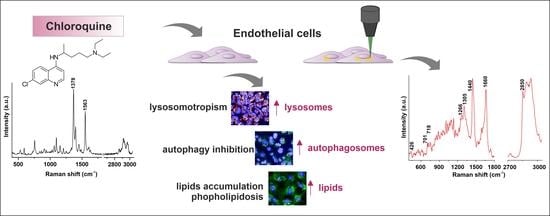
Chloroquine-Induced Accumulation of Autophagosomes and Lipids in the Endothelium
Abstract
1. Introduction
2. Results and Discussion
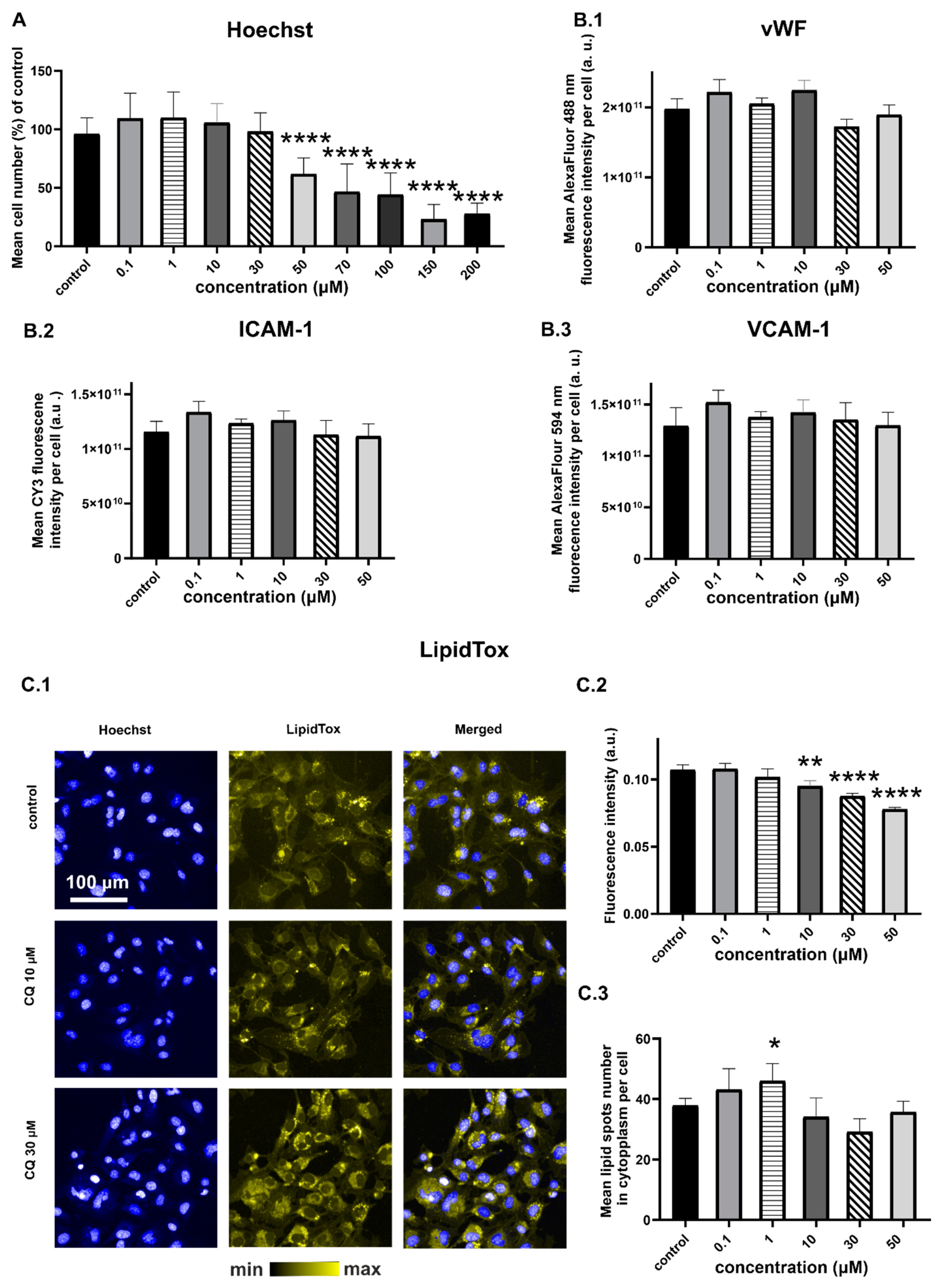
2.1. Characterization of Endothelial Responses to CQ
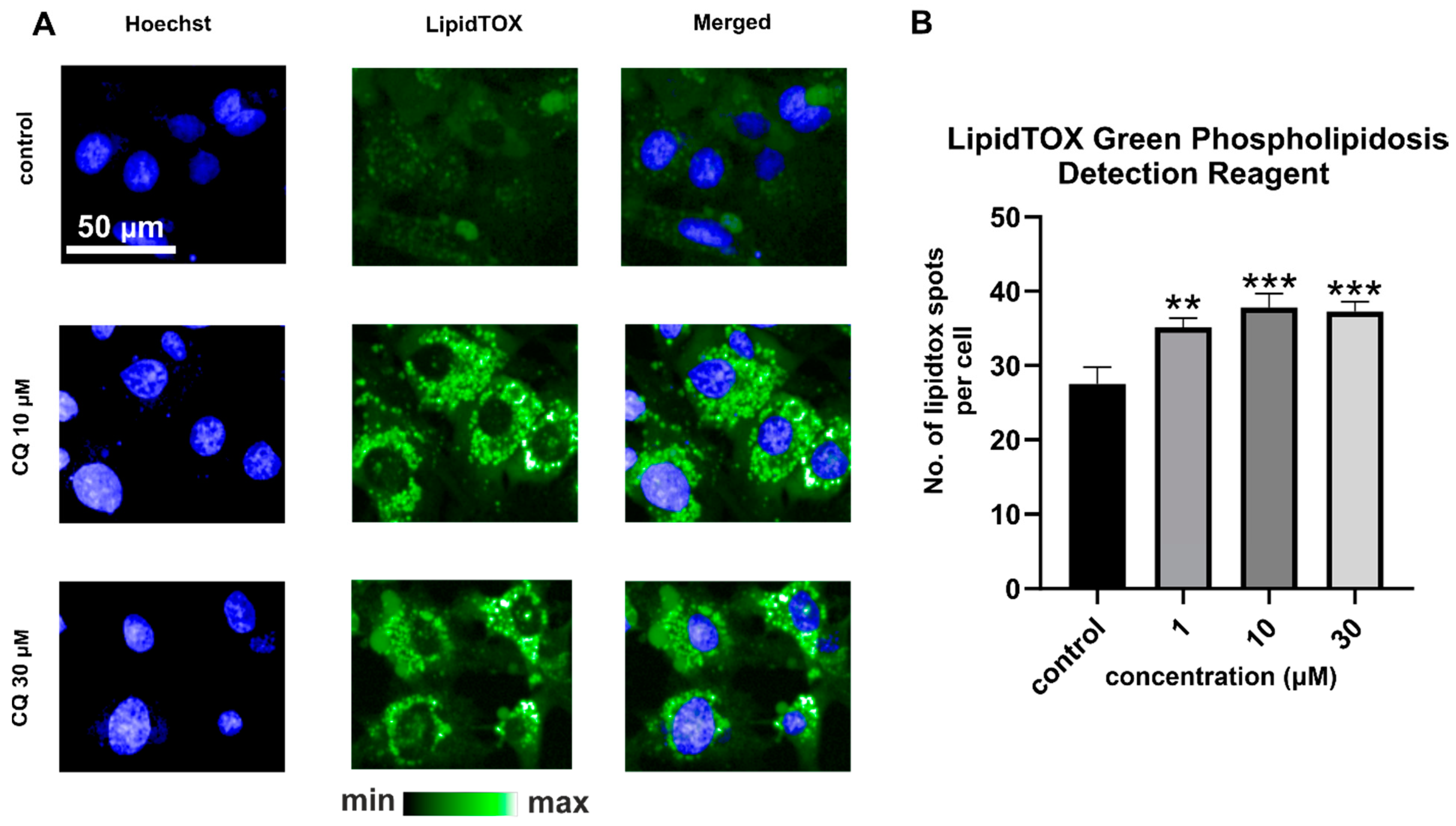
2.2. Efects of CQ on Lysosomes, Phospholipidosis, and Autophagy
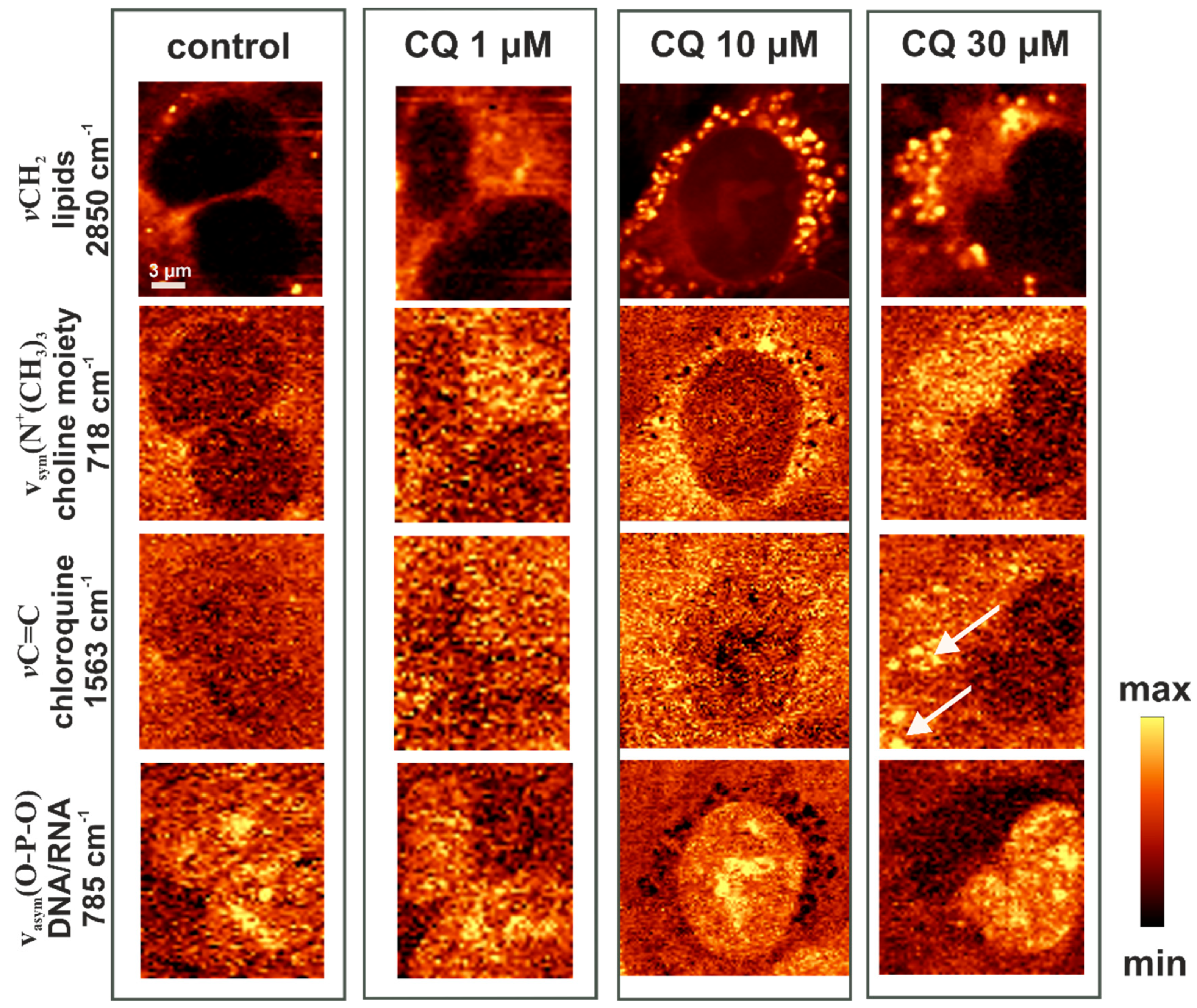
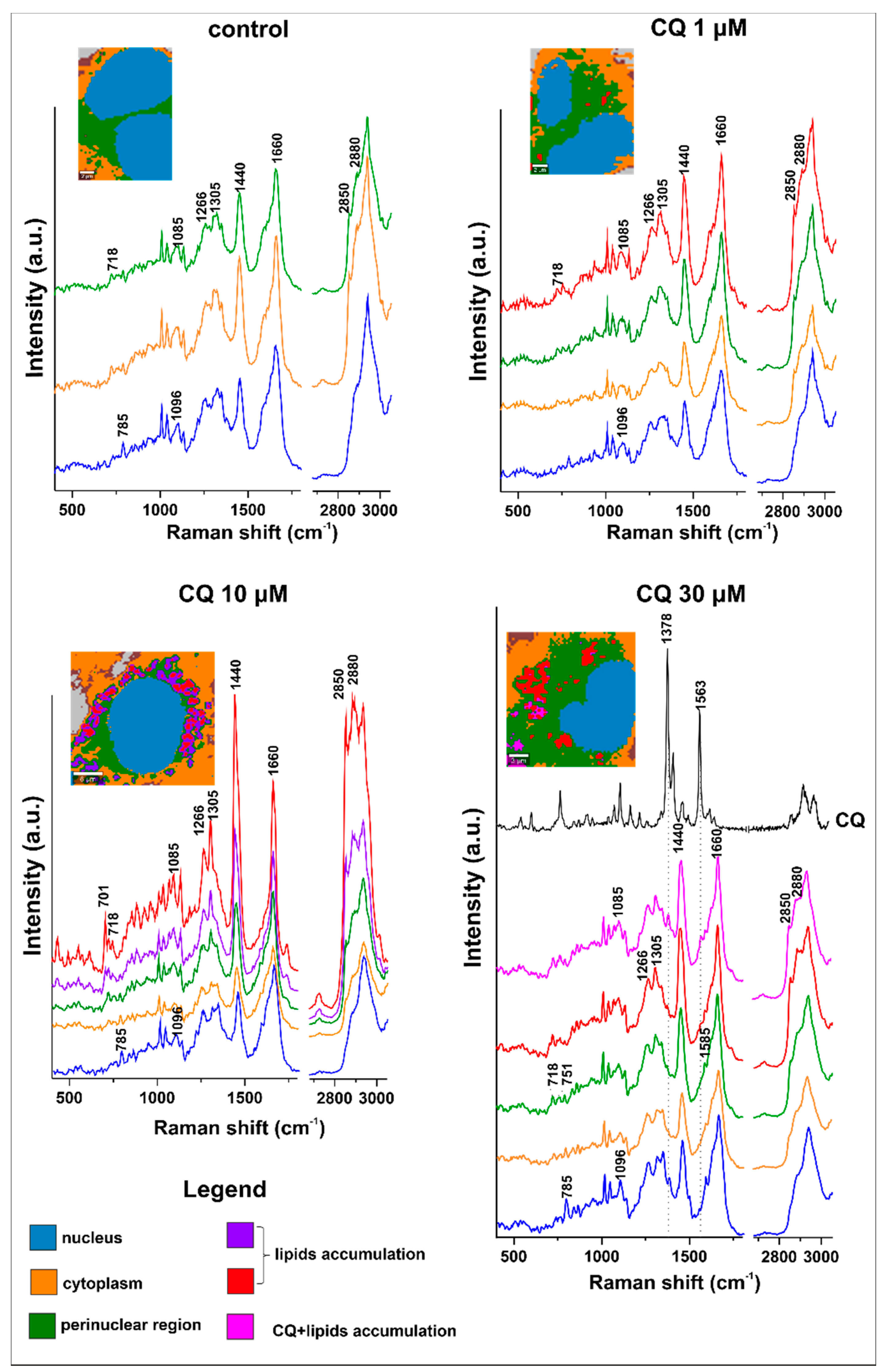
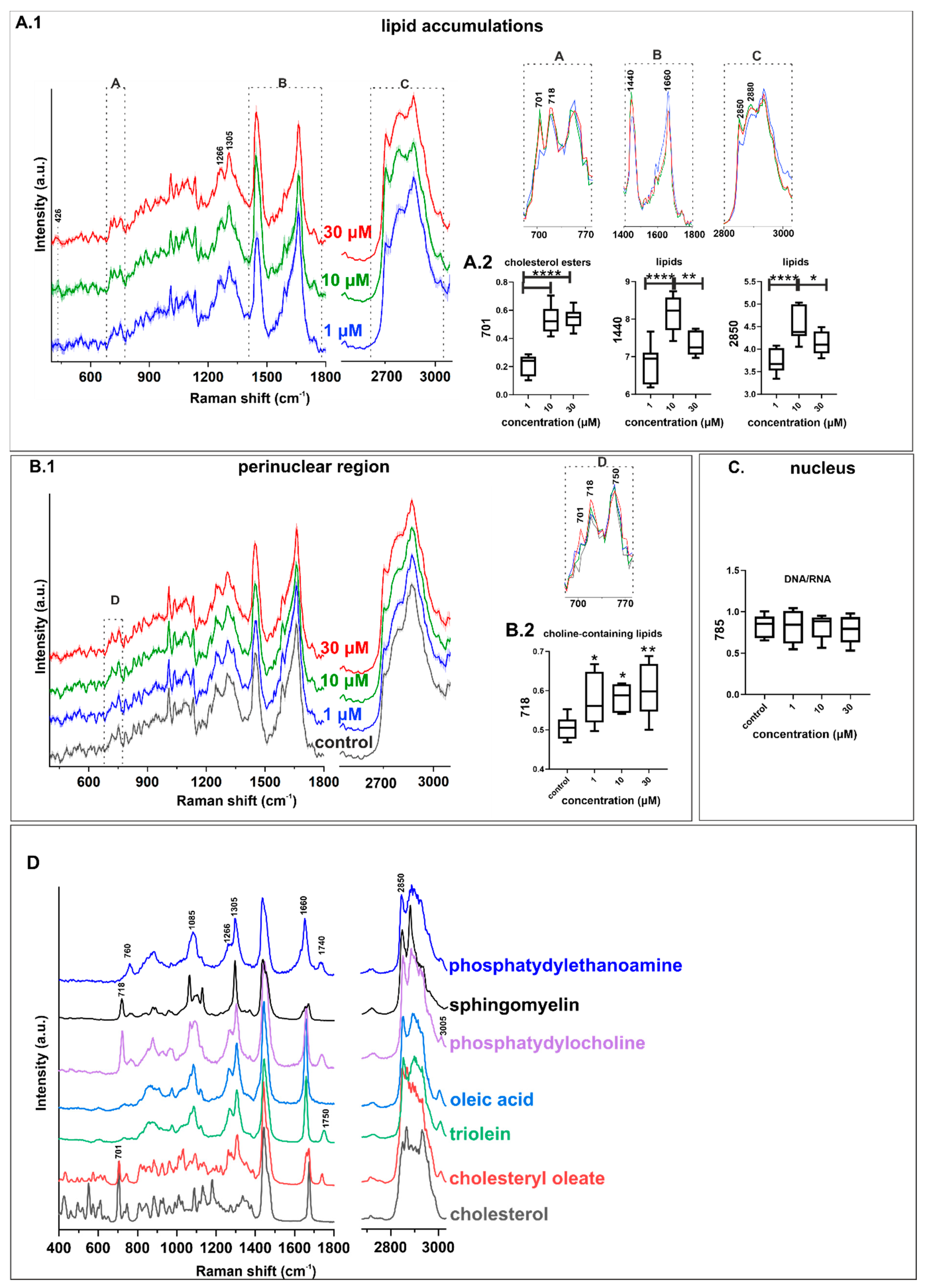
2.3. Characterization of Lipid Changes Induced by CQ Using RAMAN Imaging
3. Materials and Methods
3.1. Cell Culture
3.2. Raman Measurements with Data Analysis and Processing
3.3. Fluorescent and Immunofluorescent Staining
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gao, J.; Tian, Z.; Yang, X. Breakthrough: Chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci. Trends 2020, 14, 72–73. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Cao, R.; Xu, M.; Wang, X.; Zhang, H.; Hu, H.; Li, Y.; Hu, Z.; Zhong, W.; Wang, M. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov. 2020, 6, 6–9. [Google Scholar] [CrossRef]
- Wang, M.; Cao, R.; Zhang, L.; Yang, X.; Liu, J.; Xu, M.; Shi, Z.; Hu, Z.; Zhong, W.; Xiao, G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020, 30, 269–271. [Google Scholar] [CrossRef]
- Thelen, A.M.; Zoncu, R. Emerging Roles for the Lysosome in Lipid Metabolism. Trends Cell Biol. 2017, 27, 833–850. [Google Scholar] [CrossRef] [PubMed]
- Torisu, K.; Singh, K.K.; Torisu, T.; Lovren, F.; Liu, J.; Pan, Y.; Quan, A.; Ramadan, A.; Al-Omran, M.; Pankova, N.; et al. Intact endothelial autophagy is required to maintain vascular lipid homeostasis. Aging Cell 2016, 15, 187–191. [Google Scholar] [CrossRef]
- Mauthe, M.; Orhon, I.; Rocchi, C.; Zhou, X.; Luhr, M.; Hijlkema, K.J.; Coppes, R.P.; Engedal, N.; Mari, M.; Reggiori, F. Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy 2018, 14, 1435–1455. [Google Scholar] [CrossRef] [PubMed]
- Neill, T.; Chen, C.G.; Buraschi, S.; Iozzo, R.V. Catabolic degradation of endothelial VEGFA via autophagy. J. Biol. Chem. 2020, 295, 6064–6079. [Google Scholar] [CrossRef]
- Harvey, K.A.; Xu, Z.; Pavlina, T.M.; Zaloga, G.P.; Siddiqui, R.A. Modulation of endothelial cell integrity and inflammatory activation by commercial lipid emulsions. Lipids Health Dis. 2015, 14, 1–16. [Google Scholar] [CrossRef]
- Harvey, K.A.; Walker, C.L.; Xu, Z.; Whitley, P.; Siddiqui, R.A. Trans fatty acids: Induction of a pro-inflammatory phenotype in the endothelial cells. Lipids 2012, 47, 647–657. [Google Scholar] [CrossRef]
- Whatley, R.E.; Satoh, K.; Zimmerman, G.A.; McIntyre, T.M.; Prescott, S.M. Proliferation-dependent changes in release of arachidonic acid from endothelial cells. J. Clin. Investig. 1994, 94, 1889–1900. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Scarfo, L.M.; Weller, P.F.; Farber, H.W. Induction of endothelial cell cytoplasmic lipid bodies during hypoxia. Am. J. Physiol. Circ. Physiol. 2001, 280, H294–H301. [Google Scholar] [CrossRef]
- Ibrahim, A.; Arany, Z. Does Endothelium Buffer Fat? Circ. Res. 2017, 120, 1219–1221. [Google Scholar] [CrossRef] [PubMed]
- Colombo, S.; Melo, T.; Martínez-López, M.; Carrasco, M.J.; Domingues, M.R.; Pérez-Sala, D.; Domingues, P. Phospholipidome of endothelial cells shows a different adaptation response upon oxidative, glycative and lipoxidative stress. Sci. Rep. 2018, 8, 12365. [Google Scholar] [CrossRef]
- Magtanong, L.; Ko, P.J.; Dixon, S.J. Emerging roles for lipids in non-apoptotic cell death. Cell Death Differ. 2016, 23, 1099–1109. [Google Scholar] [CrossRef]
- Campia, U.; Moslehi, J.J.; Kordestani, L.A.; Barac, A.; Beckman, J.A.; Chism, D.D.; Cohen, P.; Groarke, J.D.; Herrmann, J.; Reilly, C.M.; et al. Cardio-Oncology: Vascular and Metabolic Perspectives: A Scientific Statement From the American Heart Association. Circulation 2019, 139, 838–839. [Google Scholar] [CrossRef]
- Lu, S.; Sung, T.; Lin, N.; Abraham, R.T.; Jessen, B.A. Lysosomal adaptation: How cells respond to lysosomotropic compounds. PLoS ONE 2017, 12, e173771. [Google Scholar] [CrossRef]
- Halliwell, W.H. Cationic Amphiphilic Drug-Induced Phospholipidosis. Toxicol. Pathol. 1997, 25, 53–60. [Google Scholar] [CrossRef]
- Nonoyama, T.; Fukuda, R. Drug-induced phospholipidosis—Pathological aspects and its prediction. J. Toxicol. Pathol. 2008, 21, 9–24. [Google Scholar] [CrossRef]
- Schmitz, G.; Muller, G. Structure and function of lamellar bodies, lipid-protein complexes involved in storage and secretion of cellular lipids. J. Lipid Res. 1991, 32, 1539–1570. [Google Scholar] [CrossRef]
- Czamara, K.; Adamczyk, A.; Stojak, M.; Radwan, B.; Baranska, M. Astaxanthin as a new Raman probe for biosensing of specific subcellular lipidic structures: Can we detect lipids in cells under resonance conditions? Cell. Mol. Life Sci. 2020. [Google Scholar] [CrossRef]
- Maulucci, G.; Chiarpotto, M.; Papi, M.; Samengo, D.; Pani, G.; De Spirito, M. Quantitative analysis of autophagic flux by confocal pH-imaging of autophagic intermediates. Autophagy 2015, 11, 1905–1916. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Zhang, Q.; Wu, X.; Lu, W.; Tang, H.; Liang, Z.; Gu, Y.; Song, S.; Ayon, R.J.; Wang, Z.; et al. Chloroquine is a potent pulmonary vasodilator that attenuates hypoxia-induced pulmonary hypertension. Br. J. Pharmacol. 2017, 174, 4155–4172. [Google Scholar] [CrossRef] [PubMed]
- Pestana, C.R.; Oishi, J.C.; Sobreiro, H.; Gerson, S.A.; Rodrigues, J. Inhibition of autophagy by chloroquine stimulates nitric oxide production and protects endothelial function during serum deprivation. Cell. Physiol. Biochem. 2015, 37, 1168–1177. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.H.; Weiss, E.; Mkaddem, S.B.; Mabire, M.; Choinier, P.M.; Picq, O.; Thibault-Sogorb, T.; Hegde, P.; Pishvaie, D.; Bens, M.; et al. LC3-associated phagocytosis protects against inflammation and liver fibrosis via immunoreceptor inhibitory signaling. Sci. Transl. Med. 2020, 12, 1–13. [Google Scholar] [CrossRef]
- Yamamoto, A.; Tagawa, Y.; Yoshimori, T.; Moriyama, Y.; Masaki, R.; Tashiro, Y. Bafilomycin A1 Prevents Maturation of Autophagic Vacuoles by Inhibiting Fusion between Autophagosomes and Lysosomes in Rat Hepatoma Cell Line, H-4-II-E Cells. Cell Struct. Funct. 1998, 23, 33–42. [Google Scholar] [CrossRef]
- Frosch, T.; Schmitt, M.; Bringmann, G.; Kiefer, W.; Popp, J. Structural Analysis of the Anti-Malaria Active Agent Chloroquine under Physiological Conditions. J. Phys. Chem. B 2007, 111, 1815–1822. [Google Scholar] [CrossRef]
- Steinbauer, B.; Mehnert, T.; Beyer, K. Hydration and lateral organization in phospholipid bilayers containing sphingomyelin: A 2H-NMR study. Biophys. J. 2003, 85, 1013–1024. [Google Scholar] [CrossRef][Green Version]
- Bik, E.; Mielniczek, N.; Jarosz, M.; Denbigh, J.; Budzynska, R.; Baranska, M.; Majzner, K. Tunicamycin induced endoplasmic reticulum changes in the endothelial cells investigated: In vitro by confocal Raman imaging. Analyst 2019, 144, 6561–6569. [Google Scholar] [CrossRef]
- Bik, E.; Dorosz, A.; Mateuszuk, L.; Baranska, M.; Majzner, K. Fixed versus live endothelial cells: The effect of glutaraldehyde fixation manifested by characteristic bands on the Raman spectra of cells. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 240, 118460. [Google Scholar] [CrossRef]
- Schwake, M.; Schröder, B.; Saftig, P. Lysosomal Membrane Proteins and Their Central Role in Physiology. Traffic 2013, 14, 739–748. [Google Scholar] [CrossRef]
- Mizushima, N.; Yoshimori, T.; Levine, B. Methods in mammalian autophagy research. Cell 2010, 140, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Czamara, K.; Majzner, K.; Pilarczyk, M.; Kochan, K.; Kaczor, A.; Baranska, M.; Czamara, K.; Majzner, K.; Pacia, M.Z.Z.; Kochan, K.; et al. Raman spectroscopy of lipids: A review. J. Raman Spectrosc. 2015, 46, 4–20. [Google Scholar] [CrossRef]
- Majzner, K.; Tott, S.; Roussille, L.; Deckert, V.; Chlopicki, S.; Baranska, M. Uptake of fatty acids by a single endothelial cell investigated by Raman spectroscopy supported by AFM. Analyst 2018, 143, 970–980. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bik, E.; Mateuszuk, L.; Orleanska, J.; Baranska, M.; Chlopicki, S.; Majzner, K. Chloroquine-Induced Accumulation of Autophagosomes and Lipids in the Endothelium. Int. J. Mol. Sci. 2021, 22, 2401. https://doi.org/10.3390/ijms22052401
Bik E, Mateuszuk L, Orleanska J, Baranska M, Chlopicki S, Majzner K. Chloroquine-Induced Accumulation of Autophagosomes and Lipids in the Endothelium. International Journal of Molecular Sciences. 2021; 22(5):2401. https://doi.org/10.3390/ijms22052401
Chicago/Turabian StyleBik, Ewelina, Lukasz Mateuszuk, Jagoda Orleanska, Malgorzata Baranska, Stefan Chlopicki, and Katarzyna Majzner. 2021. "Chloroquine-Induced Accumulation of Autophagosomes and Lipids in the Endothelium" International Journal of Molecular Sciences 22, no. 5: 2401. https://doi.org/10.3390/ijms22052401
APA StyleBik, E., Mateuszuk, L., Orleanska, J., Baranska, M., Chlopicki, S., & Majzner, K. (2021). Chloroquine-Induced Accumulation of Autophagosomes and Lipids in the Endothelium. International Journal of Molecular Sciences, 22(5), 2401. https://doi.org/10.3390/ijms22052401