How miR-31-5p and miR-33a-5p Regulates SP1/CX43 Expression in Osteoarthritis Disease: Preliminary Insights
Abstract
1. Introduction
2. Results
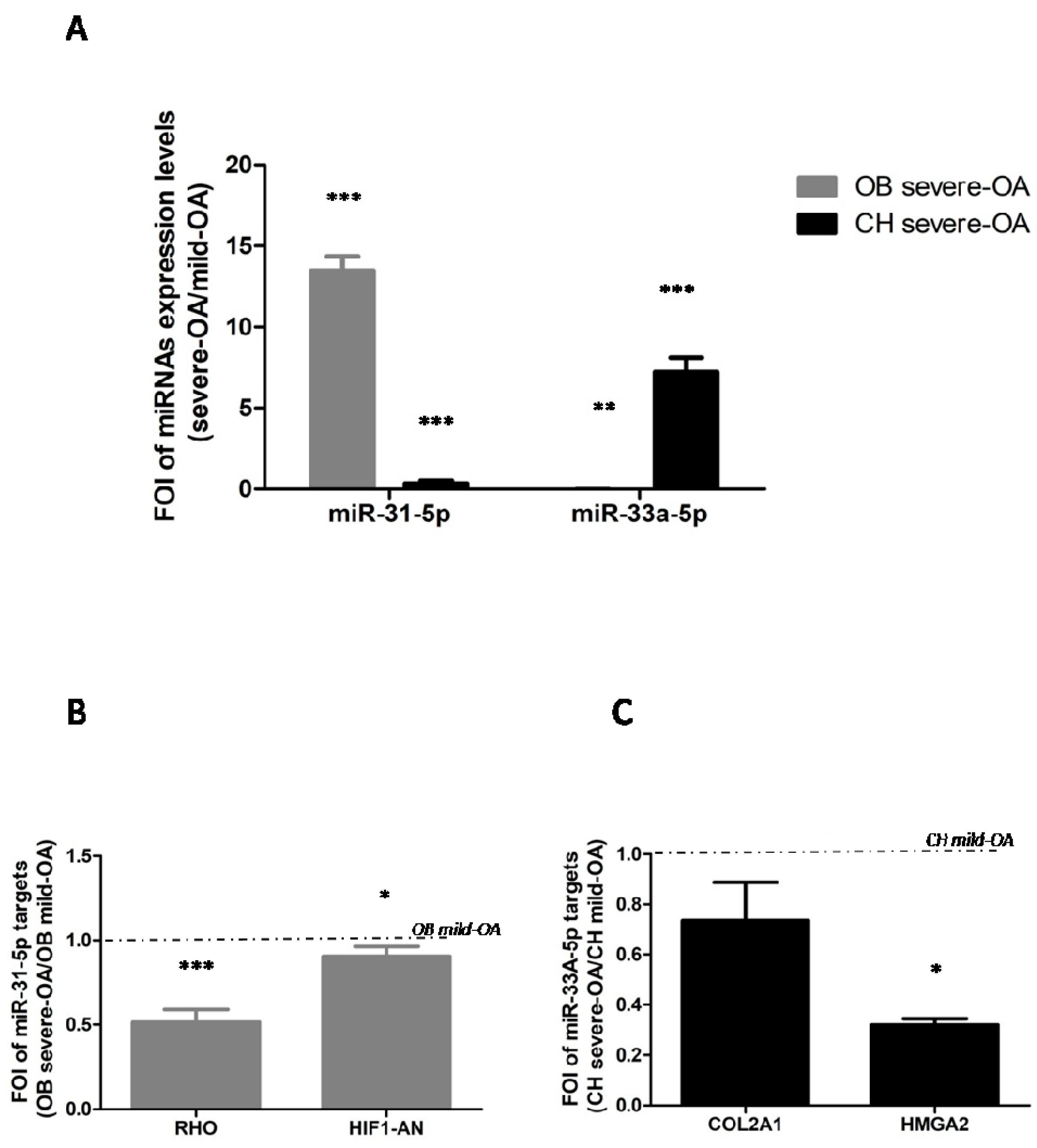
2.1. miRNAs Expression in OA Patients Derived Cells
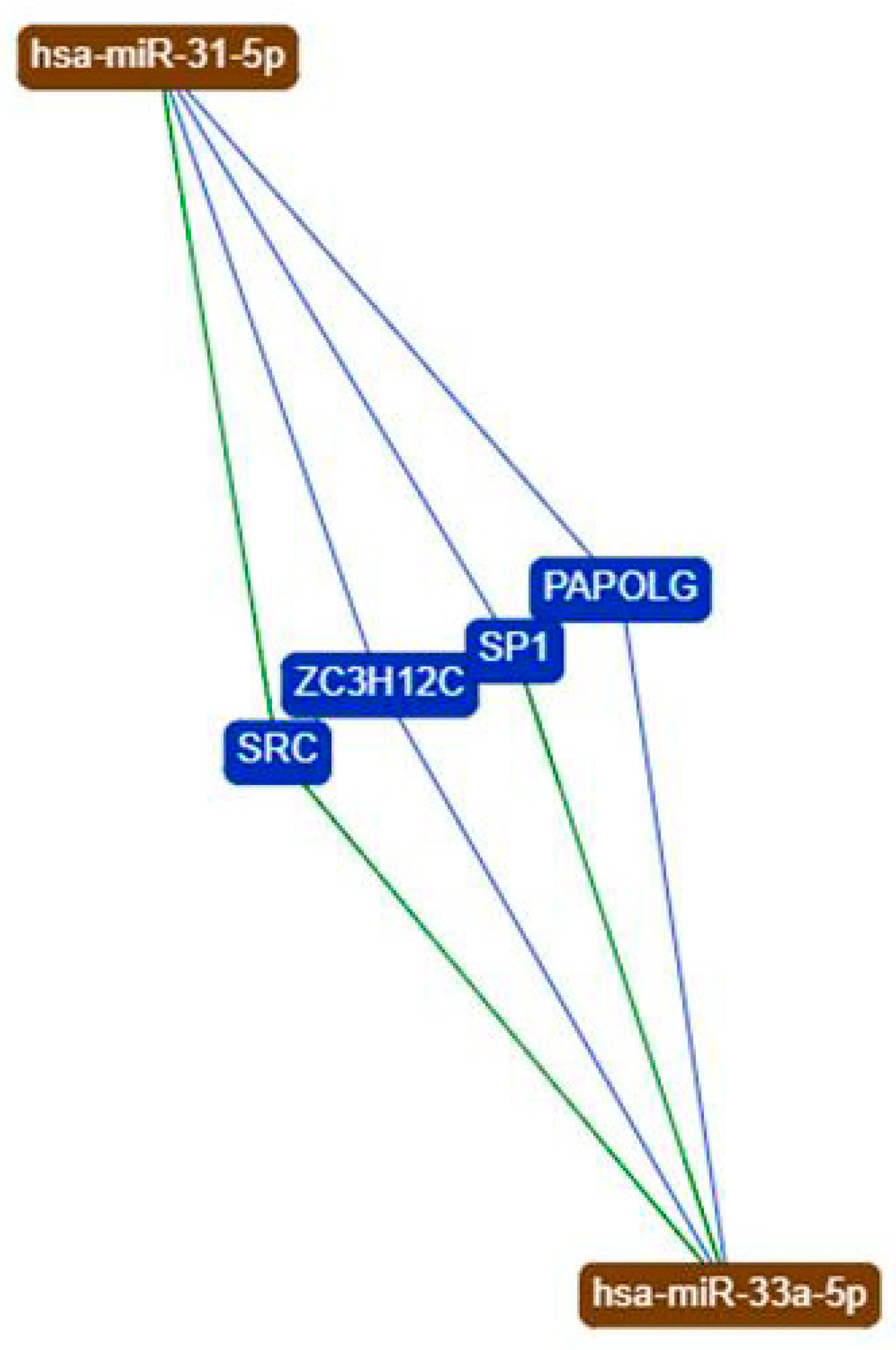
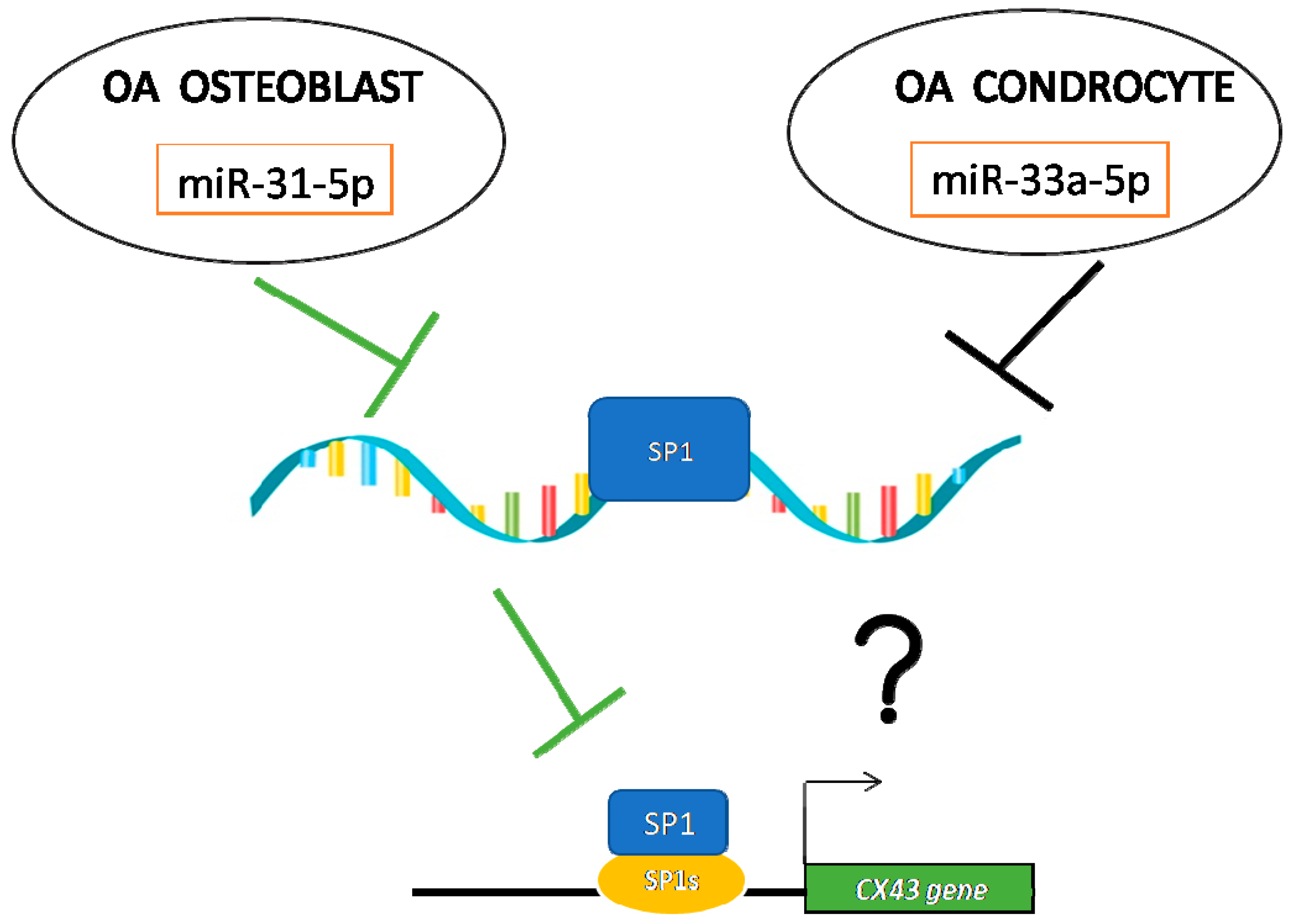
2.2. Investigation of miRNA31-5p and miR-33a-5p Common Targets
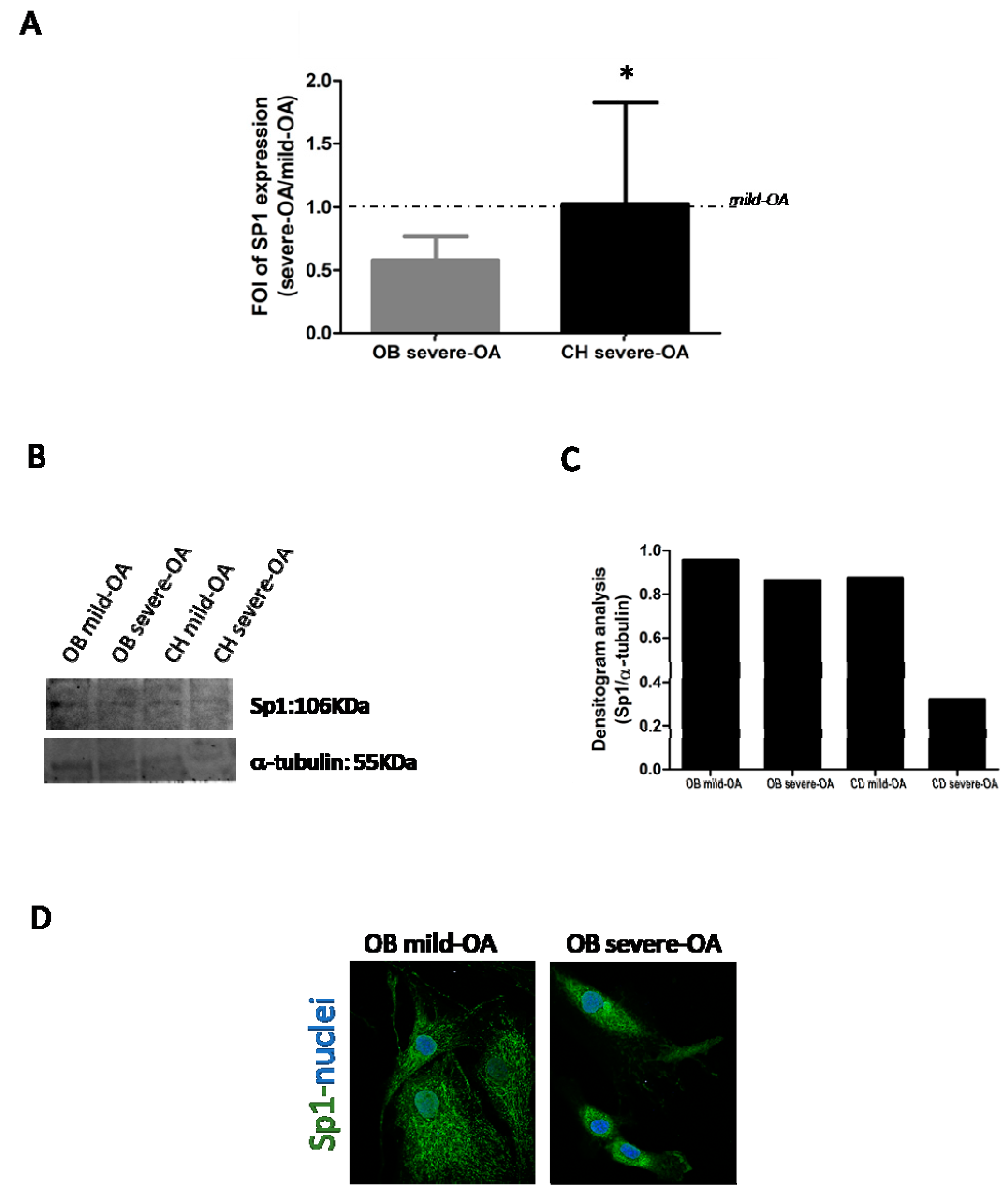
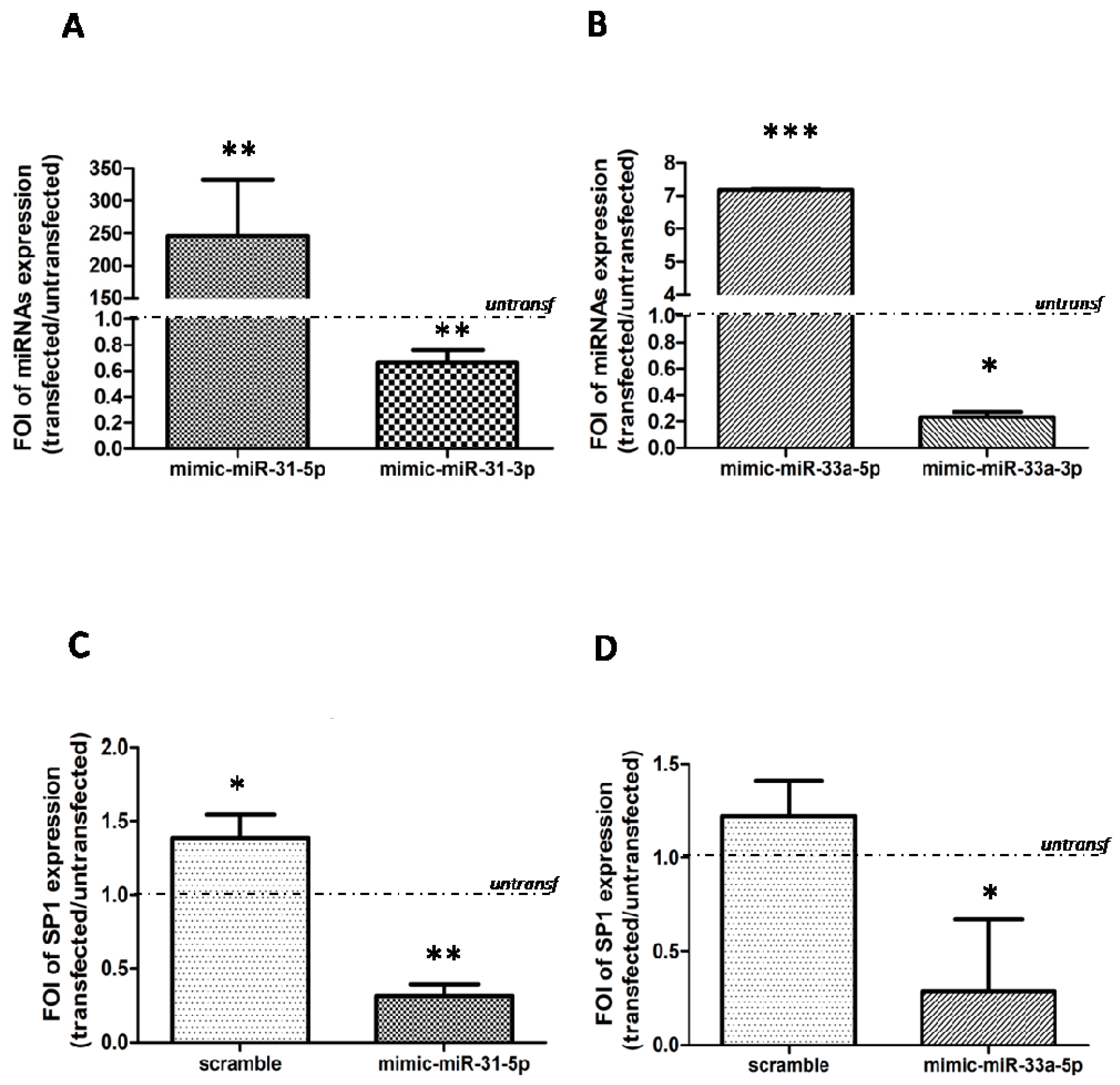
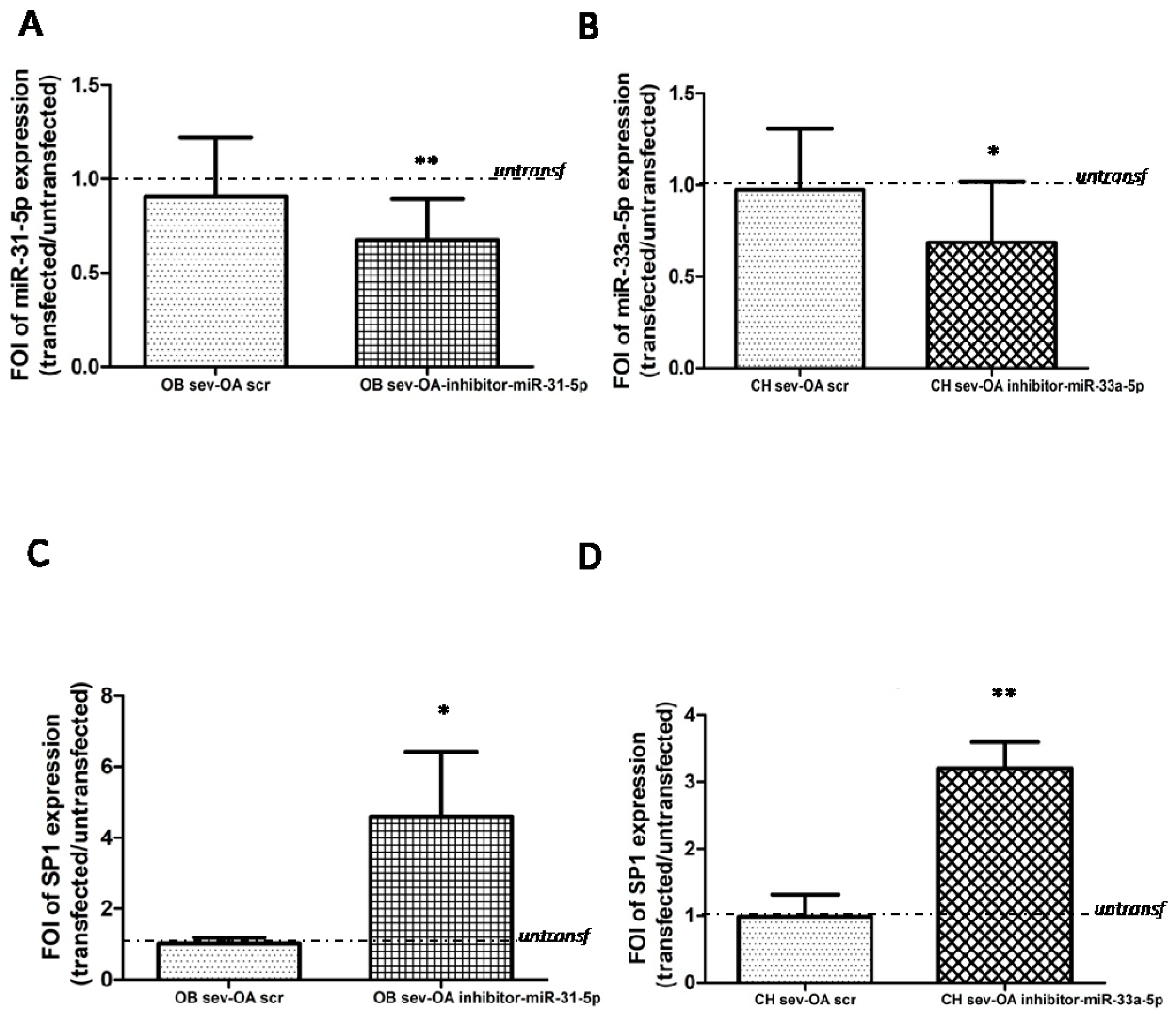
2.3. Modulation of SP-1 Expression in OA
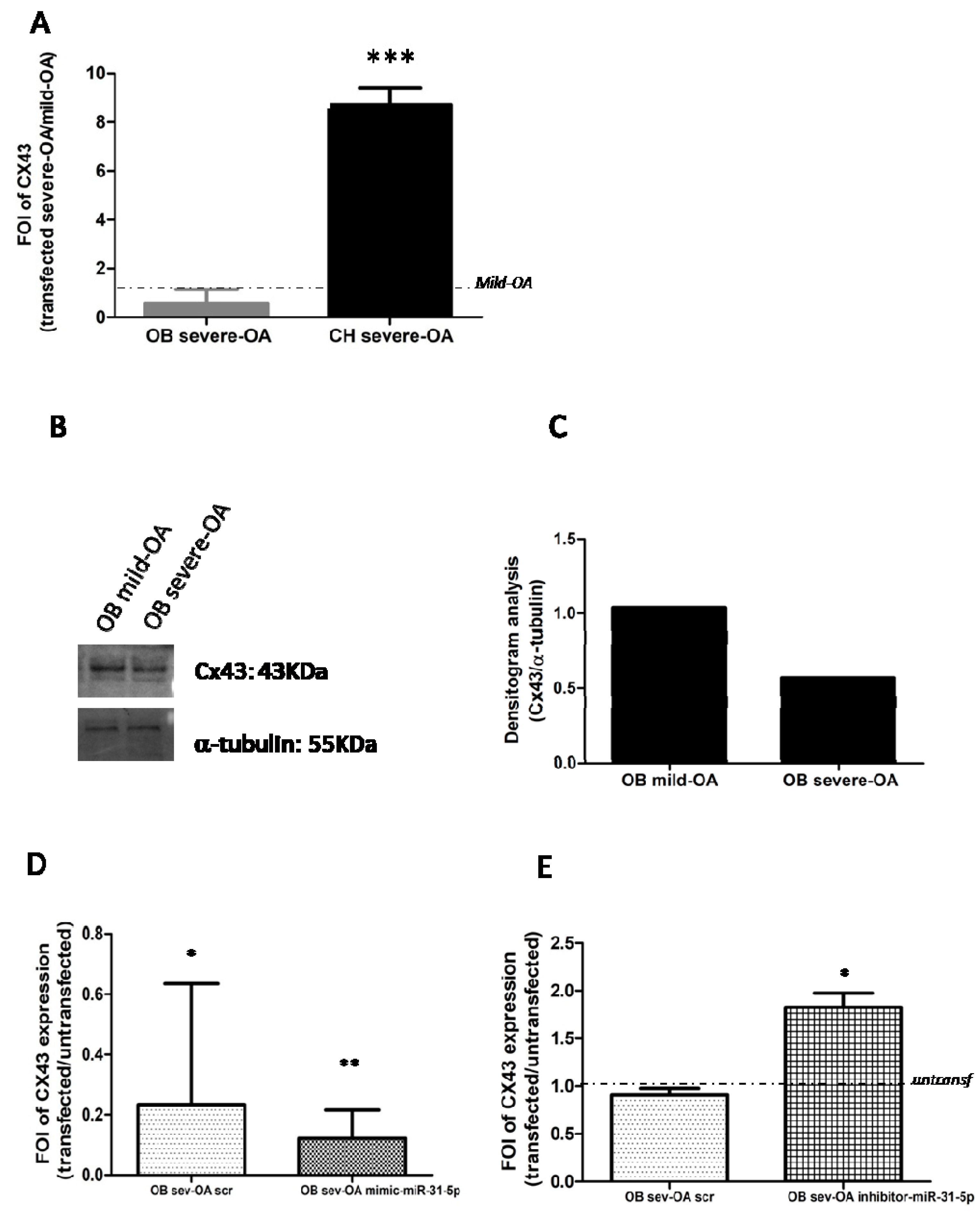
2.4. Connexin 43 Expression in OA Derived Cells
3. Discussion
4. Material and Methods
4.1. Cell Cultures and Reagents
4.2. Cell Transfection
4.3. RNA Extraction and Real-Time PCR
4.4. Western Blot Analysis
4.5. Immunofluorescence Analysis
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, S.; Wang, H.; Liu, W.; Wei, B. Identification of Key Genes and Pathways Associated with Sex Differences in Osteoarthritis Based on Bioinformatics Analysis. BioMed Res. Int. 2019, 2019, 3482751. [Google Scholar] [CrossRef]
- Hunziker, E. Articular cartilage repair: Basic science and clinical progress. A review of the current status and prospects. Osteoarthr. Cartil. 2002, 10, 432–463. [Google Scholar] [CrossRef]
- Pelletier, J.-P.; Martel-Pelletier, J.; Abramson, S.B. Osteoarthritis, an inflammatory disease: Potential implication for the selection of new therapeutic targets. Arthritis Rheum. 2001, 44, 1237–1247. [Google Scholar] [CrossRef]
- Gibon, E.; Córdova, L.A.; Lu, L.; Lin, T.-H.; Yao, Z.; Hamadouche, M.; Goodman, S.B. The biological response to orthopedic implants for joint replacement. II: Polyethylene, ceramics, PMMA, and the foreign body reaction. J. Biomed. Mater. Res. Part B Appl. Biomater. 2017, 105, 1685–1691. [Google Scholar] [CrossRef] [PubMed]
- Beavers, K.R.; Nelson, C.E.; Duvall, C.L. MiRNA inhibition in tissue engineering and regenerative medicine. Adv. Drug Deliv. Rev. 2015, 88, 123–137. [Google Scholar] [CrossRef] [PubMed]
- Murata, K.; Yoshitomi, H.; Tanida, S.; Ishikawa, M.; Nishitani, K.; Ito, H.; Nakamura, T. Plasma and synovial fluid microRNAs as potential biomarkers of rheumatoid arthritis and osteoarthritis. Arthritis Res. Ther. 2010, 12, R86. [Google Scholar] [CrossRef] [PubMed]
- Costa, V.; Raimondi, L.; Conigliaro, A.; Salamanna, F.; Carina, V.; De Luca, A.; Bellavia, D.; Alessandro, R.; Fini, M.; Giavaresi, G. Hypoxia-inducible factor 1Α may regulate the commitment of mesenchymal stromal cells toward angio-osteogenesis by mirna-675-5P. Cytotherapy 2017, 19, 1412–1425. [Google Scholar] [CrossRef] [PubMed]
- Costa, V.; Carina, V.; Conigliaro, A.; Raimondi, L.; De Luca, A.; Bellavia, D.; Salamanna, F.; Setti, S.; Alessandro, R.; Fini, M.; et al. miR-31-5p Is a LIPUS-Mechanosensitive MicroRNA that Targets HIF-1α Signaling and Cytoskeletal Proteins. Int. J. Mol. Sci. 2019, 20, 1569. [Google Scholar] [CrossRef]
- Costa, V.; Carina, V.; Raimondi, L.; De Luca, A.; Bellavia, D.; Conigliaro, A.; Salamanna, F.; Alessandro, R.; Fini, M.; Giavaresi, G. MiR-33a Controls hMSCS Osteoblast Commitment Modulating the Yap/Taz Expression Through EGFR Signaling Regulation. Cells 2019, 8, 1495. [Google Scholar] [CrossRef]
- Gu, H.; Wu, L.; Chen, H.; Huang, Z.; Xu, J.; Zhou, K.; Zhang, Y.; Chen, J.; Xia, J.; Yin, X. Identification of differentially expressed microRNAs in the bone marrow of osteoporosis patients. Am. J. Transl. Res. 2019, 11, 2940–2954. [Google Scholar]
- Ntoumou, E.; Tzetis, M.; Braoudaki, M.; Lambrou, G.; Poulou, M.; Malizos, K.; Stefanou, N.; Anastasopoulou, L.; Tsezou, A. Serum microRNA array analysis identifies miR-140-3p, miR-33b-3p and miR-671-3p as potential osteoarthritis biomarkers involved in metabolic processes. Clin. Epigenetics 2017, 9, 127. [Google Scholar] [CrossRef] [PubMed]
- Raimondo, S.; Urzì, O.; Conigliaro, A.; Bosco, G.L.; Parisi, S.; Carlisi, M.; Siragusa, S.; Raimondi, L.; De Luca, A.; Giavaresi, G.; et al. Extracellular Vesicle microRNAs Contribute to the Osteogenic Inhibition of Mesenchymal Stem Cells in Multiple Myeloma. Cancers 2020, 12, 449. [Google Scholar] [CrossRef]
- Yu, S.; Yerges-Armstrong, L.M.; Chu, Y.; Zmuda, J.M.; Zhang, Y. Transcriptional Regulation of Frizzled-1 in Human Osteoblasts by Sp1. PLoS ONE 2016, 11, e0163277. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Zhou, X.; Kunkel, G.; Zhang, Z.; Deng, J.M.; Behringer, R.R.; de Crombrugghe, B. The Novel Zinc Finger-Containing Transcription Factor Osterix Is Required for Osteoblast Differentiation and Bone Formation. Cell 2002, 108, 17–29. [Google Scholar] [CrossRef]
- Ortuño, M.J.; Susperregui, A.R.; Artigas, N.; Rosa, J.L.; Ventura, F. Osterix induces Col1a1 gene expression through binding to Sp1 sites in the bone enhancer and proximal promoter regions. Bone 2013, 52, 548–556. [Google Scholar] [CrossRef]
- Hantusch, B.; Kalt, R.; Krieger, S.; Puri, C.; Kerjaschki, D. Sp1/Sp3 and DNA-methylation contribute to basal transcriptional activation of human podoplanin in MG63 versus Saos-2 osteoblastic cells. BMC Mol. Biol. 2007, 8, 20. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yang, H.; Liu, W.; Cao, X.; Feng, X. Sp1 and Sp3 regulate the basal transcription of receptor activator of nuclear factor kappa B ligand gene in osteoblasts and bone marrow stromal cells. J. Cell. Biochem. 2005, 96, 716–727. [Google Scholar] [CrossRef]
- Goto, T.; Matsui, Y.; Fernandes, R.J.; Hanson, D.A.; Kubo, T.; Yukata, K.; Michigami, T.; Komori, T.; Fujita, T.; Yang, L.; et al. Sp1 family of transcription factors regulates the human alpha2 (XI) collagen gene (COL11A2) in Saos-2 osteoblastic cells. J. Bone Miner. Res. 2006, 21, 661–673. [Google Scholar] [CrossRef]
- Feng, X.; Teitelbaum, S.L.; Quiroz, M.E.; Cheng, S.-L.; Lai, C.-F.; Avioli, L.V.; Ross, F. Sp1/Sp3 and PU.1 Differentially Regulate β5Integrin Gene Expression in Macrophages and Osteoblasts. J. Biol. Chem. 2000, 275, 8331–8340. [Google Scholar] [CrossRef] [PubMed]
- Maneix, L.; Servent, A.; Porée, B.; Ollitrault, D.; Branly, T.; Bigot, N.; Boujrad, N.; Flouriot, G.; Demoor, M.; Boumediene, K.; et al. Up-regulation of type II collagen gene by 17β-estradiol in articular chondrocytes involves Sp1/3, Sox-9, and estrogen receptor α. J. Mol. Med. 2014, 92, 1179–1200. [Google Scholar] [CrossRef] [PubMed]
- Garnero, P.; Borel, O.; Grant, S.F.A.; Ralston, S.H.; Delmas, P.D. Collagen Iα1 Sp1 Polymorphism, Bone Mass, and Bone Turnover in Healthy French Premenopausal Women: The OFELY Study. J. Bone Miner. Res. 1998, 13, 813–817. [Google Scholar] [CrossRef]
- Mann, V.; Hobson, E.E.; Li, B.; Stewart, T.L.; Grant, S.F.; Robins, S.P.; Aspden, R.M.; Ralston, S.H. A COL1A1 Sp1 binding site polymorphism predisposes to osteoporotic fracture by affecting bone density and quality. J. Clin. Investig. 2001, 107, 899–907. [Google Scholar] [CrossRef]
- Loiselle, A.E.; Lloyd, S.A.J.; Paul, E.M.; Lewis, G.S.; Donahue, H.J. Inhibition of GSK-3β Rescues the Impairments in Bone Formation and Mechanical Properties Associated with Fracture Healing in Osteoblast Selective Connexin 43 Deficient Mice. PLoS ONE 2013, 8, e81399. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Buo, A.M.; Tomlinson, R.E.; Eidelman, E.R.; Chason, M.; Stains, J.P. Connexin43 and Runx2 Interact to Affect Cortical Bone Geometry, Skeletal Development, and Osteoblast and Osteoclast Function. J. Bone Miner. Res. 2017, 32, 1727–1738. [Google Scholar] [CrossRef]
- Moorer, M.C.; Hebert, C.; Tomlinson, R.E.; Iyer, S.R.; Chason, M.; Stains, J.P. Defective signaling, osteoblastogenesis and bone remodeling in a mouse model of connexin 43 C-terminal truncation. J. Cell Sci. 2017, 130, 531–540. [Google Scholar] [CrossRef]
- Bivi, N.; Condon, K.W.; Allen, M.R.; Farlow, N.; Passeri, G.; Brun, L.R.; Rhee, Y.; Bellido, T.; Plotkin, L.I. Cell autonomous requirement of connexin 43 for osteocyte survival: Consequences for endocortical resorption and periosteal bone formation. J. Bone Miner. Res. 2012, 27, 374–389. [Google Scholar] [CrossRef] [PubMed]
- Gago-Fuentes, R.; Fernández-Puente, P.; Megias, D.; Carpintero-Fernández, P.; Mateos, J.; Acea, B.; Fonseca, E.; Blanco, F.J.; Mayan, M.D. Proteomic Analysis of Connexin 43 Reveals Novel Interactors Related to Osteoarthritis. Mol. Cell. Proteom. 2015, 14, 1831–1845. [Google Scholar] [CrossRef] [PubMed]
- Stains, J.P.; Lecanda, F.; Screen, J.; Towler, D.A.; Civitelli, R. Gap Junctional Communication Modulates Gene Transcription by Altering the Recruitment of Sp1 and Sp3 to Connexin-response Elements in Osteoblast Promoters. J. Biol. Chem. 2003, 278, 24377–24387. [Google Scholar] [CrossRef]
- Raimondi, L.; De Luca, A.; Costa, V.; Amodio, N.; Carina, V.; Bellavia, D.; Tassone, P.; Pagani, S.; Fini, M.; Alessandro, R.; et al. Circulating biomarkers in osteosarcoma: New translational tools for diagnosis and treatment. Oncotarget 2017, 8, 100831–100851. [Google Scholar] [CrossRef] [PubMed]
- Bellavia, D.; De Luca, A.; Carina, V.; Costa, V.; Raimondi, L.; Salamanna, F.; Alessandro, R.; Fini, M.; Giavaresi, G. Deregulated miRNAs in bone health: Epigenetic roles in osteoporosis. Bone 2019, 122, 52–75. [Google Scholar] [CrossRef]
- Costa, V.; Dico, A.L.; Rizzo, A.; Rajata, F.; Tripodi, M.; Alessandro, R.; Conigliaro, A. MiR-675-5p supports hypoxia induced epithelial to mesenchymal transition in colon cancer cells. Oncotarget 2017, 8, 24292–24302. [Google Scholar] [CrossRef]
- Dico, A.L.; Costa, V.; Martelli, C.; Diceglie, C.; Rajata, F.; Rizzo, A.; Mancone, C.; Tripodi, M.; Ottobrini, L.; Alessandro, R.; et al. MiR675-5p Acts on HIF-1α to Sustain Hypoxic Responses: A New Therapeutic Strategy for Glioma. Theranostics 2016, 6, 1105–1118. [Google Scholar] [CrossRef]
- Negoro, H.; Okinami, T.; Kanematsu, A.; Imamura, M.; Tabata, Y.; Ogawa, O. Role of Rev-erbα domains for transactivation of the connexin43 promoter with Sp1. FEBS Lett. 2013, 587, 98–103. [Google Scholar] [CrossRef]
- Mayan, M.D.; Carpintero-Fernandez, P.; Gago-Fuentes, R.; Martinez-de-Ilarduya, O.; Wang, H.Z.; Valiunas, V.; Brink, P.; Blanco, F.J. Human articular chondrocytes express multiple gap junction proteins: Differential expression of connexins in normal and osteoarthritic cartilage. Am. J. Pathol. 2013, 182, 1337–1346. [Google Scholar] [CrossRef] [PubMed]
- Plotkin, L.I.; Speacht, T.L.; Donahue, H.J. Cx43 and mechanotransduction in bone. Curr. Osteoporos. Rep. 2015, 13, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Kohn, M.D.; Sassoon, A.A.; Fernando, N.D. Classifications in Brief: Kellgren-Lawrence Classification of Osteoarthritis. Clin. Orthop. Relat. Res. 2016, 474, 1886–1893. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: http://www.r-project.org (accessed on 27 February 2021).
| Group Severe OA | Group Mild OA | |||
|---|---|---|---|---|
| Woman | Man | Woman | Man | |
| Age (yrs) | 74 | 64 | 62 | 63 |
| Weight (kg) | 62 | 58 | 64 | 68 |
| BMI (kg/m²) | 25.8 | 24.1 | 26.0 | 23.8 |
| Kellgren and Lawrence grading [36] | 4 | 4 | 2 | 2 |
| Other pathologies | - | - | - | Diverticulosis |
| Therapy | Atenolol 50 mg/die | - | - | Olmesartan Medoxomil/Amlodipine, 20/5 mg/die |
| WBC (×10³/μL) | 6.76 | 6.85 | 5.45 | 8.42 |
| CRP (mg/L) | 0.17 | 0.34 | 0.30 | 0.20 |
| Gene | Primer Forward | Primer Reverse |
|---|---|---|
| RHOA “Transforming protein RhoA” | GAAAACCGGTGAATCTGCGC | AGAACACATCTGTTTGCGGA |
| HIF-1AN “Hypoxia-inducible factor 1-alpha inhibitor” | TGGGGGCAGCTTACCTCTAA | TGGGTAGAGGCACTCGAAC |
| HMGA-2 “High mobility group AT-hook 2” | GCGCCTCAGAAGAGAGGAC | GTCTTCCCCTGGGTCTCTTAG |
| COL2A1 “Collagen type II alpha 1 chain” | CCTGGCAAAGATGGTGAGACAG | CCTGGTTTTCCACCTTCACCTG |
| SP-1 ”Specific protein 1” | GCCTCCAGACCATTAACCTCAGT | GCTCCATGATCACCTGGGGCAT |
| CX-43 “Connexin43” | GAACTCAAGGTTGCCCAAAC | TTAGAGATGGTGCTTCCCG |
| Reference Gene | ||
| ACTB “Beta-Actin” | ATCAAGATCATTGCTCCTCCTGA | CTGCTTGCTGATCCACATCTG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Costa, V.; De Fine, M.; Carina, V.; Conigliaro, A.; Raimondi, L.; De Luca, A.; Bellavia, D.; Salamanna, F.; Alessandro, R.; Pignatti, G.; et al. How miR-31-5p and miR-33a-5p Regulates SP1/CX43 Expression in Osteoarthritis Disease: Preliminary Insights. Int. J. Mol. Sci. 2021, 22, 2471. https://doi.org/10.3390/ijms22052471
Costa V, De Fine M, Carina V, Conigliaro A, Raimondi L, De Luca A, Bellavia D, Salamanna F, Alessandro R, Pignatti G, et al. How miR-31-5p and miR-33a-5p Regulates SP1/CX43 Expression in Osteoarthritis Disease: Preliminary Insights. International Journal of Molecular Sciences. 2021; 22(5):2471. https://doi.org/10.3390/ijms22052471
Chicago/Turabian StyleCosta, Viviana, Marcello De Fine, Valeria Carina, Alice Conigliaro, Lavinia Raimondi, Angela De Luca, Daniele Bellavia, Francesca Salamanna, Riccardo Alessandro, Giovanni Pignatti, and et al. 2021. "How miR-31-5p and miR-33a-5p Regulates SP1/CX43 Expression in Osteoarthritis Disease: Preliminary Insights" International Journal of Molecular Sciences 22, no. 5: 2471. https://doi.org/10.3390/ijms22052471
APA StyleCosta, V., De Fine, M., Carina, V., Conigliaro, A., Raimondi, L., De Luca, A., Bellavia, D., Salamanna, F., Alessandro, R., Pignatti, G., Fini, M., & Giavaresi, G. (2021). How miR-31-5p and miR-33a-5p Regulates SP1/CX43 Expression in Osteoarthritis Disease: Preliminary Insights. International Journal of Molecular Sciences, 22(5), 2471. https://doi.org/10.3390/ijms22052471











