The Role of White Matter Dysfunction and Leukoencephalopathy/Leukodystrophy Genes in the Aetiology of Frontotemporal Dementias: Implications for Novel Approaches to Therapeutics
Abstract
1. Introduction
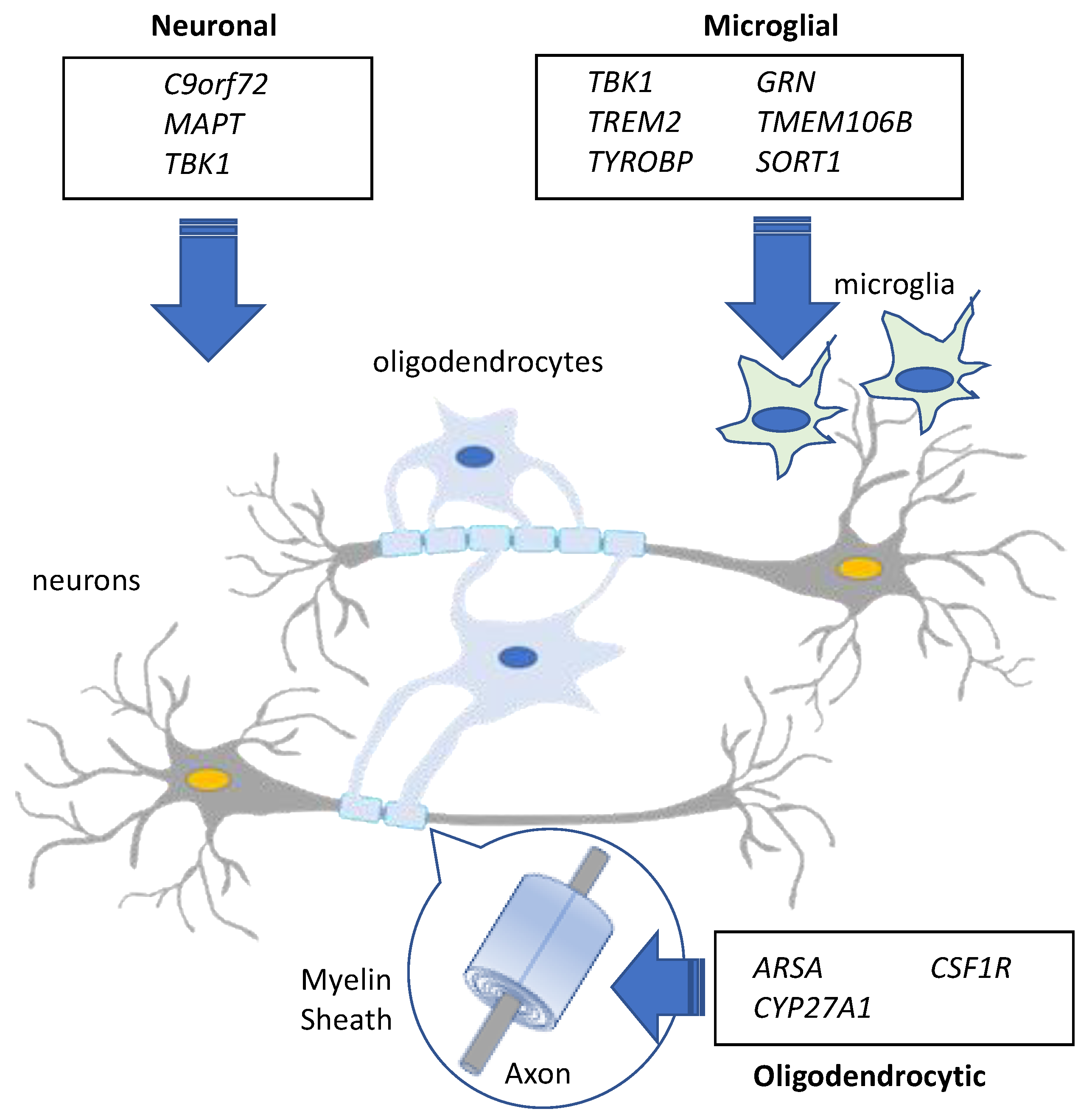
2. The Genetics of Frontotemporal Dementia and Relation to Neuropathological Subtypes
2.1. Frontotemporal Lobar Degeneration-Tau (FTLD-Tau)
MAPT
2.2. Frontotemporal Lobar Degeneration-TDP (FTLD-TDP)
2.2.1. C9orf72
2.2.2. TBK1
2.2.3. GRN
3. White Matter Changes in FTD
4. Lipid Metabolism in White Matter and Relevance to FTD
5. Granulin Mutations: A Model for Understanding the Role of Lipid Dysregulation and White Matter Changes in FTD
6. Leukoencephalopathies/Leukodystrophies as Part of the FTD Spectrum
7. GWAS and Susceptibility Loci for FTD
8. NGS and Rare Variants in FTD
8.1. TMEM106B
8.2. TREM2 and TYROBP
8.3. CSF1R
8.4. CYP27A1
9. Implications for Novel FTD Therapeutics
9.1. Novel Targets for Gene-Specific or Pharmacological Intervention
9.2. Transcranial Magnetic Stimulation for Sporadic and Genetic Forms of FTD
10. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Rascovsky, K.; Hodges, J.R.; Knopman, D.; Mendez, M.F.; Kramer, J.H.; Neuhaus, J.; Van Swieten, J.C.; Seelaar, H.; Dopper, E.G.; Onyike, C.U.; et al. Sensitivity of revised diagnostic criteria for the behavioural variant of frontotemporal dementia. Brain 2011, 134, 2456. [Google Scholar] [CrossRef]
- Gorno-Tempini, M.L.; Hillis, A.E.; Weintraub, S.; Kertesz, A.; Mendez, M.; Cappa, S.F.; Ogar, J.M.; Rohrer, J.D.; Black, S.; Boeve, B.F.; et al. Classification of primary progressive aphasia and its variants. Neurology 2011, 76, 1006–1014. [Google Scholar] [CrossRef]
- Rohrer, J.D.; Guerreiro, R.; Vandrovcova, J.; Uphill, J.; Reiman, D.; Beck, J.; Isaacs, A.M.; Authier, A.; Ferrari, R.; Fox, N.C.; et al. The heritability and genetics of frontotemporal lobar degeneration. Neurology 2009, 73, 1451–1456. [Google Scholar] [CrossRef] [PubMed]
- Hutton, M.; Lendon, C.L.; Rizzu, P.; Baker, M.; Froelich, S.; Houlden, H.; Pickering-Brown, S.; Chakraverty, S.; Isaacs, A.; Grover, A.; et al. Association of missense and 5’-splice-site mutations in tau with the inherited dementia FTDP-17. Nature 1998, 393, 702–705. [Google Scholar] [CrossRef] [PubMed]
- Baker, M.; Mackenzie, I.R.; Pickering-Brown, S.M.; Gass, J.; Rademakers, R.; Lindholm, C.; Snowden, J.; Adamson, J.; Sadovnick, A.D.; Rollinson, S.; et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature 2006, 442, 916–919. [Google Scholar] [CrossRef]
- Cruts, M.; Gijselinck, I.; Van der Zee, J.; Engelborghs, S.; Wils, H.; Pirici, D.; Rademakers, R.; Vandenberghe, R.; Dermaut, B.; Martin, J.J.; et al. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature 2006, 442, 920–924. [Google Scholar] [CrossRef]
- DeJesus-Hernandez, M.; Mackenzie, I.R.; Boeve, B.F.; Boxer, A.L.; Baker, M.; Rutherford, N.J.; Nicholson, A.M.; Finch, N.A.; Flynn, H.; Adamson, J.; et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 2011, 72, 245–256. [Google Scholar] [CrossRef]
- Renton, A.E.; Majounie, E.; Waite, A.; Simon-Sanchez, J.; Rollinson, S.; Gibbs, J.R.; Schymick, J.C.; Laaksovirta, H.; Van Swieten, J.C.; Myllykangas, L.; et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 2011, 72, 257–268. [Google Scholar] [CrossRef]
- Loy, C.T.; Schofield, P.R.; Turner, A.M.; Kwok, J.B. Genetics of dementia. Lancet 2014, 383, 828–840. [Google Scholar] [CrossRef]
- Bott, N.T.; Radke, A.; Stephens, M.L.; Kramer, J.H. Frontotemporal dementia: Diagnosis, deficits and management. Neurodegener. Dis. Manag. 2014, 4, 439–454. [Google Scholar] [CrossRef] [PubMed]
- De Majo, M.; Topp, S.D.; Smith, B.N.; Nishimura, A.L.; Chen, H.J.; Gkazi, A.S.; Miller, J.; Wong, C.H.; Vance, C.; Baas, F.; et al. ALS-associated missense and nonsense TBK1 mutations can both cause loss of kinase function. Neurobiol. Aging 2018, 71, e1–e10. [Google Scholar] [CrossRef]
- Gijselinck, I.; Van Mossevelde, S.; Van der Zee, J.; Sieben, A.; Philtjens, S.; Heeman, B.; Engelborghs, S.; Van den Bulcke, M.; De Baets, G.; Baumer, V.; et al. Loss of TBK1 is a frequent cause of frontotemporal dementia in a Belgian cohort. Neurology 2015, 85, 2116–2125. [Google Scholar] [CrossRef]
- Greaves, C.V.; Rohrer, J.D. An update on genetic frontotemporal dementia. J. Neurol. 2019, 266, 2075–2086. [Google Scholar] [CrossRef]
- Goldman, J.S.; Van Deerlin, V.M. Alzheimer’s Disease and Frontotemporal Dementia: The Current State of Genetics and Genetic Testing Since the Advent of Next-Generation Sequencing. Mol. Diagn. Ther. 2018, 22, 505–513. [Google Scholar] [CrossRef] [PubMed]
- Dobson-Stone, C.; Hallupp, M.; Shahheydari, H.; Ragagnin, A.M.G.; Chatterton, Z.; Carew-Jones, F.; Shepherd, C.E.; Stefen, H.; Paric, E.; Fath, T.; et al. CYLD is a causative gene for frontotemporal dementia-amyotrophic lateral sclerosis. Brain 2020, 143, 783–799. [Google Scholar] [CrossRef] [PubMed]
- Oyston, L.J.; Chatterton, Z.; Hallupp, M.; Rajan, N.; Kwok, J.B.; Dobson-Stone, C. Reply: CYLD variants in frontotemporal dementia associated with severe memory impairment in a Portuguese cohort. Brain 2020, 143, e68. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Landin-Romero, R.; Kumfor, F.; Irish, M.; Hodges, J.R.; Piguet, O. Cerebellar structural connectivity and contributions to cognition in frontotemporal dementias. Cortex 2020, 129, 57–67. [Google Scholar] [CrossRef]
- Hasegawa, M.; Smith, M.J.; Iijima, M.; Tabira, T.; Goedert, M. FTDP-17 mutations N279K and S305N in tau produce increased splicing of exon 10. FEBS Lett. 1999, 443, 93–96. [Google Scholar] [CrossRef]
- Hong, M.; Zhukareva, V.; Vogelsberg-Ragaglia, V.; Wszolek, Z.; Reed, L.; Miller, B.I.; Geschwind, D.H.; Bird, T.D.; McKeel, D.; Goate, A.; et al. Mutation-specific functional impairments in distinct tau isoforms of hereditary FTDP-17. Science 1998, 282, 1914–1917. [Google Scholar] [CrossRef]
- Josephs, K.A.; Hodges, J.R.; Snowden, J.S.; Mackenzie, I.R.; Neumann, M.; Mann, D.M.; Dickson, D.W. Neuropathological background of phenotypical variability in frontotemporal dementia. Acta Neuropathol. 2011, 122, 137–153. [Google Scholar] [CrossRef]
- Forrest, S.L.; Halliday, G.M.; Shepherd, C.E.; Kwok, J.B.; Hallupp, M.; Kril, J.J. Are mutations in MAPT associated with GGT type III? Neuropathol. Appl. Neurobiol. 2020, 46, 406–409. [Google Scholar] [CrossRef]
- Boeve, B.F.; Hutton, M. Refining frontotemporal dementia with parkinsonism linked to chromosome 17: Introducing FTDP-17 (MAPT) and FTDP-17 (PGRN). Arch. Neurol. 2008, 65, 460–464. [Google Scholar] [CrossRef]
- Rohrer, J.D.; Warren, J.D. Phenotypic signatures of genetic frontotemporal dementia. Curr. Opin. Neurol. 2011, 24, 542–549. [Google Scholar] [CrossRef] [PubMed]
- Pickering-Brown, S.M.; Rollinson, S.; Du Plessis, D.; Morrison, K.E.; Varma, A.; Richardson, A.M.; Neary, D.; Snowden, J.S.; Mann, D.M. Frequency and clinical characteristics of progranulin mutation carriers in the Manchester frontotemporal lobar degeneration cohort: Comparison with patients with MAPT and no known mutations. Brain 2008, 131, 721–731. [Google Scholar] [CrossRef]
- Rohrer, J.D.; Ridgway, G.R.; Modat, M.; Ourselin, S.; Mead, S.; Fox, N.C.; Rossor, M.N.; Warren, J.D. Distinct profiles of brain atrophy in frontotemporal lobar degeneration caused by progranulin and tau mutations. Neuroimage 2010, 53, 1070–1076. [Google Scholar] [CrossRef]
- Whitwell, J.L.; Avula, R.; Senjem, M.L.; Kantarci, K.; Weigand, S.D.; Samikoglu, A.; Edmonson, H.A.; Vemuri, P.; Knopman, D.S.; Boeve, B.F.; et al. Gray and white matter water diffusion in the syndromic variants of frontotemporal dementia. Neurology 2010, 74, 1279–1287. [Google Scholar] [CrossRef]
- Bang, J.; Spina, S.; Miller, B.L. Frontotemporal dementia. Lancet 2015, 386, 1672–1682. [Google Scholar] [CrossRef]
- Mackenzie, I.R.; Rademakers, R. The role of transactive response DNA-binding protein-43 in amyotrophic lateral sclerosis and frontotemporal dementia. Curr. Opin. Neurol. 2008, 21, 693–700. [Google Scholar] [CrossRef]
- Mackenzie, I.R.; Neumann, M.; Cairns, N.J.; Munoz, D.G.; Isaacs, A.M. Novel types of frontotemporal lobar degeneration: Beyond tau and TDP-43. J. Mol. Neurosci. 2011, 45, 402–408. [Google Scholar] [CrossRef]
- Tan, R.H.; Shepherd, C.E.; Kril, J.J.; McCann, H.; McGeachie, A.; McGinley, C.; Affleck, A.; Halliday, G.M. Classification of FTLD-TDP cases into pathological subtypes using antibodies against phosphorylated and non-phosphorylated TDP43. Acta Neuropathol. Commun. 2013, 1, 33. [Google Scholar] [CrossRef] [PubMed]
- Van Langenhove, T.; Van der Zee, J.; Gijselinck, I.; Engelborghs, S.; Vandenberghe, R.; Vandenbulcke, M.; De Bleecker, J.; Sieben, A.; Versijpt, J.; Ivanoiu, A.; et al. Distinct clinical characteristics of C9orf72 expansion carriers compared with GRN, MAPT, and nonmutation carriers in a Flanders-Belgian FTLD cohort. JAMA Neurol. 2013, 70, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Van Mossevelde, S.; Van der Zee, J.; Gijselinck, I.; Engelborghs, S.; Sieben, A.; Van Langenhove, T.; De Bleecker, J.; Baets, J.; Vandenbulcke, M.; Van Laere, K.; et al. Clinical features of TBK1 carriers compared with C9orf72, GRN and non-mutation carriers in a Belgian cohort. Brain 2016, 139, 452–467. [Google Scholar] [CrossRef]
- Mahoney, C.J.; Downey, L.E.; Ridgway, G.R.; Beck, J.; Clegg, S.; Blair, M.; Finnegan, S.; Leung, K.K.; Yeatman, T.; Golden, H.; et al. Longitudinal neuroimaging and neuropsychological profiles of frontotemporal dementia with C9ORF72 expansions. Alzheimer’s Res. Ther. 2012, 4, 41. [Google Scholar] [CrossRef]
- Majounie, E.; Renton, A.E.; Mok, K.; Dopper, E.G.; Waite, A.; Rollinson, S.; Chio, A.; Restagno, G.; Nicolaou, N.; Simon-Sanchez, J.; et al. Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: A cross-sectional study. Lancet Neurol. 2012, 11, 323–330. [Google Scholar] [CrossRef]
- He, J.; Tang, L.; Benyamin, B.; Shah, S.; Hemani, G.; Liu, R.; Ye, S.; Liu, X.; Ma, Y.; Zhang, H.; et al. C9orf72 hexanucleotide repeat expansions in Chinese sporadic amyotrophic lateral sclerosis. Neurobiol. Aging 2015, 36, 1–8. [Google Scholar] [CrossRef]
- Ogaki, K.; Li, Y.; Takanashi, M.; Ishikawa, K.; Kobayashi, T.; Nonaka, T.; Hasegawa, M.; Kishi, M.; Yoshino, H.; Funayama, M.; et al. Analyses of the MAPT, PGRN, and C9orf72 mutations in Japanese patients with FTLD, PSP, and CBS. Parkinsonism Relat. Disord. 2013, 19, 15–20. [Google Scholar] [CrossRef]
- Kim, E.J.; Kim, Y.E.; Jang, J.H.; Cho, E.H.; Na, D.L.; Seo, S.W.; Jung, N.Y.; Jeong, J.H.; Kwon, J.C.; Park, K.H.; et al. Analysis of frontotemporal dementia, amyotrophic lateral sclerosis, and other dementia-related genes in 107 Korean patients with frontotemporal dementia. Neurobiol. Aging 2018, 72, 186. [Google Scholar] [CrossRef] [PubMed]
- Dobson-Stone, C.; Hallupp, M.; Bartley, L.; Shepherd, C.E.; Halliday, G.M.; Schofield, P.R.; Hodges, J.R.; Kwok, J.B. C9ORF72 repeat expansion in clinical and neuropathologic frontotemporal dementia cohorts. Neurology 2012, 79, 995–1001. [Google Scholar] [CrossRef]
- Dobson-Stone, C.; Hallupp, M.; Loy, C.T.; Thompson, E.M.; Haan, E.; Sue, C.M.; Panegyres, P.K.; Razquin, C.; Seijo-Martinez, M.; Rene, R.; et al. C9ORF72 repeat expansion in Australian and Spanish frontotemporal dementia patients. PLoS ONE 2013, 8, e56899. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie, I.R.; Arzberger, T.; Kremmer, E.; Troost, D.; Lorenzl, S.; Mori, K.; Weng, S.M.; Haass, C.; Kretzschmar, H.A.; Edbauer, D.; et al. Dipeptide repeat protein pathology in C9ORF72 mutation cases: Clinico-pathological correlations. Acta Neuropathol. 2013, 126, 859–879. [Google Scholar] [CrossRef] [PubMed]
- Mann, D.M.; Rollinson, S.; Robinson, A.; Bennion Callister, J.; Thompson, J.C.; Snowden, J.S.; Gendron, T.; Petrucelli, L.; Masuda-Suzukake, M.; Hasegawa, M.; et al. Dipeptide repeat proteins are present in the p62 positive inclusions in patients with frontotemporal lobar degeneration and motor neurone disease associated with expansions in C9ORF72. Acta Neuropathol. Commun. 2013, 1, 68. [Google Scholar] [CrossRef]
- Snowden, J.S.; Rollinson, S.; Thompson, J.C.; Harris, J.M.; Stopford, C.L.; Richardson, A.M.; Jones, M.; Gerhard, A.; Davidson, Y.S.; Robinson, A.; et al. Distinct clinical and pathological characteristics of frontotemporal dementia associated with C9ORF72 mutations. Brain 2012, 135, 693–708. [Google Scholar] [CrossRef]
- Lee, S.M.; Asress, S.; Hales, C.M.; Gearing, M.; Vizcarra, J.C.; Fournier, C.N.; Gutman, D.A.; Chin, L.S.; Li, L.; Glass, J.D. TDP-43 cytoplasmic inclusion formation is disrupted in C9orf72-associated amyotrophic lateral sclerosis/frontotemporal lobar degeneration. Brain Commun. 2019, 1, 14. [Google Scholar] [CrossRef]
- Cook, C.N.; Wu, Y.; Odeh, H.M.; Gendron, T.F.; Jansen-West, K.; Del Rosso, G.; Yue, M.; Jiang, P.; Gomes, E.; Tong, J.; et al. C9orf72 poly (GR) aggregation induces TDP-43 proteinopathy. Sci. Transl. Med. 2020, 12, 559. [Google Scholar] [CrossRef]
- Van der Zee, J.; Gijselinck, I.; Van Mossevelde, S.; Perrone, F.; Dillen, L.; Heeman, B.; Baumer, V.; Engelborghs, S.; De Bleecker, J.; Baets, J.; et al. TBK1 Mutation Spectrum in an Extended European Patient Cohort with Frontotemporal Dementia and Amyotrophic Lateral Sclerosis. Hum. Mutat. 2017, 38, 297–309. [Google Scholar] [CrossRef]
- Lamb, R.; Rohrer, J.D.; Real, R.; Lubbe, S.J.; Waite, A.J.; Blake, D.J.; Walters, R.J.; Lashley, T.; Revesz, T.; Holton, J.L.; et al. A novel TBK1 mutation in a family with diverse frontotemporal dementia spectrum disorders. Cold Spring Harb. Mol. Case Stud. 2019, 5, 3. [Google Scholar] [CrossRef] [PubMed]
- Koriath, C.A.; Bocchetta, M.; Brotherhood, E.; Woollacott, I.O.; Norsworthy, P.; Simon-Sanchez, J.; Blauwendraat, C.; Dick, K.M.; Gordon, E.; Harding, S.R.; et al. The clinical, neuroanatomical, and neuropathologic phenotype of TBK1-associated frontotemporal dementia: A longitudinal case report. Alzheimer’s Dement. 2017, 6, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Hirsch-Reinshagen, V.; Alfaify, O.A.; Hsiung, G.R.; Pottier, C.; Baker, M.; Perkerson, R.B., 3rd; Rademakers, R.; Briemberg, H.; Foti, D.J.; Mackenzie, I.R. Clinicopathologic correlations in a family with a TBK1 mutation presenting as primary progressive aphasia and primary lateral sclerosis. Amyotroph. Lateral Scler. Frontotemporal. Degener. 2019, 20, 568–575. [Google Scholar] [CrossRef]
- Mackenzie, I.R. The neuropathology and clinical phenotype of FTD with progranulin mutations. Acta Neuropathol. 2007, 114, 49–54. [Google Scholar] [CrossRef]
- Mackenzie, I.R.; Baker, M.; Pickering-Brown, S.; Hsiung, G.Y.; Lindholm, C.; Dwosh, E.; Gass, J.; Cannon, A.; Rademakers, R.; Hutton, M.; et al. The neuropathology of frontotemporal lobar degeneration caused by mutations in the progranulin gene. Brain 2006, 129, 3081–3090. [Google Scholar] [CrossRef] [PubMed]
- Hosokawa, M.; Kondo, H.; Serrano, G.E.; Beach, T.G.; Robinson, A.C.; Mann, D.M.; Akiyama, H.; Hasegawa, M.; Arai, T. Accumulation of multiple neurodegenerative disease-related proteins in familial frontotemporal lobar degeneration associated with granulin mutation. Sci. Rep. 2017, 7, 1513. [Google Scholar] [CrossRef]
- Zhang, Y.J.; Xu, Y.F.; Dickey, C.A.; Buratti, E.; Baralle, F.; Bailey, R.; Pickering-Brown, S.; Dickson, D.; Petrucelli, L. Progranulin mediates caspase-dependent cleavage of TAR DNA binding protein-43. J. Neurosci. 2007, 27, 10530–10534. [Google Scholar] [CrossRef]
- Hu, F.; Padukkavidana, T.; Vaegter, C.B.; Brady, O.A.; Zheng, Y.; Mackenzie, I.R.; Feldman, H.H.; Nykjaer, A.; Strittmatter, S.M. Sortilin-mediated endocytosis determines levels of the frontotemporal dementia protein, progranulin. Neuron 2010, 68, 654–667. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.C.; Srinivasan, K.; Friedman, B.A.; Suto, E.; Modrusan, Z.; Lee, W.P.; Kaminker, J.S.; Hansen, D.V.; Sheng, M. Progranulin deficiency causes impairment of autophagy and TDP-43 accumulation. J. Exp. Med. 2017, 214, 2611–2628. [Google Scholar] [CrossRef]
- Beck, J.; Rohrer, J.D.; Campbell, T.; Isaacs, A.; Morrison, K.E.; Goodall, E.F.; Warrington, E.K.; Stevens, J.; Revesz, T.; Holton, J.; et al. A distinct clinical, neuropsychological and radiological phenotype is associated with progranulin gene mutations in a large UK series. Brain 2008, 131, 706–720. [Google Scholar] [CrossRef]
- Boeve, B.F.; Baker, M.; Dickson, D.W.; Parisi, J.E.; Giannini, C.; Josephs, K.A.; Hutton, M.; Pickering-Brown, S.M.; Rademakers, R.; Tang-Wai, D.; et al. Frontotemporal dementia and parkinsonism associated with the IVS1+1G->A mutation in progranulin: A clinicopathologic study. Brain 2006, 129, 3103–3114. [Google Scholar] [CrossRef]
- Leverenz, J.B.; Yu, C.E.; Montine, T.J.; Steinbart, E.; Bekris, L.M.; Zabetian, C.; Kwong, L.K.; Lee, V.M.; Schellenberg, G.D.; Bird, T.D. A novel progranulin mutation associated with variable clinical presentation and tau, TDP43 and alpha-synuclein pathology. Brain 2007, 130, 1360–1374. [Google Scholar] [CrossRef] [PubMed]
- Petkau, T.L.; Leavitt, B.R. Progranulin in neurodegenerative disease. Trends Neurosci. 2014, 37, 388–398. [Google Scholar] [CrossRef] [PubMed]
- Van Swieten, J.C.; Heutink, P. Mutations in progranulin (GRN) within the spectrum of clinical and pathological phenotypes of frontotemporal dementia. Lancet Neurol. 2008, 7, 965–974. [Google Scholar] [CrossRef]
- Le Ber, I.; Camuzat, A.; Hannequin, D.; Pasquier, F.; Guedj, E.; Rovelet-Lecrux, A.; Hahn-Barma, V.; Van der Zee, J.; Clot, F.; Bakchine, S.; et al. Phenotype variability in progranulin mutation carriers: A clinical, neuropsychological, imaging and genetic study. Brain 2008, 131, 732–746. [Google Scholar] [CrossRef]
- Mackenzie, I.R.; Neumann, M. Molecular neuropathology of frontotemporal dementia: Insights into disease mechanisms from postmortem studies. J. Neurochem. 2016, 138, 54–70. [Google Scholar] [CrossRef]
- Paternico, D.; Premi, E.; Gazzina, S.; Cosseddu, M.; Alberici, A.; Archetti, S.; Cotelli, M.S.; Micheli, A.; Turla, M.; Gasparotti, R.; et al. White matter hyperintensities characterize monogenic frontotemporal dementia with granulin mutations. Neurobiol. Aging 2016, 38, 176–180. [Google Scholar] [CrossRef] [PubMed]
- Sudre, C.H.; Bocchetta, M.; Heller, C.; Convery, R.; Neason, M.; Moore, K.M.; Cash, D.M.; Thomas, D.L.; Woollacott, I.O.C.; Foiani, M.; et al. White matter hyperintensities in progranulin-associated frontotemporal dementia: A longitudinal GENFI study. Neuroimage Clin. 2019, 24, 102077. [Google Scholar] [CrossRef]
- Woollacott, I.O.C.; Bocchetta, M.; Sudre, C.H.; Ridha, B.H.; Strand, C.; Courtney, R.; Ourselin, S.; Cardoso, M.J.; Warren, J.D.; Rossor, M.N.; et al. Pathological correlates of white matter hyperintensities in a case of progranulin mutation associated frontotemporal dementia. Neurocase 2018, 24, 166–174. [Google Scholar] [CrossRef] [PubMed]
- Sudre, C.H.; Bocchetta, M.; Cash, D.; Thomas, D.L.; Woollacott, I.; Dick, K.M.; Van Swieten, J.; Borroni, B.; Galimberti, D.; Masellis, M.; et al. White matter hyperintensities are seen only in GRN mutation carriers in the GENFI cohort. Neuroimage Clin. 2017, 15, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Caroppo, P.; Le Ber, I.; Camuzat, A.; Clot, F.; Naccache, L.; Lamari, F.; De Septenville, A.; Bertrand, A.; Belliard, S.; Hannequin, D.; et al. Extensive white matter involvement in patients with frontotemporal lobar degeneration: Think progranulin. JAMA Neurol. 2014, 71, 1562–1566. [Google Scholar] [CrossRef]
- Elahi, F.M.; Marx, G.; Cobigo, Y.; Staffaroni, A.M.; Kornak, J.; Tosun, D.; Boxer, A.L.; Kramer, J.H.; Miller, B.L.; Rosen, H.J. Longitudinal white matter change in frontotemporal dementia subtypes and sporadic late onset Alzheimer’s disease. Neuroimage Clin. 2017, 16, 595–603. [Google Scholar] [CrossRef] [PubMed]
- Frings, L.; Yew, B.; Flanagan, E.; Lam, B.Y.; Hull, M.; Huppertz, H.J.; Hodges, J.R.; Hornberger, M. Longitudinal grey and white matter changes in frontotemporal dementia and Alzheimer’s disease. PLoS ONE 2014, 9, e90814. [Google Scholar] [CrossRef]
- Zhang, Y.; Schuff, N.; Du, A.T.; Rosen, H.J.; Kramer, J.H.; Gorno-Tempini, M.L.; Miller, B.L.; Weiner, M.W. White matter damage in frontotemporal dementia and Alzheimer’s disease measured by diffusion MRI. Brain 2009, 132, 2579–2592. [Google Scholar] [CrossRef]
- Mahoney, C.J.; Simpson, I.J.; Nicholas, J.M.; Fletcher, P.D.; Downey, L.E.; Golden, H.L.; Clark, C.N.; Schmitz, N.; Rohrer, J.D.; Schott, J.M.; et al. Longitudinal diffusion tensor imaging in frontotemporal dementia. Ann. Neurol. 2015, 77, 33–46. [Google Scholar] [CrossRef]
- Galantucci, S.; Tartaglia, M.C.; Wilson, S.M.; Henry, M.L.; Filippi, M.; Agosta, F.; Dronkers, N.F.; Henry, R.G.; Ogar, J.M.; Miller, B.L.; et al. White matter damage in primary progressive aphasias: A diffusion tensor tractography study. Brain 2011, 134, 3011–3029. [Google Scholar] [CrossRef]
- Acosta-Cabronero, J.; Patterson, K.; Fryer, T.D.; Hodges, J.R.; Pengas, G.; Williams, G.B.; Nestor, P.J. Atrophy, hypometabolism and white matter abnormalities in semantic dementia tell a coherent story. Brain 2011, 134, 2025–2035. [Google Scholar] [CrossRef] [PubMed]
- Agosta, F.; Galantucci, S.; Svetel, M.; Lukic, M.J.; Copetti, M.; Davidovic, K.; Tomic, A.; Spinelli, E.G.; Kostic, V.S.; Filippi, M. Clinical, cognitive, and behavioural correlates of white matter damage in progressive supranuclear palsy. J. Neurol. 2014, 261, 913–924. [Google Scholar] [CrossRef] [PubMed]
- Borroni, B.; Brambati, S.M.; Agosti, C.; Gipponi, S.; Bellelli, G.; Gasparotti, R.; Garibotto, V.; Di Luca, M.; Scifo, P.; Perani, D.; et al. Evidence of white matter changes on diffusion tensor imaging in frontotemporal dementia. Arch. Neurol. 2007, 64, 246–251. [Google Scholar] [CrossRef]
- Matsuo, K.; Mizuno, T.; Yamada, K.; Akazawa, K.; Kasai, T.; Kondo, M.; Mori, S.; Nishimura, T.; Nakagawa, M. Cerebral white matter damage in frontotemporal dementia assessed by diffusion tensor tractography. Neuroradiology 2008, 50, 605–611. [Google Scholar] [CrossRef]
- Schwindt, G.C.; Graham, N.L.; Rochon, E.; Tang-Wai, D.F.; Lobaugh, N.J.; Chow, T.W.; Black, S.E. Whole-brain white matter disruption in semantic and nonfluent variants of primary progressive aphasia. Hum. Brain Mapp. 2013, 34, 973–984. [Google Scholar] [CrossRef]
- Daianu, M.; Mendez, M.F.; Baboyan, V.G.; Jin, Y.; Melrose, R.J.; Jimenez, E.E.; Thompson, P.M. An advanced white matter tract analysis in frontotemporal dementia and early-onset Alzheimer’s disease. Brain Imaging Behav. 2016, 10, 1038–1053. [Google Scholar] [CrossRef]
- Fields, R.D. White matter in learning, cognition and psychiatric disorders. Trends Neurosci. 2008, 31, 361–370. [Google Scholar] [CrossRef]
- Filley, C.M.; Fields, R.D. White matter and cognition: Making the connection. J. Neurophysiol. 2016, 116, 2093–2104. [Google Scholar] [CrossRef] [PubMed]
- Olsen, A.S.B.; Faergeman, N.J. Sphingolipids: Membrane microdomains in brain development, function and neurological diseases. Open Biol. 2017, 7, 5. [Google Scholar] [CrossRef]
- Mahoney, C.J.; Ridgway, G.R.; Malone, I.B.; Downey, L.E.; Beck, J.; Kinnunen, K.M.; Schmitz, N.; Golden, H.L.; Rohrer, J.D.; Schott, J.M.; et al. Profiles of white matter tract pathology in frontotemporal dementia. Hum. Brain Mapp. 2014, 35, 4163–4179. [Google Scholar] [CrossRef]
- Jiskoot, L.C.; Bocchetta, M.; Nicholas, J.M.; Cash, D.M.; Thomas, D.; Modat, M.; Ourselin, S.; Rombouts, S.; Dopper, E.G.P.; Meeter, L.H.; et al. Presymptomatic white matter integrity loss in familial frontotemporal dementia in the GENFI cohort: A cross-sectional diffusion tensor imaging study. Ann. Clin. Transl. Neurol. 2018, 5, 1025–1036. [Google Scholar] [CrossRef] [PubMed]
- Panman, J.L.; Jiskoot, L.C.; Bouts, M.; Meeter, L.H.H.; Van der Ende, E.L.; Poos, J.M.; Feis, R.A.; Kievit, A.J.A.; Van Minkelen, R.; Dopper, E.G.P.; et al. Gray and white matter changes in presymptomatic genetic frontotemporal dementia: A longitudinal MRI study. Neurobiol. Aging 2019, 76, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Rohrer, J.D.; Nicholas, J.M.; Cash, D.M.; van Swieten, J.; Dopper, E.; Jiskoot, L.; Van Minkelen, R.; Rombouts, S.A.; Cardoso, M.J.; Clegg, S.; et al. Presymptomatic cognitive and neuroanatomical changes in genetic frontotemporal dementia in the Genetic Frontotemporal dementia Initiative (GENFI) study: A cross-sectional analysis. Lancet Neurol. 2015, 14, 253–262. [Google Scholar] [CrossRef]
- Olm, C.A.; McMillan, C.T.; Irwin, D.J.; Van Deerlin, V.M.; Cook, P.A.; Gee, J.C.; Grossman, M. Longitudinal structural gray matter and white matter MRI changes in presymptomatic progranulin mutation carriers. Neuroimage Clin. 2018, 19, 497–506. [Google Scholar] [CrossRef]
- Chao, L.L.; Schuff, N.; Clevenger, E.M.; Mueller, S.G.; Rosen, H.J.; Gorno-Tempini, M.L.; Kramer, J.H.; Miller, B.L.; Weiner, M.W. Patterns of white matter atrophy in frontotemporal lobar degeneration. Arch. Neurol. 2007, 64, 1619–1624. [Google Scholar] [CrossRef][Green Version]
- Agosta, F.; Scola, E.; Canu, E.; Marcone, A.; Magnani, G.; Sarro, L.; Copetti, M.; Caso, F.; Cerami, C.; Comi, G.; et al. White matter damage in frontotemporal lobar degeneration spectrum. Cereb. Cortex 2012, 22, 2705–2714. [Google Scholar] [CrossRef] [PubMed]
- Lansdall, C.J.; Coyle-Gilchrist, I.T.S.; Jones, P.S.; Vazquez Rodriguez, P.; Wilcox, A.; Wehmann, E.; Dick, K.M.; Robbins, T.W.; Rowe, J.B. White matter change with apathy and impulsivity in frontotemporal lobar degeneration syndromes. Neurology 2018, 90, e1066–e1076. [Google Scholar] [CrossRef] [PubMed]
- Tovar-Moll, F.; De Oliveira-Souza, R.; Bramati, I.E.; Zahn, R.; Cavanagh, A.; Tierney, M.; Moll, J.; Grafman, J. White matter tract damage in the behavioral variant of frontotemporal and corticobasal dementia syndromes. PLoS ONE 2014, 9, e102656. [Google Scholar] [CrossRef][Green Version]
- Gopalakrishnan, G.; Awasthi, A.; Belkaid, W.; De Faria, O., Jr.; Liazoghli, D.; Colman, D.R.; Dhaunchak, A.S. Lipidome and proteome map of myelin membranes. J. Neurosci. Res. 2013, 91, 321–334. [Google Scholar] [CrossRef]
- Cermenati, G.; Mitro, N.; Audano, M.; Melcangi, R.C.; Crestani, M.; De Fabiani, E.; Caruso, D. Lipids in the nervous system: From biochemistry and molecular biology to patho-physiology. Biochim. Biophys. Acta 2015, 1851, 51–60. [Google Scholar] [CrossRef]
- Korade, Z.; Kenworthy, A.K. Lipid rafts, cholesterol, and the brain. Neuropharmacology 2008, 55, 1265–1273. [Google Scholar] [CrossRef] [PubMed]
- Jana, A.; Hogan, E.L.; Pahan, K. Ceramide and neurodegeneration: Susceptibility of neurons and oligodendrocytes to cell damage and death. J. Neurol. Sci. 2009, 278, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Soderberg, M.; Edlund, C.; Alafuzoff, I.; Kristensson, K.; Dallner, G. Lipid composition in different regions of the brain in Alzheimer’s disease/senile dementia of Alzheimer’s type. J. Neurochem. 1992, 59, 1646–1653. [Google Scholar] [CrossRef]
- Pettegrew, J.W.; Panchalingam, K.; Hamilton, R.L.; McClure, R.J. Brain membrane phospholipid alterations in Alzheimer’s disease. Neurochem. Res. 2001, 26, 771–782. [Google Scholar] [CrossRef]
- Couttas, T.A.; Kain, N.; Daniels, B.; Lim, X.Y.; Shepherd, C.; Kril, J.; Pickford, R.; Li, H.; Garner, B.; Don, A.S. Loss of the neuroprotective factor Sphingosine 1-phosphate early in Alzheimer’s disease pathogenesis. Acta Neuropathol. Commun. 2014, 2, 9. [Google Scholar] [CrossRef]
- Han, X.; McKeel, D.W., Jr.; Kelley, J.; Morris, J.C. Substantial sulfatide deficiency and ceramide elevation in very early Alzheimer’s disease: Potential role in disease pathogenesis. J. Neurochem. 2002, 82, 809–818. [Google Scholar] [CrossRef]
- Brekk, O.R.; Honey, J.R.; Lee, S.; Hallett, P.J.; Isacson, O. Cell type-specific lipid storage changes in Parkinson’s disease patient brains are recapitulated by experimental glycolipid disturbance. Proc. Natl. Acad. Sci. USA 2020, 117, 27646–27654. [Google Scholar] [CrossRef] [PubMed]
- Chan, R.B.; Perotte, A.J.; Zhou, B.; Liong, C.; Shorr, E.J.; Marder, K.S.; Kang, U.J.; Waters, C.H.; Levy, O.A.; Xu, Y.; et al. Elevated GM3 plasma concentration in idiopathic Parkinson’s disease: A lipidomic analysis. PLoS ONE 2017, 12, e0172348. [Google Scholar] [CrossRef] [PubMed]
- Di Pardo, A.; Amico, E.; Basit, A.; Armirotti, A.; Joshi, P.; Neely, M.D.; Vuono, R.; Castaldo, S.; Digilio, A.F.; Scalabri, F.; et al. Defective Sphingosine-1-phosphate metabolism is a druggable target in Huntington’s disease. Sci. Rep. 2017, 7, 5280. [Google Scholar] [CrossRef]
- Kreilaus, F.; Spiro, A.S.; McLean, C.A.; Garner, B.; Jenner, A.M. Evidence for altered cholesterol metabolism in Huntington’s disease post mortem brain tissue. Neuropathol. Appl. Neurobiol. 2016, 42, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Leoni, V.; Mariotti, C.; Nanetti, L.; Salvatore, E.; Squitieri, F.; Bentivoglio, A.R.; Bandettini di Poggio, M.; Piacentini, S.; Monza, D.; Valenza, M.; et al. Whole body cholesterol metabolism is impaired in Huntington’s disease. Neurosci. Lett. 2011, 494, 245–249. [Google Scholar] [CrossRef] [PubMed]
- Karasinska, J.M.; Hayden, M.R. Cholesterol metabolism in Huntington disease. Nat. Rev. Neurol. 2011, 7, 561–572. [Google Scholar] [CrossRef]
- Cutler, R.G.; Pedersen, W.A.; Camandola, S.; Rothstein, J.D.; Mattson, M.P. Evidence that accumulation of ceramides and cholesterol esters mediates oxidative stress-induced death of motor neurons in amyotrophic lateral sclerosis. Ann. Neurol. 2002, 52, 448–457. [Google Scholar] [CrossRef]
- Blasco, H.; Veyrat-Durebex, C.; Bocca, C.; Patin, F.; Vourc’h, P.; Kouassi Nzoughet, J.; Lenaers, G.; Andres, C.R.; Simard, G.; Corcia, P.; et al. Lipidomics Reveals Cerebrospinal-Fluid Signatures of ALS. Sci. Rep. 2017, 7, 17652. [Google Scholar] [CrossRef]
- Del Boccio, P.; Pieragostino, D.; Di Ioia, M.; Petrucci, F.; Lugaresi, A.; De Luca, G.; Gambi, D.; Onofrj, M.; Di Ilio, C.; Sacchetta, P.; et al. Lipidomic investigations for the characterization of circulating serum lipids in multiple sclerosis. J. Proteomics 2011, 74, 2826–2836. [Google Scholar] [CrossRef]
- Kim, W.S.; He, Y.; Phan, K.; Ahmed, R.M.; Rye, K.A.; Piguet, O.; Hodges, J.R.; Halliday, G.M. Altered High Density Lipoprotein Composition in Behavioral Variant Frontotemporal Dementia. Front. Neurosci. 2018, 12, 847. [Google Scholar] [CrossRef]
- Ahmed, R.M.; MacMillan, M.; Bartley, L.; Halliday, G.M.; Kiernan, M.C.; Hodges, J.R.; Piguet, O. Systemic metabolism in frontotemporal dementia. Neurology 2014, 83, 1812–1818. [Google Scholar] [CrossRef]
- Phan, K.; He, Y.; Pickford, R.; Bhatia, S.; Katzeff, J.S.; Hodges, J.R.; Piguet, O.; Halliday, G.M.; Kim, W.S. Uncovering pathophysiological changes in frontotemporal dementia using serum lipids. Sci. Rep. 2020, 10, 3640. [Google Scholar] [CrossRef] [PubMed]
- Behrens, M.I.; Mukherjee, O.; Tu, P.H.; Liscic, R.M.; Grinberg, L.T.; Carter, D.; Paulsmeyer, K.; Taylor-Reinwald, L.; Gitcho, M.; Norton, J.B.; et al. Neuropathologic heterogeneity in HDDD1: A familial frontotemporal lobar degeneration with ubiquitin-positive inclusions and progranulin mutation. Alzheimer Dis. Assoc. Disord. 2007, 21, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Snowden, J.S.; Pickering-Brown, S.M.; Mackenzie, I.R.; Richardson, A.M.; Varma, A.; Neary, D.; Mann, D.M. Progranulin gene mutations associated with frontotemporal dementia and progressive non-fluent aphasia. Brain 2006, 129, 3091–3102. [Google Scholar] [CrossRef] [PubMed]
- Evers, B.M.; Rodriguez-Navas, C.; Tesla, R.J.; Prange-Kiel, J.; Wasser, C.R.; Yoo, K.S.; McDonald, J.; Cenik, B.; Ravenscroft, T.A.; Plattner, F.; et al. Lipidomic and Transcriptomic Basis of Lysosomal Dysfunction in Progranulin Deficiency. Cell Rep. 2017, 20, 2565–2574. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.R.; Damiano, J.; Franceschetti, S.; Carpenter, S.; Canafoglia, L.; Morbin, M.; Rossi, G.; Pareyson, D.; Mole, S.E.; Staropoli, J.F.; et al. Strikingly different clinicopathological phenotypes determined by progranulin-mutation dosage. Am. J. Hum. Genet. 2012, 90, 1102–1107. [Google Scholar] [CrossRef]
- Canafoglia, L.; Morbin, M.; Scaioli, V.; Pareyson, D.; D’Incerti, L.; Fugnanesi, V.; Tagliavini, F.; Berkovic, S.F.; Franceschetti, S. Recurrent generalized seizures, visual loss, and palinopsia as phenotypic features of neuronal ceroid lipofuscinosis due to progranulin gene mutation. Epilepsia 2014, 55, e56–e59. [Google Scholar] [CrossRef]
- Yin, F.; Dumont, M.; Banerjee, R.; Ma, Y.; Li, H.; Lin, M.T.; Beal, M.F.; Nathan, C.; Thomas, B.; Ding, A. Behavioral deficits and progressive neuropathology in progranulin-deficient mice: A mouse model of frontotemporal dementia. FASEB J. 2010, 24, 4639–4647. [Google Scholar]
- Paushter, D.H.; Du, H.; Feng, T.; Hu, F. The lysosomal function of progranulin, a guardian against neurodegeneration. Acta Neuropathol. 2018, 136, 1–17. [Google Scholar] [CrossRef]
- Strong, A.; Rader, D.J. Sortilin as a regulator of lipoprotein metabolism. Curr. Atheroscler. Rep. 2012, 14, 211–218. [Google Scholar] [CrossRef]
- Zhou, X.; Sun, L.; Bracko, O.; Choi, J.W.; Jia, Y.; Nana, A.L.; Brady, O.A.; Hernandez, J.C.C.; Nishimura, N.; Seeley, W.W.; et al. Impaired prosaposin lysosomal trafficking in frontotemporal lobar degeneration due to progranulin mutations. Nat. Commun. 2017, 8, 15277. [Google Scholar] [CrossRef]
- Arrant, A.E.; Roth, J.R.; Boyle, N.R.; Kashyap, S.N.; Hoffmann, M.Q.; Murchison, C.F.; Ramos, E.M.; Nana, A.L.; Spina, S.; Grinberg, L.T.; et al. Impaired beta-glucocerebrosidase activity and processing in frontotemporal dementia due to progranulin mutations. Acta Neuropathol. Commun. 2019, 7, 218. [Google Scholar] [CrossRef]
- Van der Knaap, M.S.; Bugiani, M. Leukodystrophies: A proposed classification system based on pathological changes and pathogenetic mechanisms. Acta Neuropathol. 2017, 134, 351–382. [Google Scholar] [CrossRef] [PubMed]
- Van der Knaap, M.S.; Schiffmann, R.; Mochel, F.; Wolf, N.I. Diagnosis, prognosis, and treatment of leukodystrophies. Lancet Neurol. 2019, 18, 962–972. [Google Scholar] [CrossRef]
- Vanderver, A.; Prust, M.; Tonduti, D.; Mochel, F.; Hussey, H.M.; Helman, G.; Garbern, J.; Eichler, F.; Labauge, P.; Aubourg, P.; et al. Case definition and classification of leukodystrophies and leukoencephalopathies. Mol. Genet. Metab. 2015, 114, 494–500. [Google Scholar] [CrossRef] [PubMed]
- Kohler, W.; Curiel, J.; Vanderver, A. Adulthood leukodystrophies. Nat. Rev. Neurol. 2018, 14, 94–105. [Google Scholar] [CrossRef] [PubMed]
- Kevelam, S.H.; Steenweg, M.E.; Srivastava, S.; Helman, G.; Naidu, S.; Schiffmann, R.; Blaser, S.; Vanderver, A.; Wolf, N.I.; Van der Knaap, M.S. Update on Leukodystrophies: A Historical Perspective and Adapted Definition. Neuropediatrics 2016, 47, 349–354. [Google Scholar] [CrossRef] [PubMed]
- Vanderver, A.; Simons, C.; Helman, G.; Crawford, J.; Wolf, N.I.; Bernard, G.; Pizzino, A.; Schmidt, J.L.; Takanohashi, A.; Miller, D.; et al. Whole exome sequencing in patients with white matter abnormalities. Ann. Neurol. 2016, 79, 1031–1037. [Google Scholar] [CrossRef] [PubMed]
- Penati, R.; Fumagalli, F.; Calbi, V.; Bernardo, M.E.; Aiuti, A. Gene therapy for lysosomal storage disorders: Recent advances for metachromatic leukodystrophy and mucopolysaccaridosis I. J. Inherit. Metab. Dis. 2017, 40, 543–554. [Google Scholar] [CrossRef]
- Manolio, T.A.; Collins, F.S.; Cox, N.J.; Goldstein, D.B.; Hindorff, L.A.; Hunter, D.J.; McCarthy, M.I.; Ramos, E.M.; Cardon, L.R.; Chakravarti, A.; et al. Finding the missing heritability of complex diseases. Nature 2009, 461, 747–753. [Google Scholar] [CrossRef]
- Tam, V.; Patel, N.; Turcotte, M.; Bosse, Y.; Pare, G.; Meyre, D. Benefits and limitations of genome-wide association studies. Nat. Rev. Genet. 2019, 20, 467–484. [Google Scholar] [CrossRef]
- Van Deerlin, V.M.; Sleiman, P.M.; Martinez-Lage, M.; Chen-Plotkin, A.; Wang, L.S.; Graff-Radford, N.R.; Dickson, D.W.; Rademakers, R.; Boeve, B.F.; Grossman, M.; et al. Common variants at 7p21 are associated with frontotemporal lobar degeneration with TDP-43 inclusions. Nat. Genet. 2010, 42, 234–249. [Google Scholar] [CrossRef]
- Finch, N.; Carrasquillo, M.M.; Baker, M.; Rutherford, N.J.; Coppola, G.; Dejesus-Hernandez, M.; Crook, R.; Hunter, T.; Ghidoni, R.; Benussi, L.; et al. TMEM106B regulates progranulin levels and the penetrance of FTLD in GRN mutation carriers. Neurology 2011, 76, 467–474. [Google Scholar] [CrossRef]
- Gallagher, M.D.; Posavi, M.; Huang, P.; Unger, T.L.; Berlyand, Y.; Gruenewald, A.L.; Chesi, A.; Manduchi, E.; Wells, A.D.; Grant, S.F.A.; et al. A Dementia-Associated Risk Variant near TMEM106B Alters Chromatin Architecture and Gene Expression. Am. J. Hum. Genet. 2017, 101, 643–663. [Google Scholar] [CrossRef] [PubMed]
- Pottier, C.; Zhou, X.; Perkerson, R.B., 3rd; Baker, M.; Jenkins, G.D.; Serie, D.J.; Ghidoni, R.; Benussi, L.; Binetti, G.; Lopez de Munain, A.; et al. Potential genetic modifiers of disease risk and age at onset in patients with frontotemporal lobar degeneration and GRN mutations: A genome-wide association study. Lancet Neurol. 2018, 17, 548–558. [Google Scholar] [CrossRef]
- Rhinn, H.; Abeliovich, A. Differential Aging Analysis in Human Cerebral Cortex Identifies Variants in TMEM106B and GRN that Regulate Aging Phenotypes. Cell Syst. 2017, 4, 404–415. [Google Scholar] [CrossRef]
- Ferrari, R.; Grassi, M.; Salvi, E.; Borroni, B.; Palluzzi, F.; Pepe, D.; D’Avila, F.; Padovani, A.; Archetti, S.; Rainero, I.; et al. A genome-wide screening and SNPs-to-genes approach to identify novel genetic risk factors associated with frontotemporal dementia. Neurobiol. Aging 2015, 36, 13–26. [Google Scholar] [CrossRef]
- Ferrari, R.; Hernandez, D.G.; Nalls, M.A.; Rohrer, J.D.; Ramasamy, A.; Kwok, J.B.; Dobson-Stone, C.; Brooks, W.S.; Schofield, P.R.; Halliday, G.M.; et al. Frontotemporal dementia and its subtypes: A genome-wide association study. Lancet Neurol. 2014, 13, 686–699. [Google Scholar] [CrossRef]
- Ferrari, R.; Wang, Y.; Vandrovcova, J.; Guelfi, S.; Witeolar, A.; Karch, C.M.; Schork, A.J.; Fan, C.C.; Brewer, J.B.; International FTD-Genomics Consortium (IFGC); et al. Genetic architecture of sporadic frontotemporal dementia and overlap with Alzheimer’s and Parkinson’s diseases. J. Neurol. Neurosurg. Psychiatry 2017, 88, 152–164. [Google Scholar] [CrossRef]
- Broce, I.; Karch, C.M.; Wen, N.; Fan, C.C.; Wang, Y.; Tan, C.H.; Kouri, N.; Ross, O.A.; Hoglinger, G.U.; Muller, U.; et al. Immune-related genetic enrichment in frontotemporal dementia: An analysis of genome-wide association studies. PLoS Med. 2018, 15, e1002487. [Google Scholar]
- Karch, C.M.; Wen, N.; Fan, C.C.; Yokoyama, J.S.; Kouri, N.; Ross, O.A.; Hoglinger, G.; Muller, U.; Ferrari, R.; Hardy, J.; et al. Selective Genetic Overlap Between Amyotrophic Lateral Sclerosis and Diseases of the Frontotemporal Dementia Spectrum. JAMA Neurol. 2018, 75, 860–875. [Google Scholar] [CrossRef]
- Zhang, M.; Ferrari, R.; Tartaglia, M.C.; Keith, J.; Surace, E.I.; Wolf, U.; Sato, C.; Grinberg, M.; Liang, Y.; Xi, Z.; et al. A C6orf10/LOC101929163 locus is associated with age of onset in C9orf72 carriers. Brain 2018, 141, 2895–2907. [Google Scholar] [CrossRef] [PubMed]
- Taskesen, E.; Mishra, A.; Van der Sluis, S.; Ferrari, R.; International FTD-Genomics Consortium (IFGC); Veldink, J.H.; Van Es, M.A.; Smit, A.B.; Posthuma, D.; Pijnenburg, Y. Susceptible genes and disease mechanisms identified in frontotemporal dementia and frontotemporal dementia with Amyotrophic Lateral Sclerosis by DNA-methylation and GWAS. Sci. Rep. 2017, 7, 8899. [Google Scholar] [CrossRef]
- Mishra, A.; Ferrari, R.; Heutink, P.; Hardy, J.; Pijnenburg, Y.; Posthuma, D.; International FTD-Genomics Consortium (IFGC). Gene-based association studies report genetic links for clinical subtypes of frontotemporal dementia. Brain 2017, 140, 1437–1446. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.A.; Wang, Q.; Davis-Turak, J.; Li, Y.; Karydas, A.M.; Hsu, S.C.; Sears, R.L.; Chatzopoulou, D.; Huang, A.Y.; Wojta, K.J.; et al. A multiancestral genome-wide exome array study of Alzheimer disease, frontotemporal dementia, and progressive supranuclear palsy. JAMA Neurol. 2015, 72, 414–422. [Google Scholar] [CrossRef]
- Simons, C.; Dyment, D.; Bent, S.J.; Crawford, J.; D’Hooghe, M.; Kohlschutter, A.; Venkateswaran, S.; Helman, G.; Poll-The, B.T.; Makowski, C.C.; et al. A recurrent de novo mutation in TMEM106B causes hypomyelinating leukodystrophy. Brain 2017, 140, 3105–3111. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Arshad, M.; Wang, W.; Zhao, D.; Xu, L.; Zhou, L. LRRK2 mediated Rab8a phosphorylation promotes lipid storage. Lipids Health Dis. 2018, 17, 34. [Google Scholar] [CrossRef]
- Ferrazza, R.; Cogo, S.; Melrose, H.; Bubacco, L.; Greggio, E.; Guella, G.; Civiero, L.; Plotegher, N. LRRK2 deficiency impacts ceramide metabolism in brain. Biochem. Biophys. Res. Commun. 2016, 478, 1141–1146. [Google Scholar] [CrossRef]
- Mahley, R.W. Central Nervous System Lipoproteins: ApoE and Regulation of Cholesterol Metabolism. Arterioscler. Thromb. Vasc. Biol. 2016, 36, 1305–1315. [Google Scholar] [CrossRef] [PubMed]
- Chacinska, A.; Koehler, C.M.; Milenkovic, D.; Lithgow, T.; Pfanner, N. Importing mitochondrial proteins: Machineries and mechanisms. Cell 2009, 138, 628–644. [Google Scholar] [CrossRef] [PubMed]
- Sudre, C.H.; Cardoso, M.J.; Frost, C.; Barnes, J.; Barkhof, F.; Fox, N.; Ourselin, S. APOE ε4 status is associated with white matter hyperintensities volume accumulation rate independent of AD diagnosis. Neurobiol. Aging 2017, 53, 67–75. [Google Scholar] [CrossRef]
- Miklossy, J.; Arai, T.; Guo, J.P.; Klegeris, A.; Yu, S.; McGeer, E.G.; McGeer, P.L. LRRK2 expression in normal and pathologic human brain and in human cell lines. J. Neuropathol. Exp. Neurol. 2006, 65, 953–963. [Google Scholar] [CrossRef]
- Lyall, D.M.; Harris, S.E.; Bastin, M.E.; Muñoz Maniega, S.; Murray, C.; Lutz, M.W.; Saunders, A.M.; Roses, A.D.; Valdés Hernández Mdel, C.; Royle, N.A.; et al. Alzheimer’s disease susceptibility genes APOE and TOMM40, and brain white matter integrity in the Lothian Birth Cohort 1936. Neurobiol. Aging 2014, 35, 25–33. [Google Scholar] [CrossRef]
- Ciani, M.; Benussi, L.; Bonvicini, C.; Ghidoni, R. Genome Wide Association Study and Next Generation Sequencing: A Glimmer of Light Toward New Possible Horizons in Frontotemporal Dementia Research. Front. Neurosci. 2019, 13, 506. [Google Scholar] [CrossRef]
- Xu, Y.; Liu, X.; Shen, J.; Tian, W.; Fang, R.; Li, B.; Ma, J.; Cao, L.; Chen, S.; Li, G.; et al. The Whole Exome Sequencing Clarifies the Genotype- Phenotype Correlations in Patients with Early-Onset Dementia. Aging Dis. 2018, 9, 696–705. [Google Scholar] [CrossRef]
- Boyle, E.A.; Li, Y.I.; Pritchard, J.K. An Expanded View of Complex Traits: From Polygenic to Omnigenic. Cell 2017, 169, 1177–1186. [Google Scholar] [CrossRef] [PubMed]
- Blauwendraat, C.; Wilke, C.; Simon-Sanchez, J.; Jansen, I.E.; Reifschneider, A.; Capell, A.; Haass, C.; Castillo-Lizardo, M.; Biskup, S.; Maetzler, W.; et al. The wide genetic landscape of clinical frontotemporal dementia: Systematic combined sequencing of 121 consecutive subjects. Genet. Med. 2018, 20, 240–249. [Google Scholar] [CrossRef]
- Breza, M.; Koutsis, G.; Karadima, G.; Potagas, C.; Kartanou, C.; Papageorgiou, S.G.; Paraskevas, G.P.; Kapaki, E.; Stefanis, L.; Panas, M. The different faces of the p. A53T alpha-synuclein mutation: A screening of Greek patients with parkinsonism and/or dementia. Neurosci. Lett. 2018, 672, 136–139. [Google Scholar] [CrossRef]
- Ramos-Campoy, O.; Antonell, A.; Falgas, N.; Balasa, M.; Borrego-Ecija, S.; Rodriguez-Santiago, B.; Datta, D.; Armengol, L.; Fernandez-Villullas, G.; Bosch, B.; et al. Screening of dementia genes by whole-exome sequencing in Spanish patients with early-onset dementia: Likely pathogenic, uncertain significance and risk variants. Neurobiol. Aging 2020, 93, e1–e9. [Google Scholar] [CrossRef]
- Pottier, C.; Bieniek, K.F.; Finch, N.; Van de Vorst, M.; Baker, M.; Perkersen, R.; Brown, P.; Ravenscroft, T.; Van Blitterswijk, M.; Nicholson, A.M.; et al. Whole-genome sequencing reveals important role for TBK1 and OPTN mutations in frontotemporal lobar degeneration without motor neuron disease. Acta Neuropathol. 2015, 130, 77–92. [Google Scholar] [CrossRef] [PubMed]
- Pottier, C.; Ren, Y.; Perkerson, R.B., 3rd; Baker, M.; Jenkins, G.D.; Van Blitterswijk, M.; DeJesus-Hernandez, M.; Van Rooij, J.G.J.; Murray, M.E.; Christopher, E.; et al. Genome-wide analyses as part of the international FTLD-TDP whole-genome sequencing consortium reveals novel disease risk factors and increases support for immune dysfunction in FTLD. Acta Neuropathol. 2019, 137, 879–899. [Google Scholar] [CrossRef]
- Williams, K.L.; Topp, S.; Yang, S.; Smith, B.; Fifita, J.A.; Warraich, S.T.; Zhang, K.Y.; Farrawell, N.; Vance, C.; Hu, X.; et al. CCNF mutations in amyotrophic lateral sclerosis and frontotemporal dementia. Nat. Commun. 2016, 7, 11253. [Google Scholar] [CrossRef] [PubMed]
- Saracino, D.; Clot, F.; Camuzat, A.; Anquetil, V.; Hannequin, D.; Guyant-Marechal, L.; Didic, M.; Guillot-Noel, L.; Rinaldi, D.; Latouche, M.; et al. Novel VCP mutations expand the mutational spectrum of frontotemporal dementia. Neurobiol. Aging 2018, 72, e11–e14. [Google Scholar] [CrossRef]
- Philtjens, S.; Van Mossevelde, S.; Van der Zee, J.; Wauters, E.; Dillen, L.; Vandenbulcke, M.; Vandenberghe, R.; Ivanoiu, A.; Sieben, A.; Willems, C.; et al. Rare nonsynonymous variants in SORT1 are associated with increased risk for frontotemporal dementia. Neurobiol. Aging 2018, 66, e3–e10. [Google Scholar] [CrossRef]
- Bonvicini, C.; Scassellati, C.; Benussi, L.; Di Maria, E.; Maj, C.; Ciani, M.; Fostinelli, S.; Mega, A.; Bocchetta, M.; Lanzi, G.; et al. Next Generation Sequencing Analysis in Early Onset Dementia Patients. J. Alzheimer’s Dis. 2019, 67, 243–256. [Google Scholar] [CrossRef]
- Ng, A.S.L.; Tan, Y.J.; Yi, Z.; Tandiono, M.; Chew, E.; Dominguez, J.; Macas, M.; Ng, E.; Hameed, S.; Ting, S.; et al. Targeted exome sequencing reveals homozygous TREM2 R47C mutation presenting with behavioral variant frontotemporal dementia without bone involvement. Neurobiol. Aging 2018, 68, e15–e19. [Google Scholar] [CrossRef] [PubMed]
- Van der Zee, J.; Van Langenhove, T.; Kovacs, G.G.; Dillen, L.; Deschamps, W.; Engelborghs, S.; Matej, R.; Vandenbulcke, M.; Sieben, A.; Dermaut, B.; et al. Rare mutations in SQSTM1 modify susceptibility to frontotemporal lobar degeneration. Acta Neuropathol. 2014, 128, 397–410. [Google Scholar] [CrossRef] [PubMed]
- Stockl, M.; Fischer, P.; Wanker, E.; Herrmann, A. Alpha-synuclein selectively binds to anionic phospholipids embedded in liquid-disordered domains. J. Mol. Biol. 2008, 375, 1394–1404. [Google Scholar] [CrossRef] [PubMed]
- Bodner, C.R.; Dobson, C.M.; Bax, A. Multiple tight phospholipid-binding modes of alpha-synuclein revealed by solution NMR spectroscopy. J. Mol. Biol. 2009, 390, 775–790. [Google Scholar] [CrossRef]
- Bodner, C.R.; Maltsev, A.S.; Dobson, C.M.; Bax, A. Differential phospholipid binding of alpha-synuclein variants implicated in Parkinson’s disease revealed by solution NMR spectroscopy. Biochemistry 2010, 49, 862–871. [Google Scholar] [CrossRef]
- Quazi, F.; Molday, R.S. Differential phospholipid substrates and directional transport by ATP-binding cassette proteins ABCA1, ABCA7, and ABCA4 and disease-causing mutants. J. Biol. Chem. 2013, 288, 34414–34426. [Google Scholar] [CrossRef] [PubMed]
- Lynch, D.S.; Rodrigues Brandao de Paiva, A.; Zhang, W.J.; Bugiardini, E.; Freua, F.; Tavares Lucato, L.; Macedo-Souza, L.I.; Lakshmanan, R.; Kinsella, J.A.; Merwick, A.; et al. Clinical and genetic characterization of leukoencephalopathies in adults. Brain 2017, 140, 1204–1211. [Google Scholar] [CrossRef]
- Parikh, S.; Bernard, G.; Leventer, R.J.; Van der Knaap, M.S.; Van Hove, J.; Pizzino, A.; McNeill, N.H.; Helman, G.; Simons, C.; Schmidt, J.L.; et al. A clinical approach to the diagnosis of patients with leukodystrophies and genetic leukoencephelopathies. Mol. Genet. Metab. 2015, 114, 501–515. [Google Scholar] [CrossRef]
- Schneider, A.; Hasan, A.; Hirschel, S.; Wilhelm, C.; Kohlhase, J.; Falkai, P.; Gartner, J.; Steinfeld, R.; Wobrock, T.; Degner, D. A novel mutation of the arylsulfatase A gene in late-onset metachromatic leukodystrophy. J. Clin. Psychiatry 2009, 70, 1724–1725. [Google Scholar] [CrossRef]
- Stoeck, K.; Psychogios, M.N.; Ohlenbusch, A.; Steinfeld, R.; Schmidt, J. Late-Onset Metachromatic Leukodystrophy with Early Onset Dementia Associated with a Novel Missense Mutation in the Arylsulfatase A Gene. J. Alzheimer’s Dis. 2016, 51, 683–687. [Google Scholar] [CrossRef]
- Gore, E.; Manley, A.; Dees, D.; Appleby, B.S.; Lerner, A.J. A young-onset frontal dementia with dramatic calcifications due to a novel CSF1R mutation. Neurocase 2016, 22, 257–262. [Google Scholar] [CrossRef]
- Kawakami, I.; Iseki, E.; Kasanuki, K.; Minegishi, M.; Sato, K.; Hino, H.; Shibuya, K.; Fujisawa, K.; Higashi, S.; Akiyama, H.; et al. A family with hereditary diffuse leukoencephalopathy with spheroids caused by a novel c.2442+2T>C mutation in the CSF1R gene. J. Neurol. Sci. 2016, 367, 349–355. [Google Scholar] [CrossRef] [PubMed]
- Giraldo, M.; Lopera, F.; Siniard, A.L.; Corneveaux, J.J.; Schrauwen, I.; Carvajal, J.; Munoz, C.; Ramirez-Restrepo, M.; Gaiteri, C.; Myers, A.J.; et al. Variants in triggering receptor expressed on myeloid cells 2 are associated with both behavioral variant frontotemporal lobar degeneration and Alzheimer’s disease. Neurobiol. Aging 2013, 34, e11–e18. [Google Scholar] [CrossRef] [PubMed]
- Guerreiro, R.J.; Lohmann, E.; Bras, J.M.; Gibbs, J.R.; Rohrer, J.D.; Gurunlian, N.; Dursun, B.; Bilgic, B.; Hanagasi, H.; Gurvit, H.; et al. Using exome sequencing to reveal mutations in TREM2 presenting as a frontotemporal dementia-like syndrome without bone involvement. JAMA Neurol. 2013, 70, 78–84. [Google Scholar] [CrossRef] [PubMed]
- Borroni, B.; Ferrari, F.; Galimberti, D.; Nacmias, B.; Barone, C.; Bagnoli, S.; Fenoglio, C.; Piaceri, I.; Archetti, S.; Bonvicini, C.; et al. Heterozygous TREM2 mutations in frontotemporal dementia. Neurobiol. Aging 2014, 35, e7–e10. [Google Scholar] [CrossRef] [PubMed]
- Giannoccaro, M.P.; Bartoletti-Stella, A.; Piras, S.; Pession, A.; De Massis, P.; Oppi, F.; Stanzani-Maserati, M.; Pasini, E.; Baiardi, S.; Avoni, P.; et al. Multiple variants in families with amyotrophic lateral sclerosis and frontotemporal dementia related to C9orf72 repeat expansion: Further observations on their oligogenic nature. J. Neurol. 2017, 264, 1426–1433. [Google Scholar] [CrossRef] [PubMed]
- Alexander, S.K.; Brown, J.M.; Graham, A.; Nestor, P.J. CADASIL presenting with a behavioural variant frontotemporal dementia phenotype. J. Clin. Neurosci. 2014, 21, 165–167. [Google Scholar] [CrossRef]
- Sugama, S.; Kimura, A.; Chen, W.; Kubota, S.; Seyama, Y.; Taira, N.; Eto, Y. Frontal lobe dementia with abnormal cholesterol metabolism and heterozygous mutation in sterol 27-hydroxylase gene (CYP27). J. Inherit. Metab. Dis. 2001, 24, 379–392. [Google Scholar] [CrossRef]
- Sirkis, D.W.; Geier, E.G.; Bonham, L.W.; Karch, C.M.; Yokoyama, J.S. Recent advances in the genetics of frontotemporal dementia. Curr. Genet. Med. Rep. 2019, 7, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Lang, C.M.; Fellerer, K.; Schwenk, B.M.; Kuhn, P.H.; Kremmer, E.; Edbauer, D.; Capell, A.; Haass, C. Membrane orientation and subcellular localization of transmembrane protein 106B (TMEM106B), a major risk factor for frontotemporal lobar degeneration. J. Biol. Chem. 2012, 287, 19355–19365. [Google Scholar] [CrossRef]
- Brady, O.A.; Zheng, Y.; Murphy, K.; Huang, M.; Hu, F. The frontotemporal lobar degeneration risk factor, TMEM106B, regulates lysosomal morphology and function. Hum. Mol. Genet. 2013, 22, 685–695. [Google Scholar] [CrossRef]
- Chen-Plotkin, A.S.; Unger, T.L.; Gallagher, M.D.; Bill, E.; Kwong, L.K.; Volpicelli-Daley, L.; Busch, J.I.; Akle, S.; Grossman, M.; Van Deerlin, V.; et al. TMEM106B, the risk gene for frontotemporal dementia, is regulated by the microRNA-132/212 cluster and affects progranulin pathways. J. Neurosci. 2012, 32, 11213–11227. [Google Scholar] [CrossRef]
- Cruchaga, C.; Graff, C.; Chiang, H.H.; Wang, J.; Hinrichs, A.L.; Spiegel, N.; Bertelsen, S.; Mayo, K.; Norton, J.B.; Morris, J.C.; et al. Association of TMEM106B gene polymorphism with age at onset in granulin mutation carriers and plasma granulin protein levels. Arch. Neurol. 2011, 68, 581–586. [Google Scholar] [CrossRef] [PubMed]
- Van Blitterswijk, M.; Mullen, B.; Nicholson, A.M.; Bieniek, K.F.; Heckman, M.G.; Baker, M.C.; DeJesus-Hernandez, M.; Finch, N.A.; Brown, P.H.; Murray, M.E.; et al. TMEM106B protects C9ORF72 expansion carriers against frontotemporal dementia. Acta Neuropathol. 2014, 127, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Kubisiak, T.; Ji, H.; Xiao, J.; Wang, J.; Burmeister, M. The recurrent mutation in TMEM106B also causes hypomyelinating leukodystrophy in China and is a CpG hotspot. Brain 2018, 141, e36. [Google Scholar] [CrossRef]
- Ito, Y.; Hartley, T.; Baird, S.; Venkateswaran, S.; Simons, C.; Wolf, N.I.; Boycott, K.M.; Dyment, D.A.; Kernohan, K.D. Lysosomal dysfunction in TMEM106B hypomyelinating leukodystrophy. Neurol. Genet. 2018, 4, e288. [Google Scholar] [CrossRef]
- Luningschror, P.; Werner, G.; Stroobants, S.; Kakuta, S.; Dombert, B.; Sinske, D.; Wanner, R.; Lullmann-Rauch, R.; Wefers, B.; Wurst, W.; et al. The FTLD Risk Factor TMEM106B Regulates the Transport of Lysosomes at the Axon Initial Segment of Motoneurons. Cell Rep. 2020, 30, 3506–3519. [Google Scholar] [CrossRef]
- Zhou, X.; Nicholson, A.M.; Ren, Y.; Brooks, M.; Jiang, P.; Zuberi, A.; Phuoc, H.N.; Perkerson, R.B.; Matchett, B.; Parsons, T.M.; et al. Loss of TMEM106B leads to myelination deficits: Implications for frontotemporal dementia treatment strategies. Brain 2020, 143, 1905–1919. [Google Scholar] [CrossRef]
- Urbik, V.M.; Schmiedel, M.; Soderholm, H.; Bonkowsky, J.L. Expanded Phenotypic Definition Identifies Hundreds of Potential Causative Genes for Leukodystrophies and Leukoencephalopathies. Child Neurol. Open 2020, 7, 2329048. [Google Scholar]
- Bannwarth, S.; Ait-El-Mkadem, S.; Chaussenot, A.; Genin, E.C.; Lacas-Gervais, S.; Fragaki, K.; Berg-Alonso, L.; Kageyama, Y.; Serre, V.; Moore, D.G.; et al. A mitochondrial origin for frontotemporal dementia and amyotrophic lateral sclerosis through CHCHD10 involvement. Brain 2014, 137, 2329–2345. [Google Scholar] [CrossRef] [PubMed]
- Rademakers, R.; Baker, M.; Nicholson, A.M.; Rutherford, N.J.; Finch, N.; Soto-Ortolaza, A.; Lash, J.; Wider, C.; Wojtas, A.; DeJesus-Hernandez, M.; et al. Mutations in the colony stimulating factor 1 receptor (CSF1R) gene cause hereditary diffuse leukoencephalopathy with spheroids. Nat. Genet. 2011, 44, 200–205. [Google Scholar] [CrossRef]
- Tábuas-Pereira, M.; Santana, I.; Kun-Rodrigues, C.; Bras, J.; Guerreiro, R. CYLD variants in frontotemporal dementia associated with severe memory impairment in a Portuguese cohort. Brain 2020, 143, e67. [Google Scholar] [CrossRef]
- Ryan, N.S.; Biessels, G.J.; Kim, L.; Nicholas, J.M.; Barber, P.A.; Walsh, P.; Gami, P.; Morris, H.R.; Bastos-Leite, A.J.; Schott, J.M.; et al. Genetic determinants of white matter hyperintensities and amyloid angiopathy in familial Alzheimer’s disease. Neurobiol. Aging 2015, 36, 3140–3151. [Google Scholar] [CrossRef]
- Tu, P.H.; Galvin, J.E.; Baba, M.; Giasson, B.; Tomita, T.; Leight, S.; Nakajo, S.; Iwatsubo, T.; Trojanowski, J.Q.; Lee, V.M. Glial cytoplasmic inclusions in white matter oligodendrocytes of multiple system atrophy brains contain insoluble alpha-synuclein. Ann. Neurol. 1998, 44, 415–422. [Google Scholar] [CrossRef] [PubMed]
- Felsky, D.; Szeszko, P.; Yu, L.; Honer, W.G.; De Jager, P.L.; Schneider, J.A.; Malhotra, A.K.; Lencz, T.; Ikuta, T.; Pipitone, J.; et al. The SORL1 gene and convergent neural risk for Alzheimer’s disease across the human lifespan. Mol. Psychiatry 2014, 19, 1125–1132. [Google Scholar] [CrossRef][Green Version]
- Kovacs, G.G.; Van der Zee, J.; Hort, J.; Kristoferitsch, W.; Leitha, T.; Höftberger, R.; Ströbel, T.; Van Broeckhoven, C.; Matej, R. Clinicopathological description of two cases with SQSTM1 gene mutation associated with frontotemporal dementia. Neuropathology 2016, 36, 27–38. [Google Scholar] [CrossRef]
- Borroni, B.; Bonvicini, C.; Alberici, A.; Buratti, E.; Agosti, C.; Archetti, S.; Papetti, A.; Stuani, C.; Di Luca, M.; Gennarelli, M.; et al. Mutation within TARDBP leads to frontotemporal dementia without motor neuron disease. Hum. Mutat. 2009, 30, e974–e983. [Google Scholar] [CrossRef]
- Synofzik, M.; Maetzler, W.; Grehl, T.; Prudlo, J.; Vom Hagen, J.M.; Haack, T.; Rebassoo, P.; Munz, M.; Schöls, L.; Biskup, S. Screening in ALS and FTD patients reveals 3 novel UBQLN2 mutations outside the PXX domain and a pure FTD phenotype. Neurobiol. Aging 2012, 33, e13–e17. [Google Scholar] [CrossRef]
- Fahed, A.C.; McDonough, B.; Gouvion, C.M.; Newell, K.L.; Dure, L.S.; Bebin, M.; Bick, A.G.; Seidman, J.G.; Harter, D.H.; Seidman, C.E. UBQLN2 mutation causing heterogeneous X-linked dominant neurodegeneration. Ann. Neurol. 2014, 75, 793–798. [Google Scholar] [CrossRef]
- Yeh, F.L.; Hansen, D.V.; Sheng, M. TREM2, Microglia, and Neurodegenerative Diseases. Trends Mol. Med. 2017, 23, 512–533. [Google Scholar] [CrossRef] [PubMed]
- Paloneva, J.; Manninen, T.; Christman, G.; Hovanes, K.; Mandelin, J.; Adolfsson, R.; Bianchin, M.; Bird, T.; Miranda, R.; Salmaggi, A.; et al. Mutations in two genes encoding different subunits of a receptor signaling complex result in an identical disease phenotype. Am. J. Hum. Genet. 2002, 71, 656–662. [Google Scholar] [CrossRef] [PubMed]
- Dardiotis, E.; Siokas, V.; Pantazi, E.; Dardioti, M.; Rikos, D.; Xiromerisiou, G.; Markou, A.; Papadimitriou, D.; Speletas, M.; Hadjigeorgiou, G.M. A novel mutation in TREM2 gene causing Nasu-Hakola disease and review of the literature. Neurobiol. Aging 2017, 53, e13–e22. [Google Scholar] [CrossRef]
- Kaneko, M.; Sano, K.; Nakayama, J.; Amano, N. Nasu-Hakola disease: The first case reported by Nasu and review: The 50th Anniversary of Japanese Society of Neuropathology. Neuropathology 2010, 30, 463–470. [Google Scholar] [CrossRef] [PubMed]
- Chouery, E.; Delague, V.; Bergougnoux, A.; Koussa, S.; Serre, J.L.; Megarbane, A. Mutations in TREM2 lead to pure early-onset dementia without bone cysts. Hum. Mutat. 2008, 29, e194–e204. [Google Scholar] [CrossRef] [PubMed]
- Nugent, A.A.; Lin, K.; Van Lengerich, B.; Lianoglou, S.; Przybyla, L.; Davis, S.S.; Llapashtica, C.; Wang, J.; Kim, D.J.; Xia, D.; et al. TREM2 Regulates Microglial Cholesterol Metabolism upon Chronic Phagocytic Challenge. Neuron 2020, 105, 837–854. [Google Scholar] [CrossRef]
- Konno, T.; Kasanuki, K.; Ikeuchi, T.; Dickson, D.W.; Wszolek, Z.K. CSF1R-related leukoencephalopathy: A major player in primary microgliopathies. Neurology 2018, 91, 1092–1104. [Google Scholar] [CrossRef] [PubMed]
- Oosterhof, N.; Kuil, L.E.; Van der Linde, H.C.; Burm, S.M.; Berdowski, W.; Van Ijcken, W.F.J.; Van Swieten, J.C.; Hol, E.M.; Verheijen, M.H.G.; Van Ham, T.J. Colony-Stimulating Factor 1 Receptor (CSF1R) Regulates Microglia Density and Distribution, but Not Microglia Differentiation In Vivo. Cell Rep. 2018, 24, 1203–1217. [Google Scholar] [CrossRef]
- Sundal, C.; Fujioka, S.; Van Gerpen, J.A.; Wider, C.; Nicholson, A.M.; Baker, M.; Shuster, E.A.; Aasly, J.; Spina, S.; Ghetti, B.; et al. Parkinsonian features in hereditary diffuse leukoencephalopathy with spheroids (HDLS) and CSF1R mutations. Parkinsonism Relat. Disord. 2013, 19, 869–877. [Google Scholar] [CrossRef]
- Wong, J.C.; Chow, T.W.; Hazrati, L.N. Adult-onset leukoencephalopathy with axonal spheroids and pigmented glia can present as frontotemporal dementia syndrome. Dement. Geriatr. Cogn. Disord. 2011, 32, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.L.; Suh, E.; Wood, E.M.; Lee, E.B.; Coslett, H.B.; Raible, K.; Lee, V.M.; Trojanowski, J.Q.; Van Deerlin, V.M. Common neuropathological features underlie distinct clinical presentations in three siblings with hereditary diffuse leukoencephalopathy with spheroids caused by CSF1R p. Arg782His. Acta Neuropathol. Commun. 2015, 3, 42. [Google Scholar] [CrossRef] [PubMed]
- Kraya, T.; Quandt, D.; Pfirrmann, T.; Kindermann, A.; Lampe, L.; Schroeter, M.L.; Kohlhase, J.; Stoevesandt, D.; Hoffmann, K.; Villavicencio-Lorini, P. Functional characterization of a novel CSF1R mutation causing hereditary diffuse leukoencephalopathy with spheroids. Mol. Genet. Genomic. Med. 2019, 7, e00595. [Google Scholar] [CrossRef] [PubMed]
- Nie, S.; Chen, G.; Cao, X.; Zhang, Y. Cerebrotendinous xanthomatosis: A comprehensive review of pathogenesis, clinical manifestations, diagnosis, and management. Orphanet. J. Rare Dis. 2014, 9, 179. [Google Scholar] [CrossRef] [PubMed]
- Franklin, R.J.; Ffrench-Constant, C. Remyelination in the CNS: From biology to therapy. Nat. Rev. Neurosci. 2008, 9, 839–855. [Google Scholar] [CrossRef] [PubMed]
- Gibson, E.M.; Purger, D.; Mount, C.W.; Goldstein, A.K.; Lin, G.L.; Wood, L.S.; Inema, I.; Miller, S.E.; Bieri, G.; Zuchero, J.B.; et al. Neuronal activity promotes oligodendrogenesis and adaptive myelination in the mammalian brain. Science 2014, 344, 1252304. [Google Scholar] [CrossRef] [PubMed]
- Llorens-Bobadilla, E.; Chell, J.M.; Le Merre, P.; Wu, Y.; Zamboni, M.; Bergenstråhle, J.; Stenudd, M.; Sopova, E.; Lundeberg, J.; Shupliakov, O.; et al. A latent lineage potential in resident neural stem cells enables spinal cord repair. Science 2020, 370, 6512. [Google Scholar] [CrossRef]
- Klein, Z.A.; Takahashi, H.; Ma, M.; Stagi, M.; Zhou, M.; Lam, T.T.; Strittmatter, S.M. Loss of TMEM106B Ameliorates Lysosomal and Frontotemporal Dementia-Related Phenotypes in Progranulin-Deficient Mice. Neuron 2017, 95, 281–296. [Google Scholar] [CrossRef]
- Nicholson, A.M.; Zhou, X.; Perkerson, R.B.; Parsons, T.M.; Chew, J.; Brooks, M.; DeJesus-Hernandez, M.; Finch, N.A.; Matchett, B.J.; Kurti, A.; et al. Loss of Tmem106b is unable to ameliorate frontotemporal dementia-like phenotypes in an AAV mouse model of C9ORF72-repeat induced toxicity. Acta Neuropathol. Commun. 2018, 6, 42. [Google Scholar] [CrossRef] [PubMed]
- Beckmann, N.; Giorgetti, E.; Neuhaus, A.; Zurbruegg, S.; Accart, N.; Smith, P.; Perdoux, J.; Perrot, L.; Nash, M.; Desrayaud, S.; et al. Brain region-specific enhancement of remyelination and prevention of demyelination by the CSF1R kinase inhibitor BLZ945. Acta Neuropathol. Commun. 2018, 6, 9. [Google Scholar] [CrossRef]
- Tahmasebi, F.; Pasbakhsh, P.; Mortezaee, K.; Madadi, S.; Barati, S.; Kashani, I.R. Effect of the CSF1R inhibitor PLX3397on remyelination of corpus callosum in a cuprizone-induced demyelination mouse model. J. Cell Biochem. 2019, 120, 10576–10586. [Google Scholar] [CrossRef] [PubMed]
- Cignarella, F.; Filipello, F.; Bollman, B.; Cantoni, C.; Locca, A.; Mikesell, R.; Manis, M.; Ibrahim, A.; Deng, L.; Benitez, B.A.; et al. TREM2 activation on microglia promotes myelin debris clearance and remyelination in a model of multiple sclerosis. Acta Neuropathol. 2020, 140, 513–534. [Google Scholar] [CrossRef]
- Cullen, C.L.; Young, K.M. How Does Transcranial Magnetic Stimulation Influence Glial Cells in the Central Nervous System? Front. Neural. Circuits 2016, 10, 26. [Google Scholar] [CrossRef]
- Choi, E.H.; Blasiak, A.; Lee, J.; Yang, I.H. Modulation of Neural Activity for Myelination in the Central Nervous System. Front. Neurosci. 2019, 13, 952. [Google Scholar] [CrossRef]
- Prasad, A.; Teh, D.B.L.; Blasiak, A.; Chai, C.; Wu, Y.; Gharibani, P.M.; Yang, I.H.; Phan, T.T.; Lim, K.L.; Yang, H.; et al. Static Magnetic Field Stimulation Enhances Oligodendrocyte Differentiation and Secretion of Neurotrophic Factors. Sci. Rep. 2017, 7, 6743. [Google Scholar] [CrossRef] [PubMed]
- Dolgova, N.; Wei, Z.; Spink, B.; Gui, L.; Hua, Q.; Truong, D.; Zhang, Z.; Zhang, Y. Low-Field Magnetic Stimulation Accelerates the Differentiation of Oligodendrocyte Precursor Cells via Non-canonical TGF-beta Signaling Pathways. Mol. Neurobiol. 2020, 58, 855–866. [Google Scholar] [CrossRef] [PubMed]
- Cullen, C.L.; Senesi, M.; Tang, A.D.; Clutterbuck, M.T.; Auderset, L.; O’Rourke, M.E.; Rodger, J.; Young, K.M. Low-intensity transcranial magnetic stimulation promotes the survival and maturation of newborn oligodendrocytes in the adult mouse brain. Glia 2019, 67, 1462–1477. [Google Scholar] [CrossRef] [PubMed]
- Chalfouh, C.; Guillou, C.; Hardouin, J.; Delarue, Q.; Li, X.; Duclos, C.; Schapman, D.; Marie, J.P.; Cosette, P.; Guerout, N. The Regenerative Effect of Trans-spinal Magnetic Stimulation after Spinal Cord Injury: Mechanisms and Pathways Underlying the Effect. Neurotherapeutics 2020, 17, 2069–2088. [Google Scholar] [CrossRef] [PubMed]
- Tergau, F.; Naumann, U.; Paulus, W.; Steinhoff, B.J. Low-frequency repetitive transcranial magnetic stimulation improves intractable epilepsy. Lancet 1999, 353, 2209. [Google Scholar] [CrossRef]
- Randver, R. Repetitive transcranial magnetic stimulation of the dorsolateral prefrontal cortex to alleviate depression and cognitive impairment associated with Parkinson’s disease: A review and clinical implications. J. Neurol. Sci. 2018, 393, 88–99. [Google Scholar] [CrossRef]
- Rajji, T.K. Transcranial Magnetic and Electrical Stimulation in Alzheimer’s Disease and Mild Cognitive Impairment: A Review of Randomized Controlled Trials. Clin. Pharmacol. Ther. 2019, 106, 776–780. [Google Scholar] [CrossRef]
- Fang, J.; Zhou, M.; Yang, M.; Zhu, C.; He, L. Repetitive transcranial magnetic stimulation for the treatment of amyotrophic lateral sclerosis or motor neuron disease. Cochrane Database Syst. Rev. 2013, 5, CD008554. [Google Scholar] [CrossRef] [PubMed]
- Croarkin, P.E.; MacMaster, F.P. Transcranial Magnetic Stimulation for Adolescent Depression. Child Adolesc. Psychiatr. Clin. N. Am. 2019, 28, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Gaynes, B.N.; Lloyd, S.W.; Lux, L.; Gartlehner, G.; Hansen, R.A.; Brode, S.; Jonas, D.E.; Swinson Evans, T.; Viswanathan, M.; Lohr, K.N. Repetitive transcranial magnetic stimulation for treatment-resistant depression: A systematic review and meta-analysis. J. Clin. Psychiatry 2014, 75, 477–489. [Google Scholar] [CrossRef]
- Dougall, N.; Maayan, N.; Soares-Weiser, K.; McDermott, L.M.; McIntosh, A. Transcranial Magnetic Stimulation for Schizophrenia. Schizophr. Bull. 2015, 41, 1220–1222. [Google Scholar] [CrossRef]
- Mehta, U.M.; Naik, S.S.; Thanki, M.V.; Thirthalli, J. Investigational and Therapeutic Applications of Transcranial Magnetic Stimulation in Schizophrenia. Curr. Psychiatry Rep. 2019, 21, 89. [Google Scholar] [CrossRef]
- Kozel, F.A. Clinical Repetitive Transcranial Magnetic Stimulation for Posttraumatic Stress Disorder, Generalized Anxiety Disorder, and Bipolar Disorder. Psychiatr. Clin. N. Am. 2018, 41, 433–446. [Google Scholar] [CrossRef]
- Antal, A.; Alekseichuk, I.; Bikson, M.; Brockmoller, J.; Brunoni, A.R.; Chen, R.; Cohen, L.G.; Dowthwaite, G.; Ellrich, J.; Floel, A.; et al. Low intensity transcranial electric stimulation: Safety, ethical, legal regulatory and application guidelines. Clin. Neurophysiol. 2017, 128, 1774–1809. [Google Scholar] [CrossRef]
- Hadley, D.; Anderson, B.S.; Borckardt, J.J.; Arana, A.; Li, X.; Nahas, Z.; George, M.S. Safety, tolerability, and effectiveness of high doses of adjunctive daily left prefrontal repetitive transcranial magnetic stimulation for treatment-resistant depression in a clinical setting. J. ECT 2011, 27, 18–25. [Google Scholar] [CrossRef]
- Benussi, A.; Dell’Era, V.; Cosseddu, M.; Cantoni, V.; Cotelli, M.S.; Cotelli, M.; Manenti, R.; Benussi, L.; Brattini, C.; Alberici, A.; et al. Transcranial stimulation in frontotemporal dementia: A randomized, double-blind, sham-controlled trial. Alzheimer’s Dement. 2020, 6, e12033. [Google Scholar] [CrossRef] [PubMed]
| Genes | Protein | Function(s) | White Matter Pathology/Disease |
|---|---|---|---|
| TMEM106B [129,131] | Transmembrane protein 106B | Unknown | Hypomyelinating leukodystrophy |
| APOE [136,141] | Apolipoprotein E | Lipid Metabolism | White matter hyperintensities upon MRI [148] |
| LRRK2 [137] | Leucine-rich repeat kinase | Lipid Metabolism | Occasional LRRK2-immunopositive glia [149] |
| RAB38 [135] | Ras-related protein Rab-38 | Vesicle trafficking | Not described |
| CTSC [135] | Cathespin C | Activation of serine proteases in immune/inflammation | Not described |
| TOMM40 [141] | Translocase of the outer mitochondrial membrane complex | Mitochondrial protein transport | Lower white matter integrity upon MRI [150] |
| GFRA2 [132] | GDNF Family Receptor Alpha 2 | Cell surface receptor for glial cell line-derived neurotrophic factor and neurturin | Not described |
| Genes | Protein | Function | White Matter Pathology/Disease |
|---|---|---|---|
| AARS2 [37] | Alanyl-tRNA synthetase 2 | Translation | Leukodystrophy [191] |
| ABCA7 [156] | Phospholipid-transporting ATPase ABCA7 | Lipid transporter | Not described |
| CCNF [26] | Cyclin F | Cell cycle regulation | Not described |
| CHCHD10 [192] | Coiled-coil-helix-coiled-coil helix domain containing 10 | Mitochondrial function | Not described |
| CSF1R [193] | Colony-stimulating factor 1 receptor | Microglial function | Hereditary diffuse leukoencephalopathy [191] |
| CTSF [154] | Cathepsin F | Protein degradation | Leukoencephalopathy [191] |
| CYLD [194] | Ubiquitin carboxyl-terminal hydrolase CYLD | Autophagy, neuroinflammation | Widespread glia with CYLD-immunopositivity [15] |
| CYP27A1 [154] | Cytochrome P450 family 27 subfamily A member 1 | Cholesterol metabolism | Cerebrotendinous Xanthomatosis [191] |
| LRRK2 [157] | Leucine-rich repeat kinase 2 | Lipid metabolism | Occasional glia with LRRK2-immunopositivity [149] |
| OPTN [157] | Optineurin | Autophagy, membrane trafficking, cell cycle control, vesicle transport, NF-kB regulation | Not described |
| PNF1 [157] | Profilin1 | Regulation of actin polymerisation | Not described |
| PSEN1 [154] | Presenilin 1 | Proteolysis | Increased white matter hyperintensities [195] |
| PSEN2 [154] | Presenilin 2 | Proteolysis | Not described |
| SNCA [155] | Alpha-synuclein | Neuroprotection, neuronal differentiation, dopamine biosynthesis, maintenance of polysaturated fatty acids levels | Widespread oligodendrocytic inclusions in multiple system atrophy [196] |
| SORL1 [156] | Sortilin-related receptor 1 | Sorting and trafficking of intracellular proteins | Lower integrity of white matter tracts [197] |
| SORT1 [161] | Sortilin 1 | Protein trafficking; involved in glucose and lipid metabolism. | Not described |
| SQSTM1 [164] | Sequestosome/p62 | Autophagy | Widespread oligodendroglial pTDP-43 inclusions [198] |
| TARDBP [199] | TAR DNA-binding protein 43 | Transcription and RNA splicing | - |
| TREM2 [177] | Triggering receptor expressed on myeloid cells 2 | Activation of macrophages, microglia and dendritic cell | Nasu–Hakola disease/polycystic lipomembranous osteodysplasia with sclerosing leukoencephalopathy [191] |
| TYROBP [178] | Transmembrane immune signaling adaptor | Macrophages and dendritic cells activation. Microglia activation in the brain | Nasu–Hakola disease [191] |
| UBQLN2 [200] | Ubiquilin 2 | Protein degradation, cell cycle regulation | Widespread demylination of white matter [201] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lok, H.C.; Kwok, J.B. The Role of White Matter Dysfunction and Leukoencephalopathy/Leukodystrophy Genes in the Aetiology of Frontotemporal Dementias: Implications for Novel Approaches to Therapeutics. Int. J. Mol. Sci. 2021, 22, 2541. https://doi.org/10.3390/ijms22052541
Lok HC, Kwok JB. The Role of White Matter Dysfunction and Leukoencephalopathy/Leukodystrophy Genes in the Aetiology of Frontotemporal Dementias: Implications for Novel Approaches to Therapeutics. International Journal of Molecular Sciences. 2021; 22(5):2541. https://doi.org/10.3390/ijms22052541
Chicago/Turabian StyleLok, Hiu Chuen, and John B. Kwok. 2021. "The Role of White Matter Dysfunction and Leukoencephalopathy/Leukodystrophy Genes in the Aetiology of Frontotemporal Dementias: Implications for Novel Approaches to Therapeutics" International Journal of Molecular Sciences 22, no. 5: 2541. https://doi.org/10.3390/ijms22052541
APA StyleLok, H. C., & Kwok, J. B. (2021). The Role of White Matter Dysfunction and Leukoencephalopathy/Leukodystrophy Genes in the Aetiology of Frontotemporal Dementias: Implications for Novel Approaches to Therapeutics. International Journal of Molecular Sciences, 22(5), 2541. https://doi.org/10.3390/ijms22052541







