The Biochemistry of Phytocannabinoids and Metabolic Engineering of Their Production in Heterologous Systems
Abstract
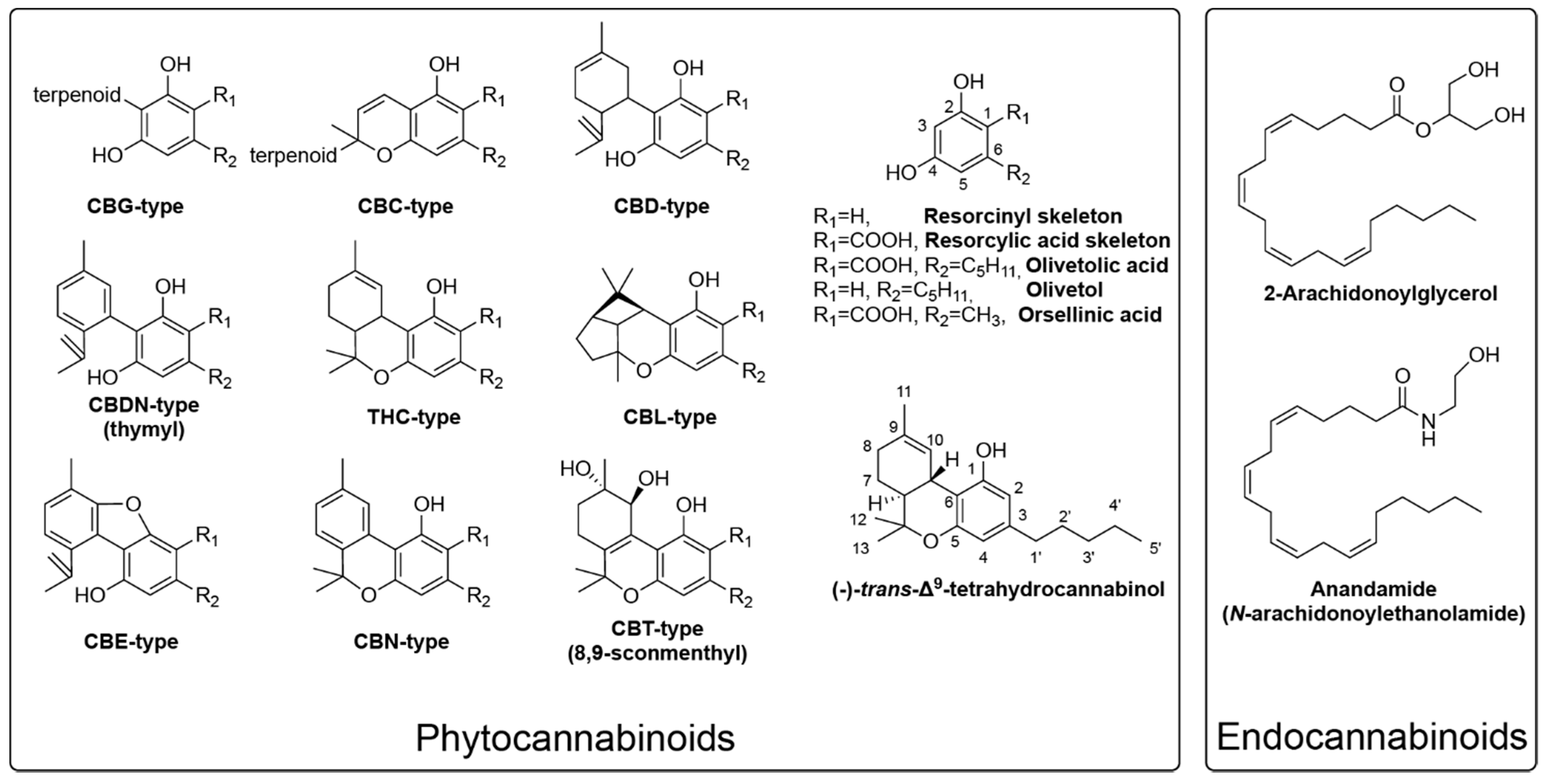
1. Introduction
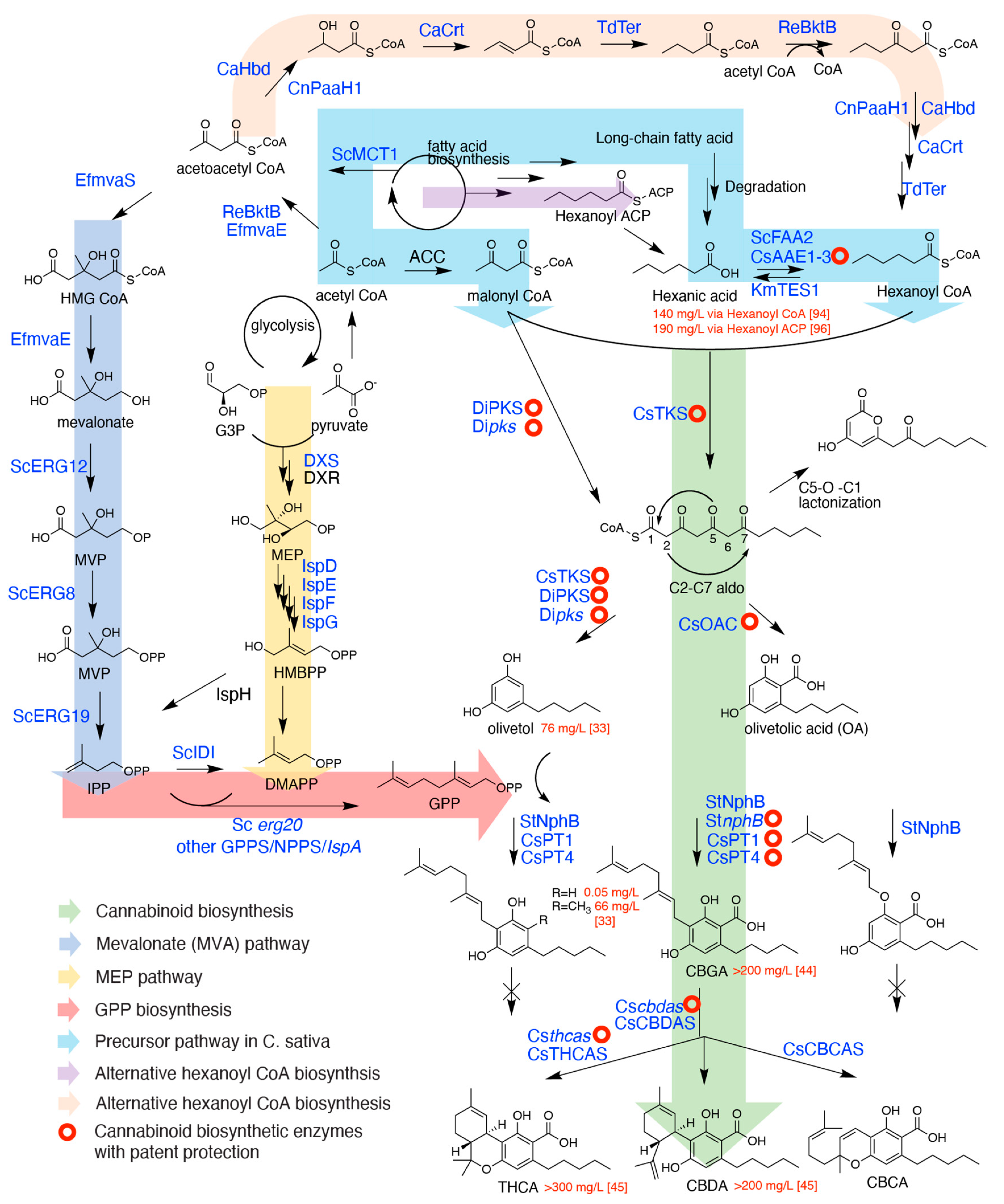
2. Critical Biosynthetic Steps for Phytocannabinoid Formation
2.1. The Formation of Olivetolic Acid
2.2. Cannabigerolic Acid Formation by Aromatic Prenyltransferases
2.3. FAD-Depended Cyclases Form Various Cannabinoids
3. Precursor Geranyl Pyrophosphate Production
4. Precursor Hexanoyl CoA Production
5. Choice of Chassis and Further Technical Development
6. Future Remarks
Author Contributions
Funding
Conflicts of Interest
References
- ElSohly, M.A. Marijuana and the Cannabinoids; Humana Press: Totowa, NJ, USA, 2007. [Google Scholar]
- Andre, C.M.; Hausman, J.-F.; Guerriero, G. Cannabis sativa: The Plant of the Thousand and One Molecules. Front. Plant Sci. 2016, 7, 19. [Google Scholar] [CrossRef]
- Bonini, S.A.; Premoli, M.; Tambaro, S.; Kumar, A.; Maccarinelli, G.; Memo, M.; Mastinu, A. Cannabis sativa: A comprehensive ethnopharmacological review of a medicinal plant with a long history. J. Ethnopharmacol. 2018, 227, 300–315. [Google Scholar] [CrossRef]
- Lynch, R.C.; Vergara, D.; Tittes, S.; White, K.; Schwartz, C.J.; Gibbs, M.J.; Ruthenburg, T.C.; DeCesare, K.; Land, D.P.; Kane, N.C. Genomic and Chemical Diversity in Cannabis. Crit. Rev. Plant Sci. 2017, 35, 349–363. [Google Scholar] [CrossRef]
- Van Bakel, H.; Stout, J.M.; Cote, A.G.; Tallon, C.M.; Sharpe, A.G.; Hughes, T.R.; Page, J.E. The draft genome and transcriptome of Cannabis sativa. Genome Biol. 2011, 12, R102–R108. [Google Scholar] [CrossRef]
- ElSohly, M.A.; Slade, D. Chemical constituents of marijuana: The complex mixture of natural cannabinoids. Life Sci. 2005, 78, 539–548. [Google Scholar] [CrossRef]
- Hanuš, L.O.; Meyer, S.M.; Muñoz, E.; Taglialatela-Scafati, O.; Appendino, G. Phytocannabinoids: A unified critical inventory. Nat. Prod. Rep. 2016, 33, 1357–1392. [Google Scholar] [CrossRef] [PubMed]
- Livingston, S.J.; Quilichini, T.D.; Booth, J.K.; Wong, D.C.J.; Rensing, K.H.; Laflamme Yonkman, J.; Castellarin, S.D.; Bohlmann, J.; Page, J.E.; Samuels, A.L. Cannabis glandular trichomes alter morphology and metabolite content during flower maturation. Plant J. 2019, 79, 37–56. [Google Scholar] [CrossRef] [PubMed]
- Citti, C.; Linciano, P.; Panseri, S.; Vezzalini, F.; Forni, F.; Vandelli, M.A.; Cannazza, G. Cannabinoid Profiling of Hemp Seed Oil by Liquid Chromatography Coupled to High-Resolution Mass Spectrometry. Front. Plant Sci. 2019, 10, 120. [Google Scholar] [CrossRef] [PubMed]
- Gülck, T.; Møller, B.L. Phytocannabinoids: Origins and Biosynthesis. Trends Plant Sci. 2020, 25, 985–1004. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Premoli, M.; Aria, F.; Bonini, S.A.; Maccarinelli, G.; Gianoncelli, A.; Memo, M.; Mastinu, A. Cannabimimetic plants: Are they new cannabinoidergic modulators? Planta 2019, 249, 1681–1694. [Google Scholar] [CrossRef] [PubMed]
- Toyota, M.; Kinugawa, T.; Asakawa, Y. Bibenzyl cannabinoid and bisbibenzyl derivative from the liverwort Radula Perrottetii. Phytochemistry 1994, 37, 859–862. [Google Scholar] [CrossRef]
- Toyota, M.; Shimamura, T.; Ishii, H.; Renner, M.; Braggins, J.; Asakawa, Y. New bibenzyl cannabinoid from the New Zealand liverwort Radula marginata. Chem. Pharm. Bull. 2002, 50, 1390–1392. [Google Scholar] [CrossRef] [PubMed]
- Taura, F.; Iijima, M.; Yamanaka, E.; Takahashi, H.; Kenmoku, H.; Saeki, H.; Morimoto, S.; Asakawa, Y.; Kurosaki, F.; Morita, H. A Novel Class of Plant Type III Polyketide Synthase Involved in Orsellinic Acid Biosynthesis from Rhododendron dauricum. Front. Plant Sci. 2016, 7, 809–815. [Google Scholar] [CrossRef]
- Saeki, H.; Hara, R.; Takahashi, H.; Iijima, M.; Munakata, R.; Kenmoku, H.; Fuku, K.; Sekihara, A.; Yasuno, Y.; Shinada, T.; et al. An Aromatic Farnesyltransferase Functions in Biosynthesis of the Anti-HIV Meroterpenoid Daurichromenic Acid. Plant Physiol. 2018, 178, 535–551. [Google Scholar] [CrossRef] [PubMed]
- Pacher, P.; Bátkai, S.; Kunos, G. The endocannabinoid system as an emerging target of pharmacotherapy. Pharmacol. Rev. 2006, 58, 389–462. [Google Scholar] [CrossRef] [PubMed]
- Cristino, L.; Bisogno, T.; Di Marzo, V. Cannabinoids and the expanded endocannabinoid system in neurological disorders. Nat. Rev. Neurol. 2019, 16, 9–29. [Google Scholar] [CrossRef] [PubMed]
- Zou, S.; Kumar, U. Cannabinoid Receptors and the Endocannabinoid System: Signaling and Function in the Central Nervous System. Int. J. Mol. Sci. 2018, 19, 833. [Google Scholar] [CrossRef]
- Yang, K.-H.; Galadari, S.; Isaev, D.; Petroianu, G.; Shippenberg, T.S.; Oz, M. The nonpsychoactive cannabinoid cannabidiol inhibits 5-hydroxytryptamine3A receptor-mediated currents in Xenopus laevis oocytes. J. Pharmacol. Exp. Ther. 2010, 333, 547–554. [Google Scholar] [CrossRef] [PubMed]
- Hebert-Chatelain, E.; Desprez, T.; Serrat, R.; Bellocchio, L.; Soria-Gómez, E.; Busquets-Garcia, A.; Pagano Zottola, A.C.; Delamarre, A.; Cannich, A.; Vincent, P.; et al. A cannabinoid link between mitochondria and memory. Nature 2016, 539, 555–559. [Google Scholar] [CrossRef] [PubMed]
- Bénard, G.; Massa, F.; Puente, N.; Lourenço, J.; Bellocchio, L.; Soria-Gómez, E.; Matias, I.; Delamarre, A.; Metna-Laurent, M.; Cannich, A.; et al. Mitochondrial CB₁ receptors regulate neuronal energy metabolism. Nat. Neurosci. 2012, 15, 558–564. [Google Scholar] [CrossRef]
- Taura, F.; Tanaka, S.; Taguchi, C.; Fukamizu, T.; Tanaka, H.; Shoyama, Y.; Morimoto, S. Characterization of olivetol synthase, a polyketide synthase putatively involved in cannabinoid biosynthetic pathway. FEBS Lett. 2009, 583, 2061–2066. [Google Scholar] [CrossRef]
- Gagne, S.J.; Stout, J.M.; Liu, E.; Boubakir, Z.; Clark, S.M.; Page, J.E. Identification of olivetolic acid cyclase from Cannabis sativa reveals a unique catalytic route to plant polyketides. Proc. Natl. Acad. Sci. USA 2012, 109, 12811–12816. [Google Scholar] [CrossRef]
- Yang, X.; Matsui, T.; Kodama, T.; Mori, T.; Zhou, X.; Taura, F.; Noguchi, H.; Abe, I.; Morita, H. Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa. FEBS J. 2016, 283, 1088–1106. [Google Scholar] [CrossRef] [PubMed]
- Page, J.E.; Gagne, S. Genes and Proteins for Aromatic Polyketide Synthesis. U.S. Patent 9611460B2, 4 April 2017. [Google Scholar]
- Shen, B. Polyketide biosynthesis beyond the type I, II and III polyketide synthase paradigms. Curr. Opin. Chem. Biol. 2003, 7, 285–295. [Google Scholar] [CrossRef]
- Austin, M.B.; Noel, J.P. The chalcone synthase superfamily of type III polyketide synthases. Nat. Prod. Rep. 2002, 20, 79–110. [Google Scholar] [CrossRef] [PubMed]
- Staunton, J.; Weissman, K.J. Polyketide biosynthesis: A millennium review. Nat. Prod. Rep. 2001, 18, 380–416. [Google Scholar] [CrossRef]
- Funa, N.; Awakawa, T.; Horinouchi, S. Pentaketide resorcylic acid synthesis by type III polyketide synthase from Neurospora crassa. J. Biol. Chem. 2007, 282, 14476–14481. [Google Scholar] [CrossRef]
- Matsuzawa, M.; Katsuyama, Y.; Funa, N.; Horinouchi, S. Alkylresorcylic acid synthesis by type III polyketide synthases from rice Oryza sativa. Phytochemistry 2010, 71, 1059–1067. [Google Scholar] [CrossRef]
- Shoyama, Y.; Tamada, T.; Kurihara, K.; Takeuchi, A.; Taura, F.; Arai, S.; Blaber, M.; Shoyama, Y.; Morimoto, S.; Kuroki, R. Structure and Function of ∆1-Tetrahydrocannabinolic Acid (THCA) Synthase, the Enzyme Controlling the Psychoactivity of Cannabis sativa. J. Mol. Biol. 2012, 423, 96–105. [Google Scholar] [CrossRef] [PubMed]
- Eckermann, C.; Schröder, G.; Eckermann, S.; Strack, D.; Schmidt, J.; Schneider, B.; Schröder, J. Stilbenecarboxylate biosynthesis: A new function in the family of chalcone synthase-related proteins. Phytochemistry 2003, 62, 271–286. [Google Scholar] [CrossRef]
- Mookerjee, S.; Campbell, A.J.; Wiltshire, Z.D.; Chen, K.J.; Inc, H.B. Method and Cell Line for Production of Phytocannabinoids and Phytocannabinoid Analogues in Yeast. U.S. Patent 0283807A1, 2020. [Google Scholar]
- Szamecz, B.K.; Varszegi, S.; Nemeth, A.; Szabo, L.; Kumar, A. Microorganisms and Methods for the Fermentation of Cannabinoids. WO Patent 2019071000, 4 November 2019. [Google Scholar]
- Keasling, J.D.; D’Espaux, L.; Wong, J.; Luo, X.; Reiter, M.; Denby, C.; Lechner, A. Recombinant Microorganisms and Methods for Producing Cannabinoids and Cannabinoid Derivatives. U.S. Patent 10563211B2, 18 February 2020. [Google Scholar]
- Horwitz, A.; D’Espaux, L.; Wong, J.; Bector, R.; Hjelmeland, A.K.; Platt, D.; Ubersax, J. Optimized Expression Systems for Producing Cannabinoid Synthase Polypeptides, Cannabinoids, and Cannabinoid Derivatives. WO Patent 2020069214A3, 7 May 2020. [Google Scholar]
- Luo, X.; Reiter, M.A.; Espaux, L.D.X.; Wong, J.; Denby, C.M.; Lechner, A.; Zhang, Y.; Grzybowski, A.T.; Harth, S.; Lin, W.; et al. Complete biosynthesis of cannabinoids and their unnatural analogues in yeast. Nature 2019, 567, 123–126. [Google Scholar] [CrossRef] [PubMed]
- Gülck, T.; Booth, J.K.; Carvalho, Â.; Khakimov, B.; Crocoll, C.; Motawia, M.S.; Møller, B.L.; Bohlmann, J.; Gallage, N.J. Synthetic Biology of Cannabinoids and Cannabinoid Glucosides in Nicotiana benthamiana and Saccharomyces cerevisiae. J. Nat. Prod. 2020, 83, 2877–2893. [Google Scholar] [CrossRef]
- Taura, F.; Dono, E.; Sirikantaramas, S.; Yoshimura, K.; Shoyama, Y.; Morimoto, S. Production of Δ1-tetrahydrocannabinolic acid by the biosynthetic enzyme secreted from transgenic Pichia pastoris. Biochem. Biophys. Res. Commun. 2007, 361, 675–680. [Google Scholar] [CrossRef]
- Zirpel, B.; Stehle, F.; Kayser, O. Production of Δ9-tetrahydrocannabinolic acid from cannabigerolic acid by whole cells of Pichia (Komagataella) pastoris expressing Δ9-tetrahydrocannabinolic acid synthase from Cannabis sativa L. Biotechnol. Lett. 2015, 37, 1869–1875. [Google Scholar] [CrossRef] [PubMed]
- Zirpel, B.; Degenhardt, F.; Martin, C.; Kayser, O.; Stehle, F. Engineering yeasts as platform organisms for cannabinoid biosynthesis. J. Biotechnol. 2017, 259, 204–212. [Google Scholar] [CrossRef]
- Zirpel, B.; Degenhardt, F.; Zammarelli, C.; Wibberg, D.; Kalinowski, J.; Stehle, F.; Kayser, O. Optimization of Δ9-tetrahydrocannabinolic acid synthase production in Komagataella phaffii via post-translational bottleneck identification. J. Biotechnol. 2018, 272, 40–47. [Google Scholar] [CrossRef]
- Ayakar, S.R.; Pawar, S.V.; Hallam, S.J.; Hossain, S.; Yadav, V.G.; Roy, P.R.; Srivastava, S.K. Metabolic Engineering of E. coli for the Biosynthesis of Cannabinoid Products. U.S. Patent 20200291434A1, 17 September 2020. [Google Scholar]
- Cheon, Y.; Kim, J.-S.; Park, J.-B.; Heo, P.; Lim, J.H.; Jung, G.Y.; Seo, J.-H.; Park, J.H.; Koo, H.M.; Cho, K.M.; et al. A biosynthetic pathway for hexanoic acid production in Kluyveromyces marxianus. J. Biotechnol. 2014, 182, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Gajewski, J.; Pavlovic, R.; Fischer, M.; Boles, E.; Grininger, M. Engineering fungal de novo fatty acid synthesis for short chain fatty acid production. Nat. Commun. 2019, 8, 14650. [Google Scholar] [CrossRef] [PubMed]
- Davidovich, A.; Chan, B.; Gupta, N.; Ladha, A. Genetically Engineered Microorganisms and Processes for the Production of Cannabinoids from a Carbon Source Precursor. U.S. Patent 10801049B2, 13 October 2020. [Google Scholar]
- Tan, Z.; Clomburg, J.M.; Gonzalez, R. Synthetic Pathway for the Production of Olivetolic Acid in Escherichia coli. ACS Synth. Biol. 2018, 7, 1886–1896. [Google Scholar] [CrossRef]
- Sayre, R.T.; Goncalves, E.C.; Zidenga, T. High Level in Vivo Biosynthesis and Isolation of Water Soluble Cannabinoids in Stably Transformed Plant Systems. U.S. Patent 0338301A1, 2019. [Google Scholar]
- Sayre, R.T.; Goncalves, E.C.; Zidenga, T. Generation of Water-Soluble Cannabinoid Compounds in Yeast and Plant Cell Suspension Cultures and Compositions of Matter. U.S. Patent 078168A1, 2019. [Google Scholar]
- Sirikantaramas, S.; Morimoto, S.; Shoyama, Y.; Ishikawa, Y.; Wada, Y.; Shoyama, Y.; Taura, F. The gene controlling marijuana psychoactivity: Molecular cloning and heterologous expression of Δ1-tetrahydrocannabinolic acid synthase from Cannabis sativa L. J. Biol. Chem. 2004, 279, 39767–39774. [Google Scholar] [CrossRef]
- Poulos, J.L.; Farnia, A.N. Production of Cannabinoids in Yeast. U.S. Patent 9822384B2, 21 November 2017. [Google Scholar]
- Beardslee, T.A. Biosynthetic Cannabinoid Production in Engineered Microorganisms. WO Patent WO2020198679, 1 October 2020. [Google Scholar]
- Laban, A. Cannabinoid Production in Algae. WO Patent 2019/202510 A1, 24 October 2019. [Google Scholar]
- Bowie, J.U.; Valliere, M.; Korman, T.P.; Woodall, N. Biosynthetic Platform for the Production of Cannabinoids and other Prenylated Compounds. WO Patent 2020028722, 6 February 2020. [Google Scholar]
- Valliere, M.A.; Korman, T.P.; Woodall, N.B.; Khitrov, G.A.; Taylor, R.E.; Baker, D.; Bowie, J.U. A cell-free platform for the prenylation of natural products and application to cannabinoid production. Nat. Commun. 2019, 10, 565. [Google Scholar] [CrossRef] [PubMed]
- Valliere, M.A.; Korman, T.P.; Arbing, M.A.; Bowie, J.U. A bio-inspired cell-free system for cannabinoid production from inexpensive inputs. Nat. Chem. Biol. 2020, 16, 1427–1433. [Google Scholar] [CrossRef]
- De Bruijn, W.J.C.; Levisson, M.; Beekwilder, J.; Van Berkel, W.J.H.; Vincken, J.-P. Plant Aromatic Prenyltransferases: Tools for Microbial Cell Factories. Trends Biotechnol. 2020, 38, 917–934. [Google Scholar] [CrossRef]
- Rea, K.A.; Casaretto, J.A.; Al-Abdul-Wahid, M.S.; Sukumaran, A.; Geddes-McAlister, J.; Rothstein, S.J.; Akhtar, T.A. Biosynthesis of cannflavins A and B from Cannabis sativa L. Phytochemistry 2019, 164, 162–171. [Google Scholar] [CrossRef]
- Fellermeier, M.; Zenk, M.H. Prenylation of olivetolate by a hemp transferase yields cannabigerolic acid, the precursor of tetrahydrocannabinol. FEBS Lett. 1998, 427, 283–285. [Google Scholar] [CrossRef]
- Page, J.E.; Boubakir, Z. Aromatic Prenyltransferase from Cannabis. U.S. Patent 9765308B2, 19 September 2017. [Google Scholar]
- Wang, J.; Luca, V.D. The biosynthesis and regulation of biosynthesis of Concord grape fruit esters, including “foxy” methylanthranilate. Plant J. 2005, 44, 606–619. [Google Scholar] [CrossRef]
- Kuzuyama, T.; Noel, J.P.; Richard, S.B. Structural basis for the promiscuous biosynthetic prenylation of aromatic natural products. Nature 2005, 435, 983–987. [Google Scholar] [CrossRef]
- Kuzuyama, T. Biosynthetic studies on terpenoids produced by Streptomyces. J. Antibiot. 2017, 70, 811–818. [Google Scholar] [CrossRef] [PubMed]
- Kumano, T.; Richard, S.B.; Noel, J.P.; Nishiyama, M.; Kuzuyama, T. Chemoenzymatic syntheses of prenylated aromatic small molecules using Streptomyces prenyltransferases with relaxed substrate specificities. Bioorg. Med. Chem. 2008, 16, 8117–8126. [Google Scholar] [CrossRef] [PubMed]
- Mendez, M.; Noel, J.; Burkart, M.; Lanoiselee, J.; Botsch, K.; Saunders, M. Compositions and Methods for Using Genetically Modified Enzymes. WO Patent 2019183152A1, 26 September 2019. [Google Scholar]
- Kayser, O.; Stehle, F.-O. Biotechnological Production of Cannabinoids. WO Patent 2020016287, 23 January 2020. [Google Scholar]
- Philippe, R.A.; Kumaran, A.P.; Santos, C.N.S. Microbial Cells and Methods for Producing Cannabinoids. WO Patent 2020102541, 22 May 2020. [Google Scholar]
- Taura, F.; Morimoto, S.; Shoyama, Y.; Mechoulam, R. First direct evidence for the mechanism of Δ1-tetrahydrocannabinolic acid biosynthesis. J. Am. Chem. Soc. 1995, 117, 9766–9767. [Google Scholar] [CrossRef]
- Taura, F.; Morimoto, S.; Shoyama, Y. Purification and Characterization of Cannabidiolic-acid Synthase from Cannabis sativa L. Biochemical analysis of a novel enzyme that catalyzes the oxidocyclization of cannabigerolic acid to cannabidiolic acid. J. Biol. Chem. 1996, 271, 17411–17416. [Google Scholar] [CrossRef]
- Morimoto, S.; Komatsu, K.; Taura, F.; Shoyama, Y. Purification and characterization of cannabichromenic acid synthase from Cannabis sativa. Phytochemistry 1998, 49, 1525–1529. [Google Scholar] [CrossRef]
- Taura, F.; Sirikantaramas, S.; Shoyama, Y.; Yoshikai, K.; Shoyama, Y.; Morimoto, S. Cannabidiolic-acid synthase, the chemotype-determining enzyme in the fiber-type Cannabis sativa. FEBS Lett. 2007, 581, 2929–2934. [Google Scholar] [CrossRef]
- Zirpel, B.; Kayser, O.; Stehle, F. Elucidation of structure-function relationship of THCA and CBDA synthase from Cannabis sativa L. J. Biotechnol. 2018, 284, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Winkler, A.; Łyskowski, A.; Riedl, S.; Puhl, M.; Kutchan, T.M.; Macheroux, P.; Gruber, K. A concerted mechanism for berberine bridge enzyme. Nat. Chem. Biol. 2008, 4, 739–741. [Google Scholar] [CrossRef]
- Geissler, M.; Volk, J.; Stehle, F.; Kayser, O.; Warzecha, H. Subcellular localization defines modification and production of Δ9-tetrahydrocannabinolic acid synthase in transiently transformed Nicotiana benthamiana. Biotechnol. Lett. 2018, 40, 913–919. [Google Scholar] [CrossRef] [PubMed]
- Laverty, K.U.; Stout, J.M.; Sullivan, M.J.; Shah, H.; Gill, N.; Holbrook, L.; Deikus, G.; Sebra, R.; Hughes, T.R.; Page, J.E.; et al. A physical and genetic map of Cannabis sativa identifies extensive rearrangements at the THC/CBD acid synthase loci. Genome Res. 2019, 29, 146–156. [Google Scholar] [CrossRef]
- Sirikantaramas, S.; Taura, F.; Tanaka, Y.; Ishikawa, Y.; Morimoto, S.; Shoyama, Y. Tetrahydrocannabinolic Acid Synthase, the Enzyme Controlling Marijuana Psychoactivity, is Secreted into the Storage Cavity of the Glandular Trichomes. Plant Cell Physiol. 2005, 46, 1578–1582. [Google Scholar] [CrossRef]
- Nagashima, Y.; Schaewen von, A.; Koiwa, H. Function of N-glycosylation in plants. Plant Sci. 2018, 274, 70–79. [Google Scholar] [CrossRef]
- Strasser, R. Plant protein glycosylation. Glycobiology 2016, 26, 926–939. [Google Scholar] [CrossRef]
- Hebert, D.N.; Lamriben, L.; Powers, E.T.; Kelly, J.W. The intrinsic and extrinsic effects of N-linked glycans on glycoproteostasis. Nat. Chem. Biol. 2014, 10, 902–910. [Google Scholar] [CrossRef]
- Ojore, B.V.O.; Bulleid, N.J. Forming disulfides in the endoplasmic reticulum. Biochem. Biophys. Acta Mol. Cell Res. 2013, 1833, 2425–2429. [Google Scholar]
- Puxbaum, V.; Mattanovich, D.; Gasser, B. Quo vadis? The challenges of recombinant protein folding and secretion in Pichia pastoris. Appl. Microbiol. Biotechnol. 2015, 99, 2925–2938. [Google Scholar] [CrossRef]
- Kirby, J.; Keasling, J.D. Biosynthesis of Plant Isoprenoids: Perspectives for Microbial Engineering. Annu. Rev. Plant Biol. 2009, 60, 335–355. [Google Scholar] [CrossRef] [PubMed]
- Ward, V.C.A.; Chatzivasileiou, A.O.; Stephanopoulos, G. Metabolic engineering of Escherichia coli for the production of isoprenoids. FEMS Microbiol. Lett. 2018, 365, 167. [Google Scholar] [CrossRef]
- Zhang, Y.; Nielsen, J.; Liu, Z. Engineering yeast metabolism for production of terpenoids for use as perfume ingredients, pharmaceuticals and biofuels. FEMS Yeast Res. 2017, 17, 1033. [Google Scholar] [CrossRef]
- Wang, C.; Liwei, M.; Park, J.-B.; Jeong, S.-H.; Wei, G.; Wang, Y.; Kim, S.-W. Microbial Platform for Terpenoid Production: Escherichia coli and Yeast. Front. Microbiol. 2018, 9, 2460. [Google Scholar] [CrossRef]
- Paddon, C.J.; Westfall, P.J.; Pitera, D.J.; Benjamin, K.; Fisher, K.; McPhee, D.; Leavell, M.D.; Tai, A.; Main, A.; Eng, D.; et al. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 2013, 496, 528–532. [Google Scholar] [CrossRef] [PubMed]
- Tsuruta, H.; Paddon, C.J.; Eng, D.; Lenihan, J.R.; Horning, T.; Anthony, L.C.; Regentin, R.; Keasling, J.D.; Renninger, N.S.; Newman, J.D. High-Level Production of Amorpha-4,11-Diene, a Precursor of the Antimalarial Agent Artemisinin, in Escherichia coli. PLoS ONE 2009, 4, e4489. [Google Scholar] [CrossRef] [PubMed]
- Ignea, C.; Raadam, M.H.; Motawia, M.S.; Makris, A.M.; Vickers, C.E.; Kampranis, S.C. Orthogonal monoterpenoid biosynthesis in yeast constructed on an isomeric substrate. Nat. Commun. 2019, 10, 3799. [Google Scholar] [CrossRef]
- Carrau, F.M.; Medina, K.; Boido, E.; Farina, L.; Gaggero, C.; Dellacassa, E.; Versini, G.; Henschke, P.A. De novo synthesis of monoterpenes by Saccharomyces cerevisiae wine yeasts. FEMS Microbiol. Lett. 2005, 243, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Ignea, C.; Pontini, M.; Maffei, M.E.; Makris, A.M.; Kampranis, S.C. Engineering Monoterpene Production in Yeast Using a Synthetic Dominant Negative Geranyl Diphosphate Synthase. ACS Synth. Biol. 2014, 3, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Fischer, M.J.C.; Meyer, S.; Claudel, P.; Bergdoll, M.; Karst, F. Metabolic engineering of monoterpene synthesis in yeast. Biotechnol. Bioeng. 2011, 108, 1883–1892. [Google Scholar] [CrossRef] [PubMed]
- Chatzivasileiou, A.O.; Ward, V.; Edgar, S.M.; Stephanopoulos, G. Two-step pathway for isoprenoid synthesis. Proc. Natl. Acad. Sci. USA 2019, 116, 506–511. [Google Scholar] [CrossRef]
- Alonso-Gutierrez, J.; Chan, R.; Batth, T.S.; Adams, P.D.; Keasling, J.D.; Petzold, C.J.; Lee, T.S. Metabolic engineering of Escherichia coli for limonene and perillyl alcohol production. Metab. Eng. 2013, 19, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Stout, J.M.; Boubakir, Z.; Ambrose, S.J.; Purves, R.W.; Page, J.E. The hexanoyl-CoA precursor for cannabinoid biosynthesis is formed by an acyl-activating enzyme in Cannabis sativa trichomes. Plant J. 2012, 71, 353–365. [Google Scholar] [CrossRef]
- Page, J.E.; Stout, J.M. Genes and Proteins for Alkanoyl-CoA Synthesis. U.S. Patent 9546362B2, 17 January 2017. [Google Scholar]
- Majidian, P.; Tabatabaei, M.; Zeinolabedini, M.; Naghshbandi, M.P.; Chisti, Y. Metabolic engineering of microorganisms for biofuel production. Renew. Sustain. Energy Rev. 2018, 82, 3863–3885. [Google Scholar] [CrossRef]
- Shen, C.R.; Lan, E.I.; Dekishima, Y.; Baez, A.; Cho, K.M.; Liao, J.C. Driving forces enable high-titer anaerobic 1-butanol synthesis in Escherichia coli. Appl. Environ. Microbiol. 2011, 77, 2905–2915. [Google Scholar] [CrossRef] [PubMed]
- Dekishima, Y.; Lan, E.I.; Shen, C.R.; Cho, K.M.; Liao, J.C. Extending carbon chain length of 1-butanol pathway for 1-hexanol synthesis from glucose by engineered Escherichia coli. J. Am. Chem. Soc. 2011, 133, 11399–11401. [Google Scholar] [CrossRef]
- Machado, H.B.; Dekishima, Y.; Luo, H.; Lan, E.I.; Liao, J.C. A selection platform for carbon chain elongation using the CoA-dependent pathway to produce linear higher alcohols. Metab. Eng. 2012, 14, 504–511. [Google Scholar] [CrossRef]
- Schmitt, P.; Taboada, A. Production of Plant-Based Active Substances (e.g., cannabinoids) by Recombinant Microorganisms. WO Patent 2020/169221A1, 2020. [Google Scholar]
- Melis, A.; Betterle, N.; Martinez, D.A.H. Production of Cannabinoids Using Genetically Engineered Photosynthetic Microorganisms. WO Patent 2020/180736A2, 2020. [Google Scholar]
- Wang, R.; Zhao, S.; Wang, Z.; Koffas, M.A.G. Recent advances in modular co-culture engineering for synthesis of natural products. Curr. Opin. Biotech. 2020, 62, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Okada, M.; Saito, K.; Wong, C.P.; Li, C.; Wang, D.; Iijima, M.; Taura, F.; Kurosaki, F.; Awakawa, T.; Abe, I. Combinatorial Biosynthesis of (+)-Daurichromenic Acid and Its Halogenated Analogue. Org. Lett. 2017, 19, 3183–3186. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, J.F.S.; Benedito, V.A.; Sandhu, D.; Marchese, J.A.; Liu, S. Seasonal and Differential Sesquiterpene Accumulation in Artemisia annua Suggest Selection Based on Both Artemisinin and Dihydroartemisinic Acid may Increase Artemisinin in planta. Front. Plant Sci. 2018, 9, 3610–3612. [Google Scholar] [CrossRef] [PubMed]
- Peplow, M. Synthetic biology’s first malaria drug meets market resistance. Nature 2016, 530, 389–390. [Google Scholar] [CrossRef] [PubMed]
- Prandi, C.; Blangetti, M.; Namdar, D.; Koltai, H. Structure-Activity Relationship of Cannabis Derived Compounds for the Treatment of Neuronal Activity-Related Diseases. Molecules 2018, 23, 1526. [Google Scholar] [CrossRef] [PubMed]
- Bow, E.W.; Rimoldi, J.M. The Structure—Function Relationships of Classical Cannabinoids: CB1/CB2 Modulation. Perspect. Med. Chem. 2016, 8, PMC.S32171. [Google Scholar] [CrossRef]
| Expression Systems | Feedstock | Product | Titer | Sources |
|---|---|---|---|---|
| Saccharomyces cerevisiae | Olivetolic acid | CBGA | 96 mg/L | [34] |
| Olivetolic acid | THCA | 84 mg/L | [34] | |
| Hexanoic acid | CBGA | 34 mg/L | [34] | |
| Hexanoic acid | THCA | 23 mg/L | [34] | |
| Olivetolic acid | CBGA | 216 mg/L | [35] | |
| Hexanoic acid | CBGA | 73 mg/L | [35] | |
| Olivetolic acid | CBDA | 234 mg/L | [36] | |
| Olivetolic acid | THCA | 320 mg/L | [36] | |
| de novo | 1-methyl-CBG | 66 mg/L | [30] | |
| de novo | Olivetol | 76 mg/L | [30] | |
| de novo | 1-methylolivetol | 42 mg/L | [30] | |
| Hexanoic acid | CBDA | 4.2 µg/L | [37] | |
| Hexanoic acid | THCA | 8.0 mg/L | [37] | |
| Olivetolic acid | CBGA | 1.0 mg/L | [38] | |
| Olivetolic acid | CBGA | 1.0 mg/L | [38] | |
| Komagataella phaffi (previously Pichia pastoris) | CBGA | THCA | 32.6 mg/L | [39] |
| CBGA | THCA | 0.36 g/L | [40] | |
| Olivetolic acid | THCA | 615 pmol/L | [41] | |
| CBGA | THCA | 3.05 g/L | [42] | |
| Escherichia coli | Olivetolic acid/GPP | CBGA | 1.2 mg/L | [43] |
| de novo | Hexanoic acid | 140 mg/L | [44] | |
| de novo | Hexanoic acid | 190 mg/L | [45] | |
| de novo | CBGA | detectable | [46] | |
| Hexanoic acid | Olivetolic acid | 0.48 mg/L | [47] | |
| Nicotiana benthamiana | CBGA | CBDA | 34ppm | [48] |
| CBDA | CBDA-glucoside | N/A | [48] | |
| CBDA | CBDA-glucoside | N/A | [49] | |
| CBGA | THCA | N/A | [38] | |
| Hexanoic acid | Olivetolic acid glucoside | N/A | [38] | |
| Hexanoic acid | Olivetolic acid | N/A | [38] | |
| CBGA | THCA | 82 µg/30 mL | [50] | |
| Kluyveromyces marxianus | de novo | CBDA, THCA | N/A | [51] |
| Candida viswanathii | oleic acid | CBGA | 0.67 mg/L supernatant 1.51 mg/L lysate | [52] |
| oleic acid | Olivetolic acid | 13.1 mg/L | [52] | |
| Chlamydomonas reinhardtii | de novo | CBGA | detectable | [53] |
| Cell free | Olivetolic acid | CBGA | 744 mg/L | [54,55] |
| Olivetolic acid | CBGA | 1.25 g/L | [54,56] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Blatt-Janmaat, K.; Qu, Y. The Biochemistry of Phytocannabinoids and Metabolic Engineering of Their Production in Heterologous Systems. Int. J. Mol. Sci. 2021, 22, 2454. https://doi.org/10.3390/ijms22052454
Blatt-Janmaat K, Qu Y. The Biochemistry of Phytocannabinoids and Metabolic Engineering of Their Production in Heterologous Systems. International Journal of Molecular Sciences. 2021; 22(5):2454. https://doi.org/10.3390/ijms22052454
Chicago/Turabian StyleBlatt-Janmaat, Kaitlyn, and Yang Qu. 2021. "The Biochemistry of Phytocannabinoids and Metabolic Engineering of Their Production in Heterologous Systems" International Journal of Molecular Sciences 22, no. 5: 2454. https://doi.org/10.3390/ijms22052454
APA StyleBlatt-Janmaat, K., & Qu, Y. (2021). The Biochemistry of Phytocannabinoids and Metabolic Engineering of Their Production in Heterologous Systems. International Journal of Molecular Sciences, 22(5), 2454. https://doi.org/10.3390/ijms22052454





