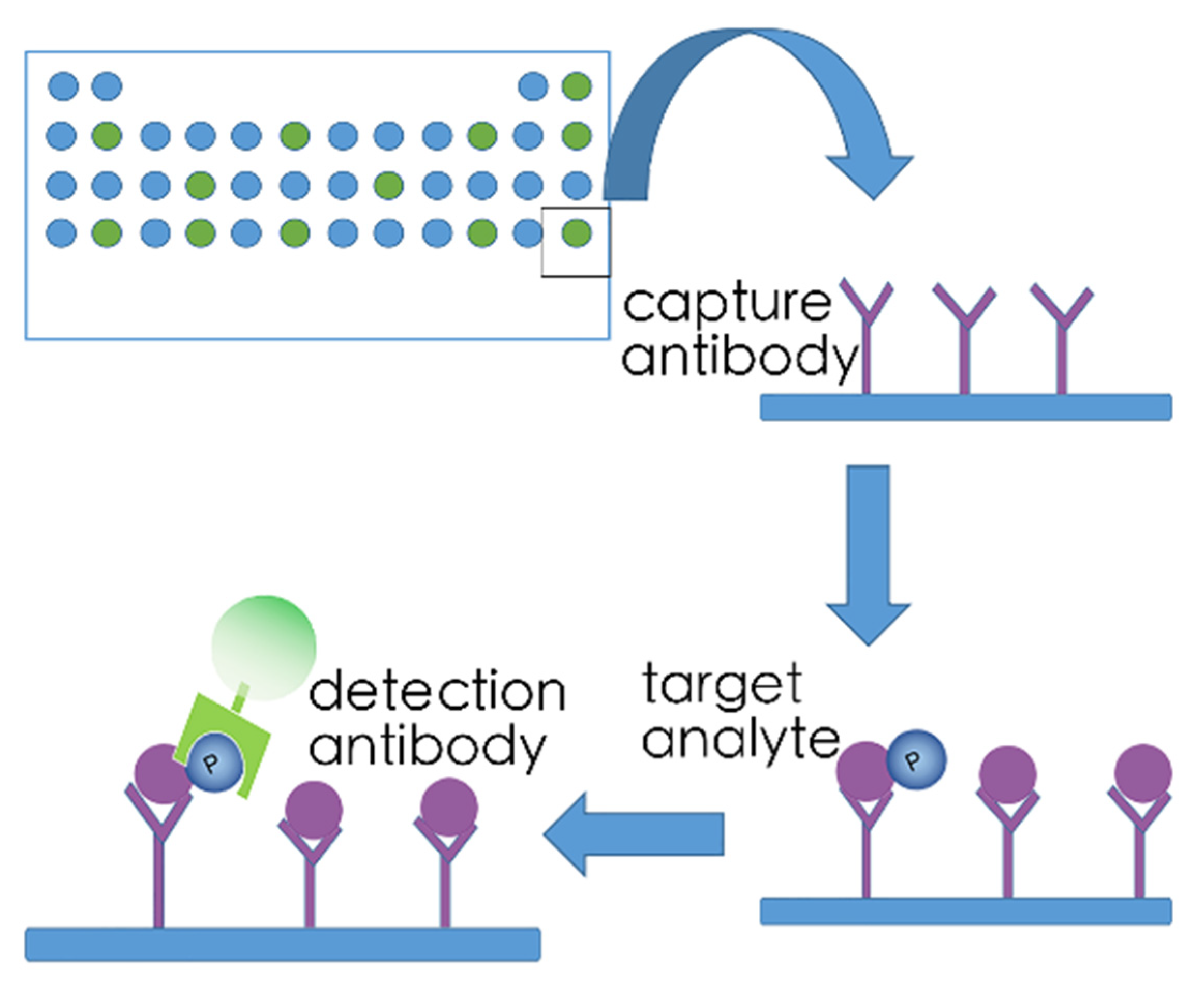
5.2. Array-Based Kinome Profiling
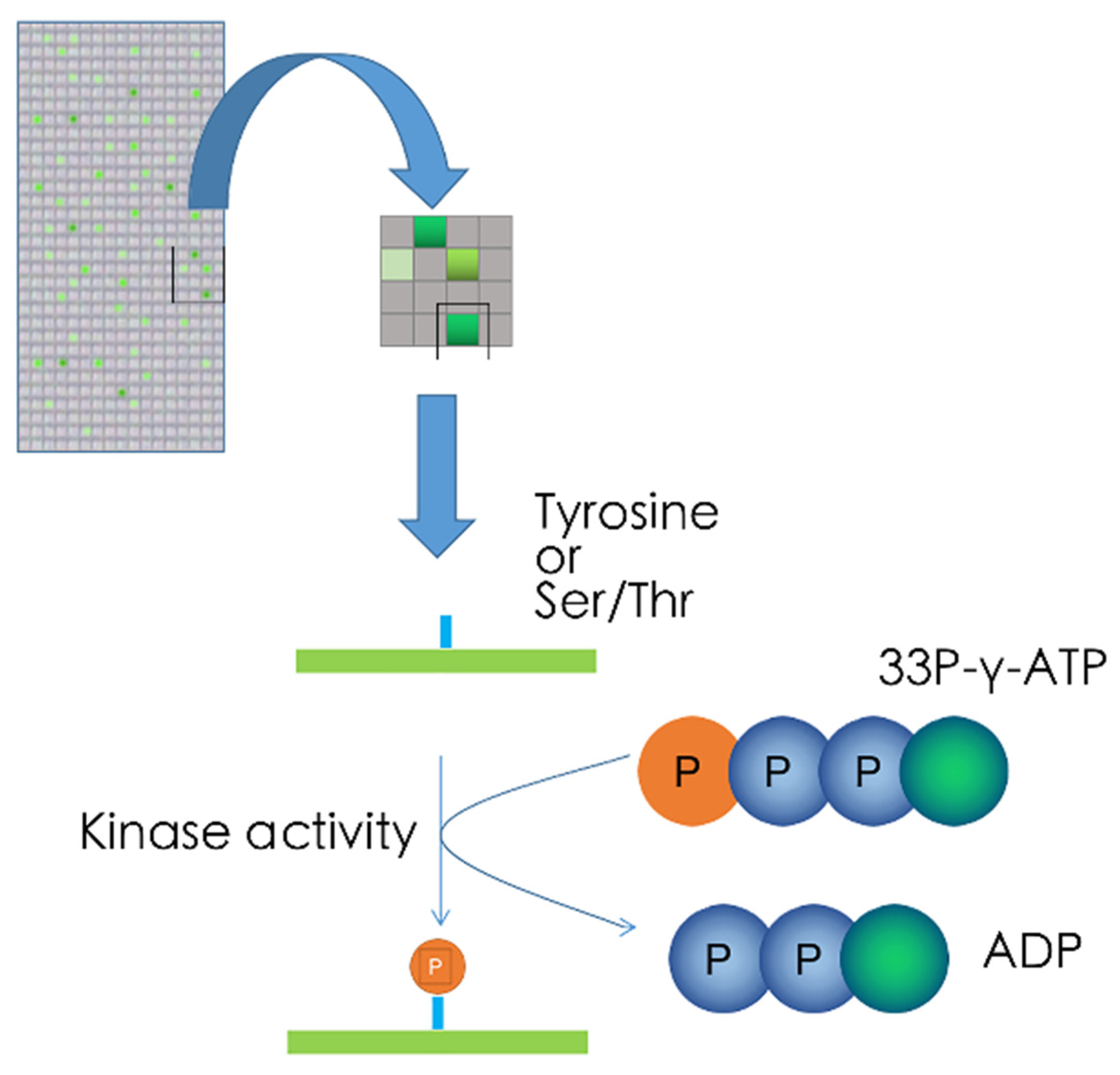
5.3. Kinase Target Sequence Arrays
5.4. Kinetic Kinase Target Sequence Arrays
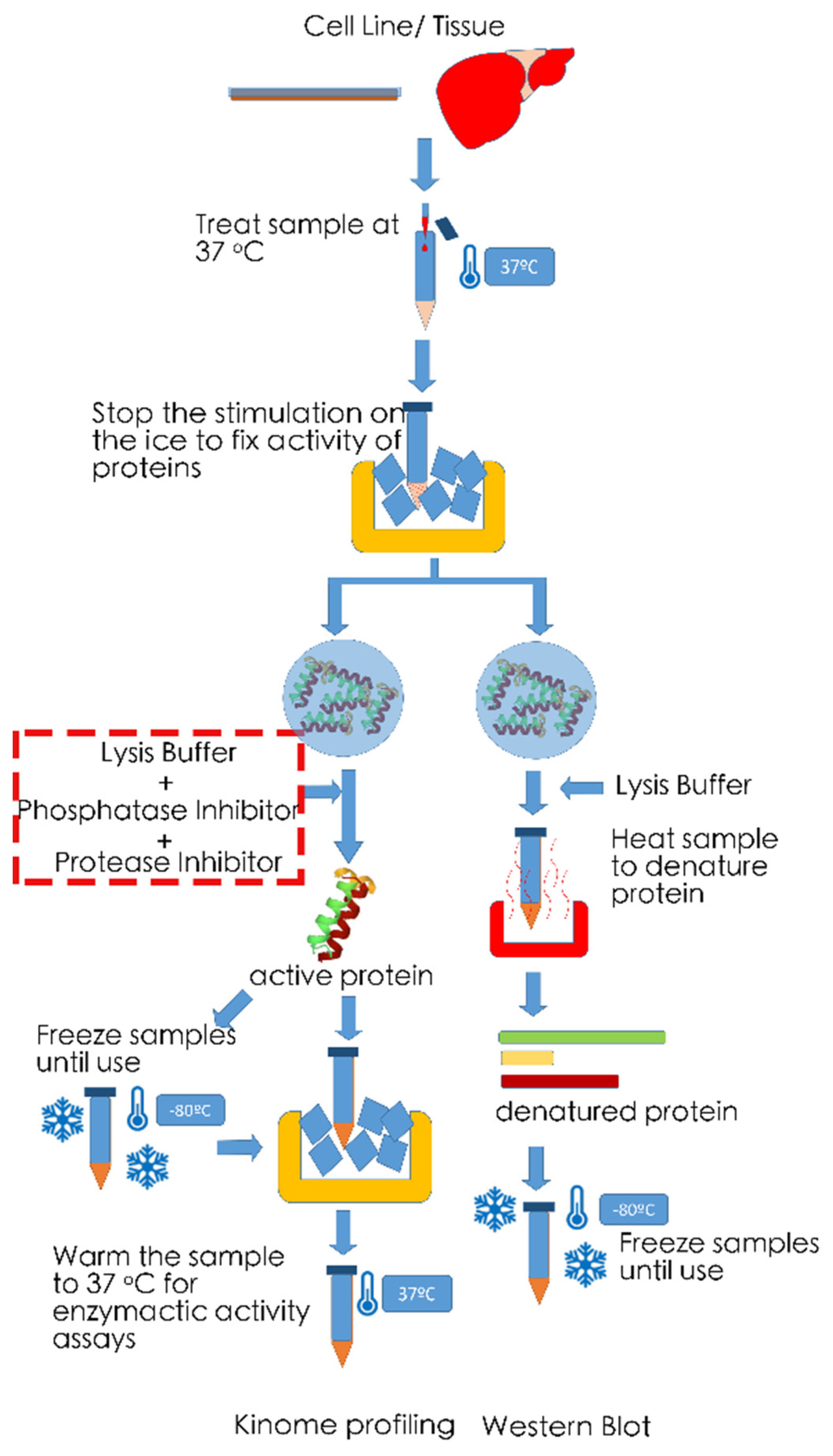
6. Technical Noted on Measurements of Phosphorylation Levels
7. Future Perspectives on the Application of Kinome Profiling in Liver Disease
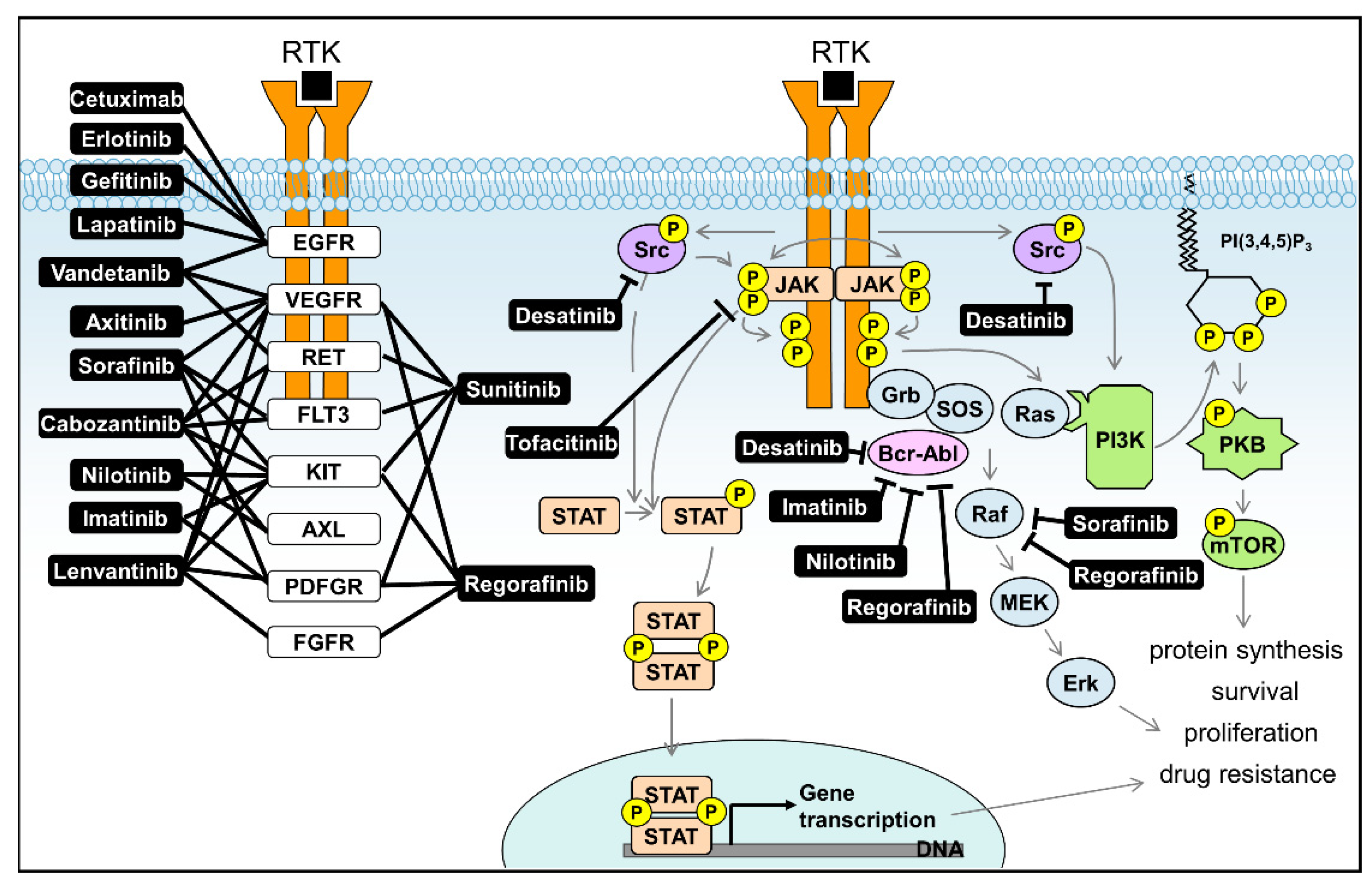
| Inhibitor | Target | Cellular Signalling | Type | FDA Approved | Studied in Liver Disease |
|---|---|---|---|---|---|
| Axitinib | VEGFR | Receptor tyrosine kinases inhibitor | Small Molecule | Approved for RCC | Phase II completed for HCC [108] |
| Cetuximab | EGFR | Receptor tyrosine kinases inhibitor | Monoclonal antibody | Approved for CRC | Phase II completed for HCC [109] |
| Cabozantinib | VEGFR/KIT/TRKB/FLT-3/AXL/RET/MET/TIE-2 | tyrosine kinase inhibitor | Small Molecule | Approved for MTC | Phase III Active Not Recruiting for HCC [110] |
| Dasatinib | BCR-ABL/Src kinase family | tyrosine kinase inhibitor | Small Molecule | Approved for CML | Phase II Terminated for HCC [111] |
| Erlotinib | EGFR tyrosine kinase | Receptor tyrosine kinases inhibitor | Small Molecule | Approved for NSCLC | Phase III completed for HCC [112,113] |
| Gefitinib | EGFR tyrosine kinase | Receptor tyrosine kinases inhibitor | Small Molecule | Approved for NSCLC | Phase II completed for HCC [114] |
| Imatinib | Bcr-Abl/KIT/PDGFR | tyrosine kinase inhibitor | Small Molecule | Approved for CML | Preclinical models for liver fibrosis [115] |
| Lapatinib | EGFR/HER2 | Receptor tyrosine kinases inhibitor | Small Molecule | Approved for Breast Cancer | Phase II completed for HCC [116] |
| Lenvatinib | VEGFR/FGFR/PDGFRα/KIT/RET | Receptor tyrosine kinases inhibitor | Small Molecule | Approved for HCC | Phase III enrolling for HCC [117] |
| Nilotinib | BCR-ABL | tyrosine kinase inhibitor | Small Molecule | Approved for CML | Preclinical models for liver fibrosis [118] |
| Sorafenib | CRAF/BRAF/KIT/FLT-3/VEGFR-2/VEGFR-3/PDGFR-ß | Raf kinase/Receptor tyrosine kinase inhibitor | Small Molecule | Approved for HCC | Phase III enrolling for HCC [119,120] |
| Sunitinib | PDGFRa/PDGFRb/VEGFR1/VEGFR2/VEGFR3/KIT/FLT3/CSF-1R/RET | Receptor tyrosine kinase inhibitor | Small Molecule | Approved for RCC and GIST | Phase III Terminated for HCC [119] |
| Vandetanib | VEGFR/EGFR/RET | Receptor tyrosine kinase inhibitor | Small Molecule | Approved for MTC | Phase II completed for HCC [121] |
| Regorafenib | RET/VEGFR1/VEGFR2/VEGFR3/KIT/PDGFR-alpha/PDGFR-beta/FGFR1/FGFR2/TIE2/DDR2/TrkA/Eph2A/RAF-1/BRAF/BRAFV600E /SAPK2/PTK5/BCR-ABL | Raf kinase/tyrosine kinase inhibitor | Small Molecule | Approved for CRC, GIST and HCC | Phase III completed for HCC [120] |
| Tofacitinib | JAK1/JAK2/JAK3/TYK2 | Janus kinases inhibitor | Small Molecule | Approved for severe rheumatoid arthritis | Phase I completed for Hepatic Insufficiency [122] |
7.1. Alcoholic Liver Disease
7.2. Liver Cancer
7.3. Hepatitis E
7.4. Wilson’s Disease
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Hall, J.E. Pocket Companion to Guyton & Hall Textbook of Medical Physiology E-Book; Elsevier Health Sciences: Amsterdam, The Netherlands, 2015. [Google Scholar]
- Thompson, F.M.; Ferguson, J.W.; Kelly, D.A.; Hirschfield, G.M. Liver disease in the young adult: The challenges and rewards. Lancet Gastroenterol. Hepatol. 2019, 4, 248–254. [Google Scholar] [CrossRef]
- Chene, P. Challenges in design of biochemical assays for the identification of small molecules to target multiple conformations of protein kinases. Drug Discov. Today 2008, 13, 522–529. [Google Scholar] [CrossRef]
- Cohen, P. The origins of protein phosphorylation. Nat. Cell Biol. 2002, 4, E127–E130. [Google Scholar] [CrossRef] [PubMed]
- Cabral, L.K.D.; Tiribelli, C.; Sukowati, C.H.C. Sorafenib resistance in hepatocellular carcinoma: The relevance of genetic heterogeneity. Cancers 2020, 12, 1576. [Google Scholar] [CrossRef] [PubMed]
- Hernanda, P.Y.; Pedroza-Gonzalez, A.; Sprengers, D.; Peppelenbosch, M.P.; Pan, Q. Multipotent mesenchymal stromal cells in liver cancer: Implications for tumor biology and therapy. Biochim. Biophys. Acta Rev. Cancer 2014, 1846, 439–445. [Google Scholar] [CrossRef]
- Parikh, K.; Peppelenbosch, M.P.; Ritsema, T. Kinome profiling using peptide arrays in eukaryotic cells. In Phospho-Proteomics; Springer: Berlin/Heidelberg, Germany, 2009; pp. 269–280. [Google Scholar]
- Stanaway, J.D.; Flaxman, A.D.; Naghavi, M.; Fitzmaurice, C.; Vos, T.; Abubakar, I.; Abu-Raddad, L.J.; Assadi, R.; Bhala, N.; Cowie, B.; et al. The global burden of viral hepatitis from 1990 to 2013: Findings from the Global Burden of Disease Study 2013. Lancet 2016, 388, 1081–1088. [Google Scholar] [CrossRef]
- World Health Organization. Global Hepatitis Report 2017; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- Miao, Z.; Zhang, S.; Ou, X.; Li, S.; Ma, Z.; Wang, W.; Peppelenbosch, M.P.; Liu, J.; Pan, Q. Estimating the Global Prevalence, Disease Progression, and Clinical Outcome of Hepatitis Delta Virus Infection. J. Infect. Dis. 2020, 221, 1677–1687. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Liu, J.; Li, Y.; Su, J.; Ma, Z.; Bramer, W.M.; Cao, W.; de Man, R.A.; Peppelenbosch, M.P.; Pan, Q. The global epidemiology of hepatitis E virus infection: A systematic review and meta-analysis. Liver Int. 2020. [Google Scholar] [CrossRef] [PubMed]
- Hoogeveen, R.C.; Boonstra, A. Checkpoint Inhibitors and Therapeutic Vaccines for the Treatment of Chronic HBV Infection. Front. Immunol. 2020, 11, 401. [Google Scholar] [CrossRef]
- Van der Meer, A.J.; Berenguer, M. Reversion of disease manifestations after HCV eradication. J. Hepatol. 2016, 65, S95–S108. [Google Scholar] [CrossRef] [PubMed]
- Debes, J.D.; De Knegt, R.J.; Boonstra, A. The path to cancer and back: Immune modulation during hepatitis C virus infection, progression to fibrosis and cancer, and unexpected roles of new antivirals. Transplantation 2017, 101, 910–915. [Google Scholar] [CrossRef]
- Wang, W.; Xu, L.; Su, J.; Peppelenbosch, M.P.; Pan, Q. Transcriptional regulation of antiviral interferon-stimulated genes. Trends Microbiol. 2017, 25, 573–584. [Google Scholar] [CrossRef]
- Ma, B.; Chen, K.; Liu, P.; Li, M.; Liu, J.; Sideras, K.; Sprengers, D.; Biermann, K.; Wang, W.; Ijzermans, J.N.M. Dichotomal functions of phosphorylated and unphosphorylated STAT1 in hepatocellular carcinoma. J. Mol. Med. 2019, 97, 77–88. [Google Scholar] [CrossRef]
- Wang, W.; Yin, Y.; Xu, L.; Su, J.; Huang, F.; Wang, Y.; Boor, P.P.C.; Chen, K.; Wang, W.; Cao, W. Unphosphorylated ISGF3 drives constitutive expression of interferon-stimulated genes to protect against viral infections. Sci. Signal. 2017, 10. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Xu, L.; Su, J.; Peppelenbosch, M.P.; Pan, Q. Biological or pharmacological activation of protein kinase C alpha constrains hepatitis E virus replication. Antivir. Res. 2017, 140, 1–12. [Google Scholar] [CrossRef]
- Qu, C.; Li, Y.; Li, Y.; Yu, P.; Li, P.; Donkers, J.M.; van de Graaf, S.F.J.; Robert, A.; Peppelenbosch, M.P.; Pan, Q. FDA-drug screening identifies deptropine inhibiting hepatitis E virus involving the NF-κB-RIPK1-caspase axis. Antivir. Res. 2019, 170, 104588. [Google Scholar] [CrossRef]
- Wang, W.; Xu, L.; Brandsma, J.H.; Wang, Y.; Hakim, M.S.; Zhou, X.; Yin, Y.; Fuhler, G.M.; Van Der Laan, L.J.W.; Van Der Woude, C.J. Convergent transcription of interferon-stimulated genes by TNF-α and IFN-α augments antiviral activity against HCV and HEV. Sci. Rep. 2016, 6, 25482. [Google Scholar] [CrossRef]
- Dauber, B.; Wolff, T. Activation of the Antiviral Kinase PKR and Viral Countermeasures. Viruses 2009, 1, 523–544. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Han, M.; Liu, S.; Yuan, X.; Zhao, J.; Lu, H.; Han, K.; Liang, P.; Cheng, J. S6K1 inhibits HBV replication through inhibiting AMPK-ULK1 pathway and disrupting acetylation modification of H3K27. Life Sci. 2021, 265, 118848. [Google Scholar] [CrossRef] [PubMed]
- Kurniawan, D.W.; Storm, G.; Prakash, J.; Bansal, R. Role of spleen tyrosine kinase in liver diseases. World J. Gastroenterol. 2020, 26, 1005. [Google Scholar] [CrossRef]
- Cheng, Y.; Sun, F.; Wang, L.; Gao, M.; Xie, Y.; Sun, Y.; Liu, H.; Yuan, Y.; Yi, W.; Huang, Z.; et al. Virus-induced p38 MAPK activation facilitates viral infection. Theranostics 2020, 10, 12223–12240. [Google Scholar] [CrossRef]
- Arvind, A.; Osganian, S.A.; Cohen, D.E.; Corey, K.E. Lipid and Lipoprotein Metabolism in Liver Disease, in Endotext [Internet]. Available online: https://www.endotext.org/chapter/lipid-and-lipoprotein-metabolism-in-liver-disease (accessed on 21 July 2019).
- Alferink, L.J.M.; Kiefte-de Jong, J.C.; Darwish Murad, S. Potential mechanisms underlying the role of coffee in liver health. Semin. Liver Dis. 2018, 38, 193–214. [Google Scholar] [CrossRef] [PubMed]
- Mahady, S.E.; George, J. Predicting the future burden of NAFLD and NASH. J. Hepatol. 2018, 69, 774–775. [Google Scholar] [CrossRef]
- Qureshi, K.; Neuschwander-Tetri, B.A. The molecular basis for current targets of NASH therapies. Expert Opin. Investig. Drugs 2020, 29, 151–161. [Google Scholar] [CrossRef]
- Reibe, S.; Febbraio, M.A. Relieving ER stress to target NASH-driven hepatocellular carcinoma. Nat. Rev. Endocrinol. 2019, 15, 73–74. [Google Scholar] [CrossRef]
- Wan, X.; Xu, C.; Yu, C.; Li, Y. Role of NLRP3 inflammasome in the progression of NAFLD to NASH. Can. J. Gastroenterol. Hepatol. 2016, 2016. [Google Scholar] [CrossRef]
- Sanderson, T.H.; Gallaway, M.; Kumar, R. Unfolding the unfolded protein response: Unique insights into brain ischemia. Int. J. Mol. Sci. 2015, 16, 7133–7142. [Google Scholar] [CrossRef] [PubMed]
- Gardner, B.M.; Pincus, D.; Gotthardt, K.; Gallagher, C.M.; Walter, P. Endoplasmic reticulum stress sensing in the unfolded protein response. Cold Spring Harb. Perspect. Biol. 2013, 5, a013169. [Google Scholar] [CrossRef]
- Urano, F.; Wang, X.; Bertolotti, A.; Zhang, Y.; Chung, P.; Harding, H.P.; Ron, D. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science 2000, 287, 664–666. [Google Scholar] [CrossRef] [PubMed]
- Song, M.J.; Malhi, H. The unfolded protein response and hepatic lipid metabolism in non alcoholic fatty liver disease. Pharmacol. Ther. 2019, 203, 107401. [Google Scholar] [CrossRef]
- Van der Giessen, J.; van der Woude, C.J.; Peppelenbosch, M.P.; Fuhler, G.M. A direct effect of sex hormones on epithelial barrier function in inflammatory bowel disease models. Cells 2019, 8, 261. [Google Scholar] [CrossRef] [PubMed]
- Grootjans, J.; Krupka, N.; Hosomi, S.; Matute, J.D.; Hanley, T.; Saveljeva, S.; Gensollen, T.; Heijmans, J.; Li, H.; Limenitakis, J.P. Epithelial endoplasmic reticulum stress orchestrates a protective IgA response. Science 2019, 363, 993–998. [Google Scholar] [CrossRef]
- Lie, M.R.K.L.; van der Giessen, J.; Fuhler, G.M.; de Lima, A.; Peppelenbosch, M.P.; van der Ent, C.; van der Woude, C.J. Low dose Naltrexone for induction of remission in inflammatory bowel disease patients. J. Transl. Med. 2018, 16, 55. [Google Scholar] [CrossRef] [PubMed]
- Tschurtschenthaler, M.; Adolph, T.E.; Ashcroft, J.W.; Niederreiter, L.; Bharti, R.; Saveljeva, S.; Bhattacharyya, J.; Flak, M.B.; Shih, D.Q.; Fuhler, G.M. Defective ATG16L1-mediated removal of IRE1α drives Crohn’s disease–like ileitis. J. Exp. Med. 2017, 214, 401–422. [Google Scholar] [CrossRef]
- Abdullah, A.; Ravanan, P. The unknown face of IRE1α–Beyond ER stress. Eur. J. Cell Biol. 2018, 97, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Szabo, G.; Csak, T. Inflammasomes in liver diseases. J. Hepatol. 2012, 57, 642–654. [Google Scholar] [CrossRef] [PubMed]
- de Sant’Ana, L.P.; Ribeiro, D.J.S.; Martins, A.M.A.; Dos Santos, F.N.; Corrêa, R.; Almeida, R.D.N.; Eberlin, M.N.; Maurice, C.F.; Magalhães, K.G. Absence of the Caspases 1/11 Modulates Liver Global Lipid Profile and Gut Microbiota in High-Fat-Diet-Induced Obese Mice. Front. Immunol. 2019, 10, 2926. [Google Scholar] [CrossRef]
- Dixon, L.J.; Flask, C.A.; Papouchado, B.G.; Feldstein, A.E.; Nagy, L.E. Caspase-1 as a central regulator of high fat diet-induced non-alcoholic steatohepatitis. PLoS ONE 2013, 8, e56100. [Google Scholar] [CrossRef] [PubMed]
- Bukong, T.N.; Iracheta-Vellve, A.; Saha, B.; Ambade, A.; Satishchandran, A.; Gyongyosi, B.; Lowe, P.; Catalano, D.; Kodys, K.; Szabo, G. Inhibition of spleen tyrosine kinase activation ameliorates inflammation, cell death, and steatosis in alcoholic liver disease. Hepatology 2016, 64, 1057–1071. [Google Scholar] [CrossRef]
- Weber, A.N.R.; Bittner, Z.A.; Shankar, S.; Liu, X.; Chang, T.-H.; Jin, T.; Tapia-Abellán, A. Recent insights into the regulatory networks of NLRP3 inflammasome activation. J. Cell Sci. 2020, 133. [Google Scholar] [CrossRef]
- Sharafi, H.; Alavian, S.M. The rising Threat of Hepatocellular Carcinoma in the Middle east and north africa region: Results From Global Burden of Disease study 2017. Clin. Liver Dis. 2019, 14, 219. [Google Scholar] [CrossRef] [PubMed]
- Reinders, M.T.M.; van Meer, S.; Burgmans, M.C.; de Jong, K.P.; Klümpen, H.-J.; Robert, A.; Ramsoekh, D.S.; Sprengers, D.; Tjwa, E.T.T.L.; de Vos-Geelen, J. Trends in incidence, diagnosis, treatment and survival of hepatocellular carcinoma in a low-incidence country: Data from the Netherlands in the period 2009–2016. Eur. J. Cancer 2020, 137, 214–223. [Google Scholar] [CrossRef]
- Sideras, K.; Robert, A.; Harrington, S.M.; Polak, W.G.; Zhou, G.; Schutz, H.M.; Pedroza-Gonzalez, A.; Biermann, K.; Mancham, S.; Hansen, B.E. Circulating levels of PD-L1 and Galectin-9 are associated with patient survival in surgically treated Hepatocellular Carcinoma independent of their intra-tumoral expression levels. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef]
- van Beek, A.A.; Zhou, G.; Doukas, M.; Boor, P.P.C.; Noordam, L.; Mancham, S.; Campos Carrascosa, L.; van der Heide-Mulder, M.; Polak, W.G.; Ijzermans, J.N.M. GITR ligation enhances functionality of tumor-infiltrating T cells in hepatocellular carcinoma. Int. J. Cancer 2019, 145, 1111–1124. [Google Scholar] [CrossRef] [PubMed]
- Sideras, K.; Biermann, K.; Verheij, J.; Takkenberg, B.R.; Mancham, S.; Hansen, B.E.; Schutz, H.M.; de Man, R.A.; Sprengers, D.; Buschow, S.I. PD-L1, Galectin-9 and CD8+ tumor-infiltrating lymphocytes are associated with survival in hepatocellular carcinoma. Oncoimmunology 2017, 6, e1273309. [Google Scholar] [CrossRef]
- Zhou, G.; Sprengers, D.; Boor, P.P.C.; Doukas, M.; Schutz, H.; Mancham, S.; Pedroza-Gonzalez, A.; Polak, W.G.; De Jonge, J.; Gaspersz, M. Antibodies against immune checkpoint molecules restore functions of tumor-infiltrating T cells in hepatocellular carcinomas. Gastroenterology 2017, 153, 1107–1119.e10. [Google Scholar] [CrossRef]
- Dhanasekaran, R.; Nault, J.-C.; Roberts, L.R.; Zucman-Rossi, J. Genomic Medicine and Implications for Hepatocellular Carcinoma Prevention and Therapy. Gastroenterology 2019, 156, 492–509. [Google Scholar] [CrossRef] [PubMed]
- Gravitz, L. Liver cancer. Nature 2014, 516, S1. [Google Scholar] [CrossRef]
- Liu, P.; Liang, B.; Liu, M.; Lebbink, J.H.G.; Li, S.; Qian, M.; Lavrijsen, M.; Peppelenbosch, M.P.; Chen, X.; Smits, R. Oncogenic Mutations in Armadillo Repeats 5 and 6 of β-Catenin Reduce Binding to APC, Increasing Signaling and Transcription of Target Genes. Gastroenterology 2020, 158, 1029–1043.e10. [Google Scholar] [CrossRef]
- Cao, W.; Li, M.; Liu, J.; Zhang, S.; Noordam, L.; Verstegen, M.M.A.; Wang, L.; Ma, B.; Li, S.; Wang, W. LGR5 marks targetable tumor-initiating cells in mouse liver cancer. Nat. Commun. 2020, 11, 1–16. [Google Scholar] [CrossRef]
- Chung, B.K.; Karlsen, T.H. Genetic Discoveries Highlight Environmental Factors as Key Drivers of Liver Disease. Dig. Dis. 2017, 35, 323–333. [Google Scholar] [CrossRef]
- Liu, J.; Li, P.; Wang, L.; Li, M.; Ge, Z.; Noordam, L.; Lieshout, R.; Verstegen, M.M.A.; Ma, B.; Su, J. Cancer-associated fibroblasts provide a stromal niche for liver cancer organoids that confers trophic effects and therapy resistance. Cell. Mol. Gastroenterol. Hepatol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Demory, A.; Nault, J.C. Molecular perspectives for the treatment of hepatocellular carcinoma. Acta Gastroenterol. Belg. 2020, 83, 309–312. [Google Scholar]
- Okuma, H.S.; Kondo, S. Trends in the development of MET inhibitors for hepatocellular carcinoma. Future Oncol. 2016, 12, 1275–1286. [Google Scholar] [CrossRef]
- Granito, A.; Guidetti, E.; Gramantieri, L. c-MET receptor tyrosine kinase as a molecular target in advanced hepatocellular carcinoma. J. Hepatocell Carcinoma 2015, 2, 29–38. [Google Scholar] [PubMed]
- Huynh, H.; Ong, R.W.; Li, P.Y.; Lee, S.S.; Yang, S.; Chong, L.W.; Luu, D.A.; Jong, C.T.; Lam, I.W. Targeting receptor tyrosine kinase pathways in hepatocellular carcinoma. Anticancer Agents Med. Chem. 2011, 11, 560–575. [Google Scholar] [CrossRef] [PubMed]
- Panera, N.; Crudele, A.; Romito, I.; Gnani, D.; Alisi, A. Focal Adhesion Kinase: Insight into Molecular Roles and Functions in Hepatocellular Carcinoma. Int. J. Mol. Sci. 2017, 18, 99. [Google Scholar] [CrossRef] [PubMed]
- Heinrich, S.; Craig, A.J.; Ma, L.; Heinrich, B.; Greten, T.F.; Wang, X.W. Understanding tumor cell heterogeneity and its implication for immunotherapy in liver cancer by single cell analysis. J. Hepatol. 2020. [Google Scholar] [CrossRef]
- Wang, C.; Vegna, S.; Jin, H.; Benedict, B.; Lieftink, C.; Ramirez, C.; de Oliveira, R.L.; Morris, B.; Gadiot, J.; Wang, W. Inducing and exploiting vulnerabilities for the treatment of liver cancer. Nature 2019, 574, 268–272. [Google Scholar] [CrossRef]
- Jiang, Y.; Sun, A.; Zhao, Y.; Ying, W.; Sun, H.; Yang, X.; Xing, B.; Sun, W.; Ren, L.; Hu, B. Proteomics identifies new therapeutic targets of early-stage hepatocellular carcinoma. Nature 2019, 567, 257–261. [Google Scholar] [CrossRef]
- Schmidlin, T.; Debets, D.O.; van Gelder, C.A.G.H.; Stecker, K.E.; Rontogianni, S.; van den Eshof, B.L.; Kemper, K.; Lips, E.H.; van den Biggelaar, M.; Peeper, D.S. High-throughput assessment of kinome-wide activation states. Cell Syst. 2019, 9, 366–374.e5. [Google Scholar] [CrossRef]
- Dillon, L.M.; Miller, T.W. Therapeutic targeting of cancers with loss of PTEN function. Curr. Drug Targets 2014, 15, 65–79. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, W.L.; Zhang, Y.; Guo, S.P.; Zhang, J.; Li, Q.L. Epigenetic and genetic alterations of PTEN in hepatocellular carcinoma. Hepatol. Res. 2007, 37, 389–396. [Google Scholar] [CrossRef] [PubMed]
- Ruela-de-Sousa, R.R.; Queiroz, K.C.; Peppelenbosch, M.P.; Fuhler, G.M. Reversible phosphorylation in haematological malignancies: Potential role for protein tyrosine phosphatases in treatment? Biochim. Biophys. Acta 2010, 1806, 287–303. [Google Scholar] [CrossRef] [PubMed]
- Hoekstra, E.; Peppelenbosch, M.P.; Fuhler, G.M. The role of protein tyrosine phosphatases in colorectal cancer. Biochim. Biophys. Acta 2012, 1826, 179–188. [Google Scholar] [CrossRef]
- Hoekstra, E.; Das, A.M.; Swets, M.; Cao, W.; van der Woude, C.J.; Bruno, M.J.; Peppelenbosch, M.P.; Kuppen, P.J.K.; Ten Hagen, T.L.M.; Fuhler, G.M. Increased PTP1B expression and phosphatase activity in colorectal cancer results in a more invasive phenotype and worse patient outcome. Oncotarget 2016, 7, 21922. [Google Scholar] [CrossRef]
- Alho, I.; Costa, L.; Bicho, M.; Coelho, C. Low Molecular Weight Protein Tyrosine Phosphatase Slow Isoform Knockdown in MDA-MB-435 Cells Decreases RAW 264.7 Osteoclastic Differentiation. Anticancer Res. 2016, 36, 2227–2232. [Google Scholar]
- Song, G.J.; Jung, M.; Kim, J.H.; Park, H.; Rahman, M.H.; Zhang, S.; Zhang, Z.Y.; Park, D.H.; Kook, H.; Lee, I.K.; et al. A novel role for protein tyrosine phosphatase 1B as a positive regulator of neuroinflammation. J. Neuroinflam. 2016, 13, 86. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.; Wu, Y.; Wang, T.; Zhang, Y.; Kong, D.; Zhang, L.; Li, X.; Wang, G.; Jin, Y.; Jin, X.; et al. Elevated Src expression associated with hepatocellular carcinoma metastasis in northern Chinese patients. Oncol. Lett. 2015, 10, 3026–3034. [Google Scholar] [CrossRef] [PubMed]
- Walker, S.; Wankell, M.; Ho, V.; White, R.; Deo, N.; Devine, C.; Dewdney, B.; Bhathal, P.; Govaere, O.; Roskams, T.; et al. Targeting mTOR and Src restricts hepatocellular carcinoma growth in a novel murine liver cancer model. PLoS ONE 2019, 14, e0212860. [Google Scholar] [CrossRef] [PubMed]
- He, R.J.; Yu, Z.H.; Zhang, R.Y.; Zhang, Z.Y. Protein tyrosine phosphatases as potential therapeutic targets. Acta Pharm. Sin. 2014, 35, 1227–1246. [Google Scholar] [CrossRef]
- Hoekstra, E.; Peppelenbosch, M.P.; Fuhler, G.M. Meeting Report Europhosphatase 2015: Phosphatases as Drug Targets in Cancer. Cancer Res. 2016, 76, 193–196. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Robinson, D.R.; Wu, Y.-M.; Lin, S.-F. The protein tyrosine kinase family of the human genome. Oncogene 2000, 19, 5548–5557. [Google Scholar] [CrossRef]
- Alonso, A.; Sasin, J.; Bottini, N.; Friedberg, I.; Friedberg, I.; Osterman, A.; Godzik, A.; Hunter, T.; Dixon, J.; Mustelin, T. Protein tyrosine phosphatases in the human genome. Cell 2004, 117, 699–711. [Google Scholar] [CrossRef] [PubMed]
- Subramani, S.; Jayapalan, S.; Kalpana, R.; Natarajan, J. HomoKinase: A curated database of human protein kinases. Int. Sch. Res. Not. 2013, 2013. [Google Scholar] [CrossRef]
- Bartsch, L.M.; Damasio, M.P.S.; Subudhi, S.; Drescher, H.K. Tissue-Resident Memory T Cells in the Liver—Unique Characteristics of Local Specialists. Cells 2020, 9, 2457. [Google Scholar] [CrossRef] [PubMed]
- Obi, S.; Sato, T.; Sato, S. Immune checkpoint inhibitor in liver cancer—Unique regional differences. Ann. Transl. Med. 2020, 8. [Google Scholar] [CrossRef] [PubMed]
- van Baal, J.W.P.M.; Diks, S.H.; Wanders, R.J.A.; Rygiel, A.M.; Milano, F.; Joore, J.; Bergman, J.J.; Peppelenbosch, M.P.; Krishnadath, K.K. Comparison of kinome profiles of Barrett’s esophagus with normal squamous esophagus and normal gastric cardia. Cancer Res. 2006, 66, 11605–11612. [Google Scholar] [CrossRef]
- Grove, J.R.; Banerjee, P.; Balasubramanyam, A.; Coffer, P.J.; Price, D.J.; Avruch, J.; Woodgett, J.R. Cloning and expression of two human p70 S6 kinase polypeptides differing only at their amino termini. Mol. Cell. Biol. 1991, 11, 5541–5550. [Google Scholar] [CrossRef]
- Song, C.; Ye, M.; Liu, Z.; Cheng, H.; Jiang, X.; Han, G.; Songyang, Z.; Tan, Y.; Wang, H.; Ren, J.; et al. Systematic analysis of protein phosphorylation networks from phosphoproteomic data. Mol. Cell Proteom. 2012, 11, 1070–1083. [Google Scholar] [CrossRef]
- Ochoa, D.; Jarnuczak, A.F.; Viéitez, C.; Gehre, M.; Soucheray, M.; Mateus, A.; Kleefeldt, A.A.; Hill, A.; Garcia-Alonso, L.; Stein, F.; et al. The functional landscape of the human phosphoproteome. Nat. Biotechnol. 2020, 38, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Dittus, L.; Werner, T.; Muelbaier, M.; Bantscheff, M. Differential kinobeads profiling for target identification of irreversible kinase inhibitors. ACS Chem. Biol. 2017, 12, 2515–2521. [Google Scholar] [CrossRef]
- Golkowski, M.; Lau, H.-T.; Chan, M.; Kenerson, H.; Vidadala, V.N.; Shoemaker, A.; Maly, D.J.; Yeung, R.S.; Gujral, T.S.; Ong, S.-E. Pharmacoproteomics Identifies Kinase Pathways that Drive the Epithelial-Mesenchymal Transition and Drug Resistance in Hepatocellular Carcinoma. Cell Syst. 2020, 11, 196–207. [Google Scholar] [CrossRef]
- Voorneveld, P.W.; Kodach, L.L.; Jacobs, R.J.; Liv, N.; Zonnevylle, A.C.; Hoogenboom, J.P.; Biemond, I.; Verspaget, H.W.; Hommes, D.W.; De Rooij, K. Loss of SMAD4 alters BMP signaling to promote colorectal cancer cell metastasis via activation of Rho and ROCK. Gastroenterology 2014, 147, 196–208. [Google Scholar] [CrossRef] [PubMed]
- Daub, H.; Olsen, J.V.; Bairlein, M.; Gnad, F.; Oppermann, F.S.; Körner, R.; Greff, Z.; Kéri, G.; Stemmann, O.; Mann, M. Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Mol. Cell 2008, 31, 438–448. [Google Scholar] [CrossRef]
- Penzo, M.; de Las Heras-Dueña, L.; Mata-Cantero, L.; Diaz-Hernandez, B.; Vazquez-Muñiz, M.-J.; Ghidelli-Disse, S.; Drewes, G.; Fernandez-Alvaro, E.; Baker, D.A. High-throughput screening of the Plasmodium falciparum cGMP-dependent protein kinase identified a thiazole scaffold which kills erythrocytic and sexual stage parasites. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Anjum, R.; Kubota, K.; Rush, J.; Villen, J.; Gygi, S.P. A site-specific, multiplexed kinase activity assay using stable-isotope dilution and high-resolution mass spectrometry. Proc. Natl. Acad. Sci. USA 2009, 106, 11606–11611. [Google Scholar] [CrossRef]
- Kubota, K.; Anjum, R.; Yu, Y.; Kunz, R.C.; Andersen, J.N.; Kraus, M.; Keilhack, H.; Nagashima, K.; Krauss, S.; Paweletz, C. Sensitive multiplexed analysis of kinase activities and activity-based kinase identification. Nat. Biotechnol. 2009, 27, 933–940. [Google Scholar] [CrossRef]
- Kunz, R.C.; McAllister, F.E.; Rush, J.; Gygi, S.P. A high-throughput, multiplexed kinase assay using a benchtop orbitrap mass spectrometer to investigate the effect of kinase inhibitors on kinase signaling pathways. Anal. Chem. 2012, 84, 6233–6239. [Google Scholar] [CrossRef]
- Melicharkova, K.; Neradil, J.; Mudry, P.; Zitterbart, K.; Obermannova, R.; Skotakova, J.; Veselská, R.; Štěrba, J. Profile of activation of tyrosine kinases and MAP kinases in therapy of maffucci syndrome. Klin. Onkol. Cas. Ceske Slov. Onkol. Spol. 2015, 28, 2S47-51. [Google Scholar] [CrossRef]
- Chung, S.; Dwabe, S.; Elshimali, Y.; Sukhija, H.; Aroh, C.; Vadgama, J.V. Identification of novel biomarkers for metastatic colorectal cancer using angiogenesis-antibody array and intracellular signaling array. PLoS ONE 2015, 10, e0134948. [Google Scholar] [CrossRef]
- Jalal, S.; Arsenault, R.; Potter, A.A.; Babiuk, L.A.; Griebel, P.J.; Napper, S. Genome to kinome: Species-specific peptide arrays for kinome analysis. Sci. Signal. 2009, 2, pl1. [Google Scholar] [CrossRef] [PubMed]
- Hoogendijk, A.J.; van Vught, L.A.; Wiewel, M.A.; Fuhler, G.M.; Belkasim-Bohoudi, H.; Horn, J.; Schultz, M.J.; Scicluna, B.P.; Peppelenbosch, M.P.; van’t Veer, C. Kinase activity is impaired in neutrophils of sepsis patients. Haematologica 2019, 104, e233. [Google Scholar] [CrossRef]
- Mondanelli, G.; Bianchi, R.; Pallotta, M.T.; Orabona, C.; Albini, E.; Iacono, A.; Belladonna, M.L.; Vacca, C.; Fallarino, F.; Macchiarulo, A. A relay pathway between arginine and tryptophan metabolism confers immunosuppressive properties on dendritic cells. Immunity 2017, 46, 233–244. [Google Scholar] [CrossRef] [PubMed]
- ter Haar, E.; Coll, J.T.; Austen, D.A.; Hsiao, H.M.; Swenson, L.; Jain, J. Structure of GSK3beta reveals a primed phosphorylation mechanism. Nat. Struct. Biol. 2001, 8, 593–596. [Google Scholar] [CrossRef] [PubMed]
- Peppelenbosch, M.P.; Frijns, N.; Fuhler, G. Systems medicine approaches for peptide array-based protein kinase profiling: Progress and prospects. Expert Rev. Proteom. 2016, 13, 571–578. [Google Scholar] [CrossRef]
- Lemeer, S.; Jopling, C.; Naji, F.; Ruijtenbeek, R.; Slijper, M.; Heck, A.J.R.; Den Hertog, J. Protein-tyrosine kinase activity profiling in knock down zebrafish embryos. PLoS ONE 2007, 2, e581. [Google Scholar] [CrossRef]
- Sikkema, A.H.; Diks, S.H.; den Dunnen, W.F.A.; ter Elst, A.; Scherpen, F.J.G.; Hoving, E.W.; Ruijtenbeek, R.; Boender, P.J.; de Wijn, R.; Kamps, W.A. Kinome profiling in pediatric brain tumors as a new approach for target discovery. Cancer Res. 2009, 69, 5987–5995. [Google Scholar] [CrossRef]
- Lemeer, S.; Jopling, C.; Gouw, J.; Mohammed, S.; Heck, A.J.R.; Slijper, M.; den Hertog, J. Comparative phosphoproteomics of zebrafish Fyn/Yes morpholino knockdown embryos. Mol. Cell. Proteom. 2008, 7, 2176–2187. [Google Scholar] [CrossRef]
- Baroncelli, M.; Fuhler, G.M.; Van de Peppel, J.; Zambuzzi, W.F.; van Leeuwen, J.P.; van der Eerden, B.C.J.; Peppelenbosch, M.P. Human mesenchymal stromal cells in adhesion to cell-derived extracellular matrix and titanium: Comparative kinome profile analysis. J. Cell. Physiol. 2019, 234, 2984–2996. [Google Scholar] [CrossRef]
- Kriegsmann, M.; Zgorzelski, C.; Casadonte, R.; Schwamborn, K.; Muley, T.; Winter, H.; Eichhorn, M.; Eichhorn, F.; Warth, A.; Deininger, S.O.; et al. Mass Spectrometry Imaging for Reliable and Fast Classification of Non-Small Cell Lung Cancer Subtypes. Cancers 2020, 12, 2704. [Google Scholar] [CrossRef]
- Panicker, R.C.; Chattopadhaya, S.; Coyne, A.G.; Srinivasan, R. Allosteric Small-Molecule Serine/Threonine Kinase Inhibitors. Adv. Exp. Med. Biol. 2019, 1163, 253–278. [Google Scholar]
- Roskoski, R., Jr. Properties of FDA-approved small molecule protein kinase inhibitors. Pharmacol. Res. 2019, 144, 19–50. [Google Scholar] [CrossRef]
- Kang, Y.K.; Yau, T.; Park, J.W.; Lim, H.Y.; Lee, T.Y.; Obi, S.; Chan, S.L.; Qin, S.K.; Kim, R.D.; Casey, M. Randomized phase II study of axitinib versus placebo plus best supportive care in second-line treatment of advanced hepatocellular carcinoma. Ann. Oncol. 2015, 26, 2457–2463. [Google Scholar] [CrossRef]
- Zhu, A.X.; Blaszkowsky, L.; Enzinger, P.C.; Bhargava, P.; Ryan, D.P.; Meyerhardt, J.; Horgan, K.; Hale, K.; Sheehan, S.; Stuart, K. Phase II study of cetuximab in patients with unresectable or metastatic hepatocellular carcinoma. J. Clin. Oncol. 2006, 24 (Suppl. 18), 14096. [Google Scholar] [CrossRef]
- Trojan, J. Cabozantinib for the Treatment of Advanced Hepatocellular Carcinoma: Current Data and Future Perspectives. Drugs 2020, 1–8. [Google Scholar] [CrossRef]
- Chang, A.Y.; Wang, M. Molecular mechanisms of action and potential biomarkers of growth inhibition of dasatinib (BMS-354825) on hepatocellular carcinoma cells. BMC Cancer 2013, 13, 1–12. [Google Scholar] [CrossRef]
- Deng, G.-L.; Zeng, S.; Shen, H. Chemotherapy and target therapy for hepatocellular carcinoma: New advances and challenges. World J. Hepatol. 2015, 7, 787. [Google Scholar] [CrossRef]
- Wu, C.-P.; Hung, T.-H.; Hsiao, S.-H.; Huang, Y.-H.; Hung, L.-C.; Yu, Y.-J.; Chang, Y.-T.; Wang, S.-P.; Wu, Y.-S. Erdafitinib Resensitizes ABCB1-Overexpressing Multidrug-Resistant Cancer Cells to Cytotoxic Anticancer Drugs. Cancers 2020, 12, 1366. [Google Scholar] [CrossRef]
- Roberts, L.R.; Gores, G.J. Hepatocellular carcinoma: Molecular pathways and new therapeutic targets. Semin. Liver Dis. 2005. [Google Scholar] [CrossRef]
- Yoshiji, H.; Noguchi, R.; Kuriyama, S.; Ikenaka, Y.; Yoshii, J.; Yanase, K.; Namisaki, T.; Kitade, M.; Masaki, T.; Fukui, H. Imatinib mesylate (STI-571) attenuates liver fibrosis development in rats. Am. J. Physiol. Gastrointest. Liver Physiol. 2005, 288, G907–G913. [Google Scholar] [CrossRef]
- Faloppi, L.; Scartozzi, M.; Maccaroni, E.; Paolo, M.D.P.; Berardi, R.; Del Prete, M.; Cascinu, S. Evolving strategies for the treatment of hepatocellular carcinoma: From clinical-guided to molecularly-taylored therapeutic options. Cancer Treat. Rev. 2011, 37, 169–177. [Google Scholar] [CrossRef]
- Kudo, M.; Finn, R.S.; Qin, S.; Han, K.-H.; Ikeda, K.; Cheng, A.-L.; Piscaglia, F.; Ueshima, K.; Aikata, H.; Vogel, A. Analysis of survival and objective response (OR) in patients with hepatocellular carcinoma in a phase III study of lenvatinib (REFLECT). J. Clin. Oncol. 2019, 37, 186. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, Z.; Kwong, S.Q.; Lui, E.L.H.; Friedman, S.L.; Li, F.R.; Lam, R.W.C.; Zhang, G.C.; Zhang, H.; Ye, T. Inhibition of PDGF, TGF-β, and Abl signaling and reduction of liver fibrosis by the small molecule Bcr-Abl tyrosine kinase antagonist Nilotinib. J. Hepatol. 2011, 55, 612–625. [Google Scholar] [CrossRef] [PubMed]
- Cheng, A.; Kang, Y.; Lin, D.; Park, J.; Kudo, M.; Qin, S.; Omata, M.; Pitman Lowenthal, S.W.; Lanzalone, S.; Yang, L. Phase III trial of sunitinib (Su) versus sorafenib (So) in advanced hepatocellular carcinoma (HCC). J. Clin. Oncol. 2011, 29 (Suppl. 15), 4000. [Google Scholar] [CrossRef]
- Finn, R.S.; Merle, P.; Granito, A.; Huang, Y.-H.; Bodoky, G.; Pracht, M.; Yokosuka, O.; Rosmorduc, O.; Gerolami, R.; Caparello, C. Outcomes of sequential treatment with sorafenib followed by regorafenib for HCC: Additional analyses from the phase III RESORCE trial. J. Hepatol. 2018, 69, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.; Yang, T.-S.; Huo, T.-I.; Hsieh, R.-K.; Yu, C.-W.; Hwang, W.-S.; Hsieh, T.-Y.; Huang, W.-T.; Chao, Y.; Meng, R. Vandetanib in patients with inoperable hepatocellular carcinoma: A phase II, randomized, double-blind, placebo-controlled study. J. Hepatol. 2012, 56, 1097–1103. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Feng, X.; Han, P.; Lei, Y.; Xia, Y.; Tian, D.; Yan, W. The JAK inhibitor tofacitinib ameliorates immune-mediated liver injury in mice. Mol. Med. Rep. 2019, 20, 4883–4892. [Google Scholar] [CrossRef]
- Dimri, M.; Satyanarayana, A. Molecular Signaling Pathways and Therapeutic Targets in Hepatocellular Carcinoma. Cancers 2020, 12, 491. [Google Scholar] [CrossRef]
- Osna, N.A.; Donohue, T.M., Jr.; Kharbanda, K.K. Alcoholic liver disease: Pathogenesis and current management. Alcohol Res. Curr. Rev. 2017, 38, 147. [Google Scholar]
- Vandenhooff, P.; Peppelenbosch, M.; Urban, I.J.A. N-methyl-d-aspartate component in hippocampal transmission to the lateral septum of the rat. Neurosci. Res. Commun. 1989, 5, 111–115. [Google Scholar]
- Bunchorntavakul, C.; Reddy, K.R. Pharmacologic Management of Portal Hypertension. Clin. Liver Dis. 2019, 23, 713–736. [Google Scholar] [CrossRef]
- Weinrieb, R.M. New Treatment Models for Alcohol Use Disorders and Alcoholic Liver Disease. Clin. Liver Dis. 2019, 13, 118. [Google Scholar] [CrossRef] [PubMed]
- Massey, V.; Parrish, A.; Argemi, J.; Moreno, M.; Mello, A.; García-Rocha, M.; Altamirano, J.; Odena, G.; Dubuquoy, L.; Louvet, A. Integrated Multi-Omics Reveals Glucose Use Reprogramming and Identifies a Novel Hexokinase in Alcoholic Hepatitis. Gastroenterology 2020. [Google Scholar] [CrossRef]
- Wang, H.; Zhou, H.; Zhang, Q.; Poulsen, K.L.; Taylor, V.; McMullen, M.R.; Czarnecki, D.; Dasarathy, D.; Yu, M.; Liao, Y. Inhibition of IRAK4 kinase activity improves ethanol-induced liver injury in mice. J. Hepatol. 2020, 73, 1470–1481. [Google Scholar] [CrossRef]
- Lee, K.J.; Jang, Y.O.; Cha, S.-K.; Kim, M.Y.; Park, K.-S.; Eom, Y.W.; Baik, S.K. Expression of fibroblast growth factor 21 and β-Klotho regulates hepatic fibrosis through the nuclear factor-κB and c-Jun N-terminal kinase pathways. Gut Liver 2018, 12, 449. [Google Scholar] [CrossRef] [PubMed]
- Afifiyan, N.; Tillman, B.; French, B.A.; Sweeny, O.; Masouminia, M.; Samadzadeh, S.; French, S.W. The role of Tec kinase signaling pathways in the development of Mallory Denk Bodies in balloon cells in alcoholic hepatitis. Exp. Mol. Pathol. 2017, 103, 191–199. [Google Scholar] [CrossRef]
- Qu, C.; Zheng, D.; Li, S.; Liu, Y.; Lidofsky, A.; Holmes, J.A.; Chen, J.; He, L.; Wei, L.; Liao, Y. Tyrosine kinase SYK is a potential therapeutic target for liver fibrosis. Hepatology 2018, 68, 1125–1139. [Google Scholar] [CrossRef] [PubMed]
- Kurniawan, D.W.; Jajoriya, A.K.; Dhawan, G.; Mishra, D.; Argemi, J.; Bataller, R.; Storm, G.; Mishra, D.P.; Prakash, J.; Bansal, R. Therapeutic inhibition of spleen tyrosine kinase in inflammatory macrophages using PLGA nanoparticles for the treatment of non-alcoholic steatohepatitis. J. Control. Release 2018, 288, 227–238. [Google Scholar] [CrossRef]
- Sansone, V.; Tovoli, F.; Casadei-Gardini, A.; Di Costanzo, G.G.; Magini, G.; Sacco, R.; Pressiani, T.; Trevisani, F.; Rimini, M.; Tortora, R.; et al. Comparison of Prognostic Scores in Patients With Hepatocellular Carcinoma Treated With Sorafenib. Clin. Transl. Gastroenterol. 2021, 12, e00286. [Google Scholar] [CrossRef]
- Jiang, X.; Tan, H.-Y.; Teng, S.; Chan, Y.-T.; Wang, D.; Wang, N. The role of AMP-activated protein kinase as a potential target of treatment of hepatocellular carcinoma. Cancers 2019, 11, 647. [Google Scholar] [CrossRef] [PubMed]
- De Rosamel, L.; Blanc, J.-F. Emerging tyrosine kinase inhibitors for the treatment of hepatocellular carcinoma. Expert Opin. Emerg. Drugs 2017, 22, 175–190. [Google Scholar] [CrossRef] [PubMed]
- Niu, Z.-S.; Niu, X.-J.; Wang, W.-H. Role of the receptor tyrosine kinase Axl in hepatocellular carcinoma and its clinical relevance. Future Oncol. 2019, 15, 653–662. [Google Scholar] [CrossRef]
- Ren, L.; Li, C.; Wang, Y.; Teng, Y.; Sun, H.; Xing, B.; Yang, X.; Jiang, Y.; He, F. In vivo phosphoproteome analysis reveals kinome reprogramming in hepatocellular carcinoma. Mol. Cell. Proteom. 2018, 17, 1067–1083. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.; van der Laan, L.J.W.; Janssen, H.L.A.; Peppelenbosch, M.P. A dynamic perspective of RNAi library development. Trends Biotechnol. 2012, 30, 206–215. [Google Scholar] [CrossRef] [PubMed]
- Shu, C.-W.; Cheng, J.-S.; Tsai, W.-L.; Liu, P.-F.; Goan, Y.-G.; Tseng, H.-H.; Lee, C.-H. The MAP3K7-mTOR axis promotes the proliferation and malignancy of hepatocellular carcinoma cells. Front. Oncol. 2019, 9, 474. [Google Scholar]
- Rodrigues, P.M.; Olaizola, P.; Paiva, N.A.; Olaizola, I.; Agirre-Lizaso, A.; Landa, A.; Bujanda, L.; Perugorria, M.J.; Banales, J.M. Pathogenesis of Cholangiocarcinoma. Annu. Rev. Pathol. 2020. [Google Scholar] [CrossRef]
- Wang, J.; Xing, X.; Li, Q.; Zhang, G.; Wang, T.; Pan, H.; Li, D. Targeting the FGFR signaling pathway in cholangiocarcinoma: Promise or delusion? Ther. Adv. Med. Oncol. 2020, 12, 1758835920940948. [Google Scholar] [CrossRef] [PubMed]
- McGrath, N.A.; Fu, J.; Gu, S.Z.; Xie, C. Targeting cancer stem cells in cholangiocarcinoma. Int. J. Oncol. 2020. [Google Scholar] [CrossRef]
- Saqub, H.; Proetsch-Gugerbauer, H.; Bezrookove, V.; Nosrati, M.; Vaquero, E.M.; de Semir, D.; Ice, R.J.; McAllister, S.; Soroceanu, L.; Kashani-Sabet, M. Dinaciclib, a cyclin-dependent kinase inhibitor, suppresses cholangiocarcinoma growth by targeting CDK2/5/9. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef]
- Lhomme, S.; Marion, O.; Abravanel, F.; Izopet, J.; Kamar, N. Clinical manifestations, pathogenesis and treatment of hepatitis E virus infections. J. Clin. Med. 2020, 9, 331. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, P.; Li, Y.; Zhang, R.; Yu, P.; Ma, Z.; Kainov, D.E.; Robert, A.; Peppelenbosch, M.P.; Pan, Q. Drug screening identified gemcitabine inhibiting hepatitis E virus by inducing interferon-like response via activation of STAT1 phosphorylation. Antivir. Res. 2020, 184, 104967. [Google Scholar] [CrossRef]
- Yin, X.; Feng, Z. Hepatitis E virus entry. Viruses 2019, 11, 883. [Google Scholar] [CrossRef] [PubMed]
- Lei, Q.; Li, L.; Zhang, S.; Li, T.; Zhang, X.; Ding, X.; Qin, B. HEV ORF3 downregulates TLR7 to inhibit the generation of type I interferon via impairment of multiple signaling pathways. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef]
- Ortiz, J.F.; Cox, Á.M.; Tambo, W.; Eskander, N.; Wirth, M.; Valdez, M.; Niño, M. Neurological Manifestations of Wilson’s Disease: Pathophysiology and Localization of Each Component. Cureus 2020, 12. [Google Scholar] [CrossRef]
- Kathawala, M.; Hirschfield, G.M. Insights into the management of Wilson’s disease. Ther. Adv. Gastroenterol. 2017, 10, 889–905. [Google Scholar] [CrossRef] [PubMed]
- Wooton-Kee, C.R.; Robertson, M.; Zhou, Y.; Dong, B.; Sun, Z.; Ho Kim, K.; Liu, H.; Xu, Y.; Putluri, N.; Saha, P.; et al. Metabolic dysregulation in the Atp7b−/− Wilson’s disease mouse model. Proc. Natl. Acad. Sci. USA 2020, 117, 2076–2083. [Google Scholar] [CrossRef] [PubMed]
- Kadowaki, S.; Meguro, S.; Imaizumi, Y.; Sakai, H.; Endoh, D.; Hayashi, M. Role of p38 Mapk in development of acute hepatic injury in Long-Evans Cinnamon (LEC) rats, an animal model of human Wilson’s disease. J. Vet. Med. Sci. 2013, 75, 1551–1556. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, B.; Mamedov, R.; Fuhler, G.M.; Peppelenbosch, M.P. Drug Discovery in Liver Disease Using Kinome Profiling. Int. J. Mol. Sci. 2021, 22, 2623. https://doi.org/10.3390/ijms22052623
Yu B, Mamedov R, Fuhler GM, Peppelenbosch MP. Drug Discovery in Liver Disease Using Kinome Profiling. International Journal of Molecular Sciences. 2021; 22(5):2623. https://doi.org/10.3390/ijms22052623
Chicago/Turabian StyleYu, Bingting, Ruslan Mamedov, Gwenny M. Fuhler, and Maikel P. Peppelenbosch. 2021. "Drug Discovery in Liver Disease Using Kinome Profiling" International Journal of Molecular Sciences 22, no. 5: 2623. https://doi.org/10.3390/ijms22052623
APA StyleYu, B., Mamedov, R., Fuhler, G. M., & Peppelenbosch, M. P. (2021). Drug Discovery in Liver Disease Using Kinome Profiling. International Journal of Molecular Sciences, 22(5), 2623. https://doi.org/10.3390/ijms22052623







