Silencing DNA Polymerase β Induces Aneuploidy as a Biomarker of Poor Prognosis in Oral Squamous Cell Cancer
Abstract
1. Introduction
2. Results
2.1. Correlations between POLB and Clinicopathological Parameters
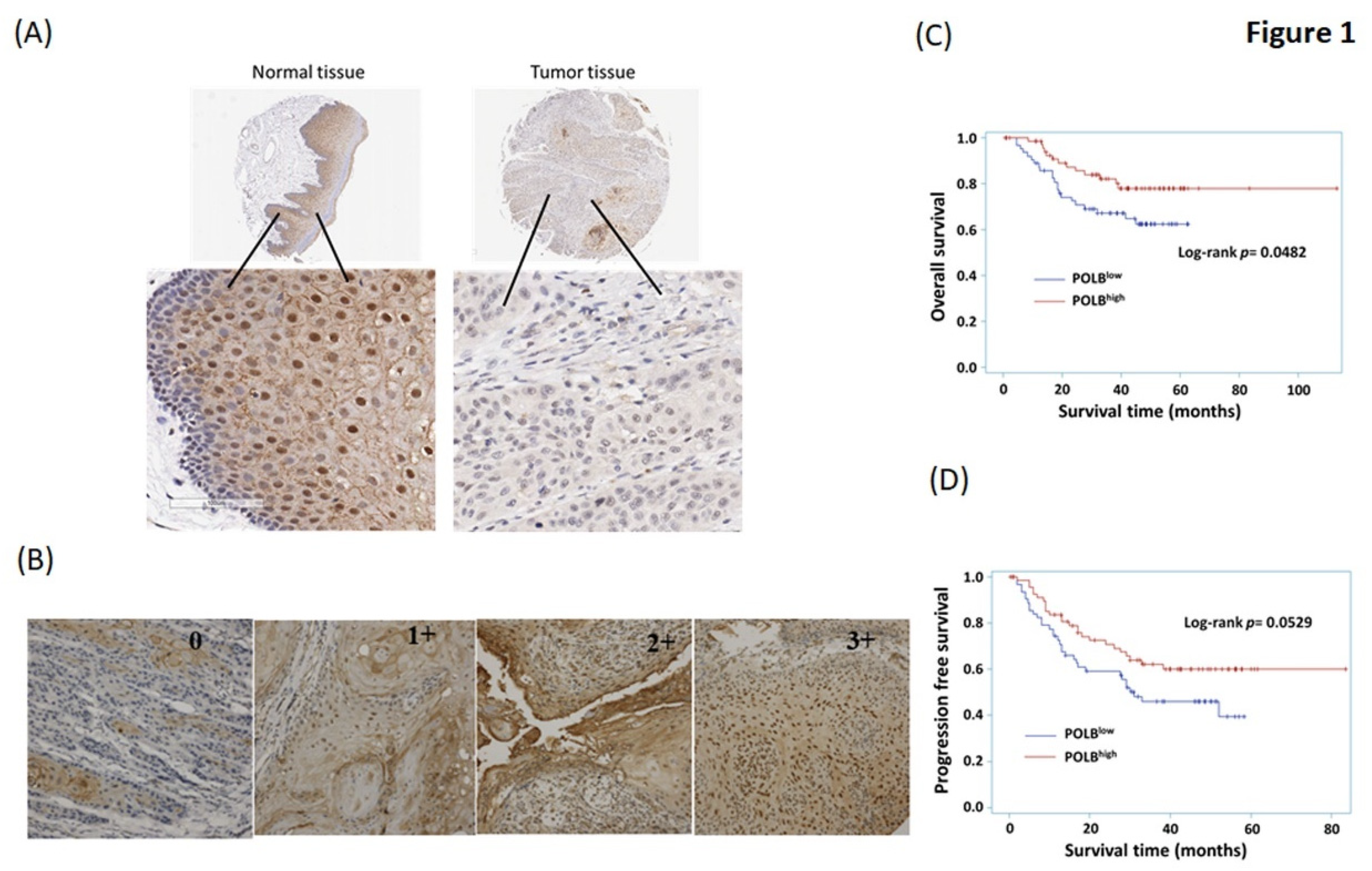
2.2. High Level of POLB Is Positive Correlated with Poor Outcome of OSCC
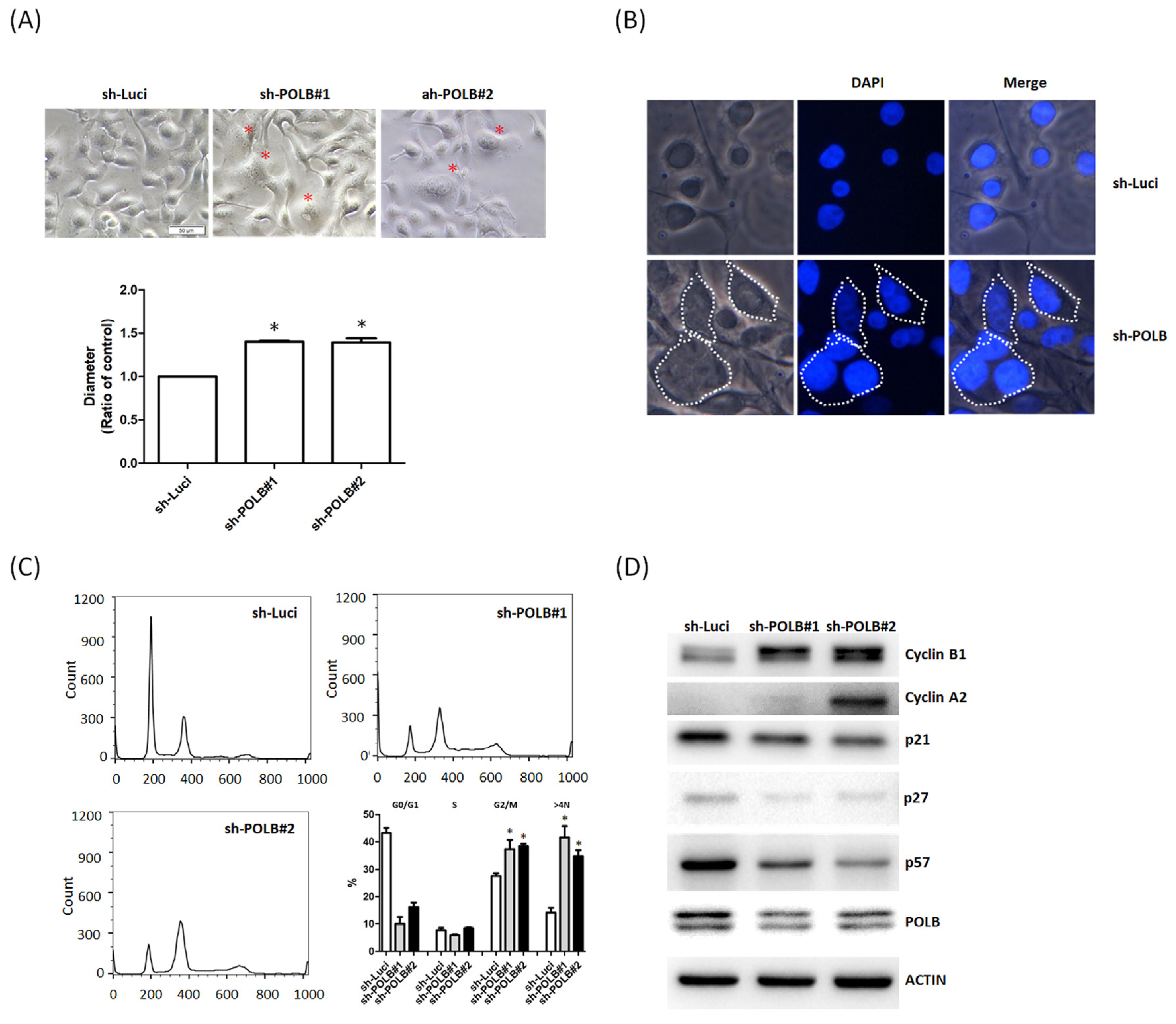
2.3. Loss of POLB Increases Cell Proliferation of OSCC Cancer
2.4. Depletion of POLB Leads to Cell-Cycle Dysregulation and Aneuploidy Formation
3. Discussion
4. Material and Methods
4.1. Specimens
4.2. Immunohistochemical Staining
4.3. Scoring
4.4. Cell Culture
4.5. Antibodies
4.6. Western Blotting
4.7. POLB Short Interfering RNA Transfection
4.8. Cell Viability Test
4.9. Flow Cytometry for Cell-Cycle Analysis
4.10. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Parkin, D.M.; Pisani, P.; Ferlay, J. Global cancer statistics. CA Cancer J. Clin. 1999, 49, 33–64. [Google Scholar] [CrossRef]
- Jemal, A.; Siegel, R.; Ward, E.; Hao, Y.; Xu, J.; Thun, M.J. Cancer Statistics, 2009. CA Cancer J. Clin. 2009, 59, 225–249. [Google Scholar] [CrossRef]
- Lin, Y.-S.; Jen, Y.-M.; Wang, B.-B.; Lee, J.-C.; Kang, B.-H. Epidemiology of Oral Cavity Cancer in Taiwan with Emphasis on the Role of Betel Nut Chewing. ORL 2005, 67, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-J.; You, S.-L.; Lin, L.-H.; Hsu, W.-L.; Yang, Y.-W. Cancer epidemiology and control in Taiwan: A brief review. Jpn. J. Clin. Oncol. 2002, 32, S66–S81. [Google Scholar] [CrossRef][Green Version]
- Chen, J.Y.-F.; Chang, Y.-L.; Yu, Y.-C.; Chao, C.-C.; Kao, H.-W.; Wu, C.-T.; Lin, W.-C.; Ko, J.-Y.; Jou, Y.-S. Specific induction of the high-molecular-weight microtubule-associated protein 2 (hmw-MAP2) by betel quid extract in cultured oral keratinocytes: Clinical implications in betel quid-associated oral squamous cell carcinoma (OSCC). Carcinology 2003, 25, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.-F.; Chuang, W.-Y.; Chen, I.-H.; Liao, C.-T.; Wang, H.-M.; Hsieh, L.-L. EGFR protein overexpression and mutation in areca quid-associated oral cavity squamous cell carcinoma in Taiwan. Head Neck 2009, 31, 1068–1077. [Google Scholar] [CrossRef]
- Zorat, P.L.; Paccagnella, A.; Cavaniglia, G.; Loreggian, L.; Gava, A.; Mione, C.A.; Boldrin, F.; Marchiori, C.; Lunghi, F.; Fede, A.; et al. Randomized Phase III Trial of Neoadjuvant Chemotherapy in Head and Neck Cancer: 10-Year Follow-Up. J. Natl. Cancer Inst. 2004, 96, 1714–1717. [Google Scholar] [CrossRef]
- Scher, E.D.; Romesser, P.B.; Chen, C.; Ho, F.; Wuu, Y.; Sherman, E.J.; Fury, M.G.; Wong, R.J.; McBride, S.; Lee, N.Y.; et al. Definitive chemoradiation for primary oral cavity carcinoma: A single institution experience. Oral Oncol. 2015, 51, 709–715. [Google Scholar] [CrossRef]
- Fan, K.-H.; Lin, C.-Y.; Kang, C.-J.; Huang, S.-F.; Wang, H.-M.; Chen, E.Y.-C.; Chen, I.-H.; Liao, C.-T.; Cheng, A.-J.; Chang, J.T.-C. Combined-modality treatment for advanced oral tongue squamous cell carcinoma. Int. J. Radiat. Oncol. 2007, 67, 453–461. [Google Scholar] [CrossRef]
- Sasahira, T.; Kirita, T. Hallmarks of Cancer-Related Newly Prognostic Factors of Oral Squamous Cell Carcinoma. Int. J. Mol. Sci. 2018, 19, 2413. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Eckert, A.W.; Kappler, M.; Große, I.; Wickenhauser, C.; Seliger, B. Current Understanding of the HIF-1-Dependent Metabolism in Oral Squamous Cell Carcinoma. Int. J. Mol. Sci. 2020, 21, 6083. [Google Scholar] [CrossRef]
- Zhou, J.; Huang, S.; Wang, L.; Yuan, X.; Dong, Q.; Zhang, D.; Wang, X. Clinical and prognostic significance of HIF-1 overex-pression in oral squamous cell carcinoma: A meta-analysis. World J. Surg. Oncol. 2017, 15, 104. [Google Scholar] [CrossRef] [PubMed]
- Tsai, Y.-S.; Lee, K.-W.; Huang, J.-L.; Liu, Y.-S.; Juo, S.-H.H.; Kuo, W.-R.; Chang, J.-G.; Lin, C.-S.; Jong, Y.-J. Arecoline, a major alkaloid of areca nut, inhibits p53, represses DNA repair, and triggers DNA damage response in human epithelial cells. Toxicology 2008, 249, 230–237. [Google Scholar] [CrossRef]
- Lee, P.-H.; Chang, M.-C.; Chang, W.-H.; Wang, T.-M.; Wang, Y.-J.; Hahn, L.-J.; Ho, Y.-S.; Lin, C.-Y.; Jeng, J.-H. Prolonged exposure to arecoline arrested human KB epithelial cell growth: Regulatory mechanisms of cell cycle and apoptosis. Toxicology 2006, 220, 81–89. [Google Scholar] [CrossRef]
- Löbrich, M.; Jeggo, P.A. The impact of a negligent G2/M checkpoint on genomic instability and cancer induction. Nat. Rev. Cancer 2007, 7, 861–869. [Google Scholar] [CrossRef]
- Vitale, I.; Galluzzi, L.; Senovilla, L.; Criollo, A.; Jemaà, M.; Castedo, M.; Kroemer, G. Illicit survival of cancer cells during polyploidization and depolyploidization. Cell Death Differ. 2010, 18, 1403–1413. [Google Scholar] [CrossRef] [PubMed]
- Pickering, C.R.; Zhang, J.; Yoo, S.Y.; Bengtsson, L.; Moorthy, S.; Neskey, D.M.; Zhao, M.; Alves, M.V.O.; Chang, K.; Drummond, J.; et al. Integrative Genomic Characterization of Oral Squamous Cell Carcinoma Identifies Frequent Somatic Drivers. Cancer Discov. 2013, 3, 770–781. [Google Scholar] [CrossRef]
- Zack, T.I.; Schumacher, S.E.; Carter, S.L.; Cherniack, A.D.; Saksena, G.; Tabak, B.; Lawrence, M.S.; Zhang, C.-Z.; Wala, J.; Mermel, C.H.; et al. Pan-cancer patterns of somatic copy number alteration. Nat. Genet. 2013, 45, 1134–1140. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, A.; Roy, R.; Snijders, A.M.; Hamilton, G.; Paquette, J.; Tokuyasu, T.; Bengtsson, H.; Jordan, R.C.K.; Olshen, A.B.; Pinkel, D.; et al. Two Distinct Routes to Oral Cancer Differing in Genome Instability and Risk for Cervical Node Metastasis. Clin. Cancer Res. 2011, 17, 7024–7034. [Google Scholar] [CrossRef]
- Mooren, J.J.; Kremer, B.; Claessen, S.M.; Voogd, A.C.; Bot, F.J.; Peter Klussmann, J.; Huebbers, C.U.; Hopman, A.H.; Ramaekers, F.C.; Speel, E.J.M. Chromosome stability in tonsillar squamous cell carcinoma is associated with HPV16 inte-gration and indicates a favorable prognosis. Int. J. Cancer 2013, 132, 1781–1789. [Google Scholar] [CrossRef]
- Mathews, L.A.; Cabarcas, S.M.; Hurt, E.M.; Zhang, X.; Jaffee, E.M.; Farrar, W.L. Increased Expression of DNA Repair Genes in Invasive Human Pancreatic Cancer Cells. Pancreas 2011, 40, 730–739. [Google Scholar] [CrossRef] [PubMed]
- Kauffmann, A.; Rosselli, F.; Lazar, V.; Winnepenninckx, V.; Mansuet-Lupo, A.; Dessen, P.; Oord, J.V.D.; Spatz, A.; Sarasin, A. High expression of DNA repair pathways is associated with metastasis in melanoma patients. Oncogene 2007, 27, 565–573. [Google Scholar] [CrossRef]
- Pitroda, S.P.; Pashtan, I.M.; Logan, H.L.; Budke, B.; Darga, T.E.; Weichselbaum, R.R.; Connell, P.P. DNA Repair Pathway Gene Expression Score Correlates with Repair Proficiency and Tumor Sensitivity to Chemotherapy. Sci. Transl. Med. 2014, 6, 229ra42. [Google Scholar] [CrossRef]
- Dabholkar, M.; Vionnet, J.; Bostick-Bruton, F.; Yu, J.J.; Reed, E. Messenger RNA levels of XPAC and ERCC1 in ovarian cancer tissue correlate with response to platinum-based chemotherapy. J. Clin. Investig. 1994, 94, 703–708. [Google Scholar] [CrossRef]
- Çağlayan, M.; Batra, V.K.; Sassa, A.; Prasad, R.; Wilson, S.H. Role of polymerase β in complementing aprataxin deficiency during abasic-site base excision repair. Nat. Struct. Mol. Biol. 2014, 21, 497–499. [Google Scholar] [CrossRef]
- Mark, W.-Y.; Liao, J.C.; Lu, Y.; Ayed, A.; Laister, R.; Szymczyna, B.; Chakrabartty, A.; Arrowsmith, C.H. Characterization of Segments from the Central Region of BRCA1: An Intrinsically Disordered Scaffold for Multiple Protein–Protein and Protein–DNA Interactions? J. Mol. Biol. 2005, 345, 275–287. [Google Scholar] [CrossRef] [PubMed]
- Sweasy, J.B.; Lang, T.; Starcevic, D.; Sun, K.-W.; Lai, C.-C.; DiMaio, D.; Dalal, S. Expression of DNA polymerase β can-cer-associated variants in mouse cells results in cellular transformation. Proc. Natl. Acad. Sci. USA 2005, 102, 14350–14355. [Google Scholar] [CrossRef] [PubMed]
- Bergoglio, V.; Pillaire, M.-J.; Lacroix-Triki, M.; Raynaud-Messina, B.; Canitrot, Y.; Bieth, A.; Garès, M.; Wright, M.; Delsol, G.; Loeb, L.A. Deregulated DNA polymerase β induces chromosome instability and tumorigenesis. Cancer Res. 2002, 62, 3511–3514. [Google Scholar] [PubMed]
- Ramakodi, M.P.; Devarajan, K.; Mph, E.B.; Bs, D.G.; Luce, D.; Deloumeaux, J.; Duflo, S.; Liu, J.C.; Mehra, R.; Kulathinal, R.J.; et al. Integrative genomic analysis identifies ancestry-related expression quantitative trait loci on DNA polymerase β and supports the association of genetic ancestry with survival disparities in head and neck squamous cell carcinoma. Cancer 2017, 123, 849–860. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Xue, P.; Li, M.; Zhao, J.; Dong, Z.; Zhao, G. DNA polymerase beta overexpression correlates with poor prognosis in esophageal cancer patients. Chin. Sci. Bull. 2013, 58, 3274–3279. [Google Scholar] [CrossRef][Green Version]
- Tan, X.; Wu, X.; Ren, S.; Wang, H.; Li, Z.; Alshenawy, W.; Li, W.; Cui, J.; Luo, G.; Siegel, R.S.; et al. A Point Mutation in DNA Polymerase β (POLB) Gene Is Associated with Increased Progesterone Receptor (PR) Expression and Intraperitoneal Metastasis in Gastric Cancer. J. Cancer 2016, 7, 1472–1480. [Google Scholar] [CrossRef][Green Version]
- Mathews, L.A.; Cabarcas, S.M.; Farrar, W.L. DNA repair: The culprit for tumor-initiating cell survival? Cancer Metastasis Rev. 2011, 30, 185–197. [Google Scholar] [CrossRef]
- Marangos, P.; Carroll, J. The dynamics of cyclin B1 distribution during meiosis I in mouse oocytes. Reproduction 2004, 128, 153–162. [Google Scholar] [CrossRef][Green Version]
- Landis, S.H.; Murray, T.; Bolden, M.S.; Wingo, P.A. Cancer statistics, 1999. CA Cancer J. Clin. 1999, 49, 8–31. [Google Scholar] [CrossRef]
- Notani, P.N. Global variation in cancer incidence and mortality. Curr. Sci. 2001, 465–474. [Google Scholar]
- Chen, T.-W.; Lee, C.-C.; Liu, H.; Wu, C.-S.; Pickering, C.R.; Huang, P.-J.; Wang, J.; Chang, I.Y.-F.; Yeh, Y.-M.; Chen, C.-D.; et al. APOBEC3A is an oral cancer prognostic biomarker in Taiwanese carriers of an APOBEC deletion polymorphism. Nat. Commun. 2017, 8, 1–13. [Google Scholar] [CrossRef]
- Zhao, X.; Sun, S.; Zeng, X.; Cui, L. Expression profiles analysis identifies a novel three-mRNA signature to predict overall survival in oral squamous cell carcinoma. Am. J. Cancer Res. 2018, 8, 450–461. [Google Scholar] [PubMed]
- Hadler-Olsen, E.; Wirsing, A.M. Tissue-infiltrating immune cells as prognostic markers in oral squamous cell carcinoma: A systematic review and meta-analysis. Br. J. Cancer 2019, 120, 714–727. [Google Scholar] [CrossRef] [PubMed]
- Lingle, W.L.; Salisbury, J.L. Altered Centrosome Structure Is Associated with Abnormal Mitoses in Human Breast Tumors. Am. J. Pathol. 1999, 155, 1941–1951. [Google Scholar] [CrossRef]
- Yamtich, J.; Nemec, A.A.; Keh, A.; Sweasy, J.B. A germline polymorphism of DNA polymerase beta induces genomic in-stability and cellular transformation. PLoS Genet. 2012, 8, e1003052. [Google Scholar] [CrossRef]
- Canitrot, Y.; Fréchet, M.; Servant, L.; Cazaux, C.; Hoffmann, J. Overexpression of DNA polymerase β: A genomic instability enhancer process. FASEB J. 1999, 13, 1107–1111. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Klattenhoff, A.W.; Thakur, M.; Sebastian, M.; Kidane, D. Mutation in DNA Polymerase Beta Causes Spontaneous Chromosomal Instability and Inflammation-Associated Carcinogenesis in Mice. Cancers 2019, 11, 1160. [Google Scholar] [CrossRef] [PubMed]
- Canitrot, Y.; Cazaux, C.; FREchet, M.; Bouayadi, K.; Lesca, C.; Salles, B.; Hoffmann, J.-S. Overexpression of DNA polymerase β in cell results in a mutator phenotype and a decreased sensitivity to anticancer drugs. Proc. Natl. Acad. Sci. USA 1998, 95, 12586–12590. [Google Scholar] [CrossRef] [PubMed]
- Canitrot, Y.; Hoffmann, J.-S.; Calsou, P.; Hayakawa, H.; Salles, B.; Cazaux, C. Nucleotide excision repair DNA synthesis by excess DNA polymerase β: A potential source of genetic instability in cancer cells. FASEB J. 2000, 14, 1765–1774. [Google Scholar] [CrossRef][Green Version]
- Srivastava, D.K.; Husain, I.; Arteaga, C.L.; Wilson, S.H. DNA polymerase β expression differences in selected human tumors and cell lines. Carcinogenesis 1999, 20, 1049–1054. [Google Scholar] [CrossRef] [PubMed]
- Hou, M.-F.; Luo, C.-W.; Chang, T.-M.; Hung, W.-C.; Chen, T.-Y.; Tsai, Y.-L.; Chai, C.-Y.; Pan, M.-R. The NuRD com-plex-mediated p21 suppression facilitates chemoresistance in BRCA-proficient breast cancer. Exp. Cell Res. 2017, 359, 458–465. [Google Scholar] [CrossRef] [PubMed]
- Su, J.-L.; Shih, J.-Y.; Yen, M.-L.; Jeng, Y.-M.; Chang, C.-C.; Hsieh, C.-Y.; Wei, L.-H.; Yang, P.-C.; Kuo, M.-L. Cyclooxygenase-2 induces EP1-and HER-2/Neu-dependent vascular endothelial growth factor-C up-regulation: A novel mechanism of lym-phangiogenesis in lung adenocarcinoma. Cancer Res. 2004, 64, 554–564. [Google Scholar] [CrossRef]
- Cepeda, V.; Fuertes, M.; Castilla, J.; Alonso, C.; Quevedo, C.; Soto, M.; Perez, J. Poly(ADP-Ribose) Polymerase-1 (PARP-1) Inhibitors in Cancer Chemotherapy. Recent Pat. Anti-Cancer Drug Discov. 2006, 1, 39–53. [Google Scholar] [CrossRef]
- Yin, H.-L.; Wu, C.-C.; Lin, C.-H.; Chai, C.-Y.; Hou, M.-F.; Chang, S.-J.; Tsai, H.-P.; Hung, W.-C.; Pan, M.-R.; Luo, C.-W. β1 Integrin as a Prognostic and Predictive Marker in Triple-Negative Breast Cancer. Int. J. Mol. Sci. 2016, 17, 1432. [Google Scholar] [CrossRef]
- Tian, Y.-F.; Wang, H.-C.; Luo, C.-W.; Hung, W.-C.; Lin, Y.-H.; Chen, T.-Y.; Li, C.-F.; Lin, C.-Y.; Pan, M.-R. Preprogramming therapeutic response of PI3K/mTOR dual inhibitor via the regulation of EHMT2 and p27 in pancreatic cancer. Am. J. Cancer Res. 2018, 8, 1812–1822. [Google Scholar] [PubMed]

| Characteristics | All (n = 133) |
|---|---|
| Age, years | 55.05 ± 10.29 |
| Sex, No. (%) | |
| Male | 125 (93.98) |
| Female | 8 (6.02) |
| Tumor site, No. (%) | |
| Buccal | 82 (61.65) |
| Non-buccal | 51 (38.35) |
| Alcohol, No. (%) | |
| Yes | 85 (63.91) |
| No | 48 (36.09) |
| Areca nuts, No. (%) | |
| Yes | 102 (76.69) |
| No | 31 (23.31) |
| Smoking, No. (%) | |
| Yes | 112 (84.21) |
| No | 21 (15.79) |
| Lymph node metastases, No. (%) | |
| Yes | 33 (24.81) |
| No | 100 (75.19) |
| Grade, No. (%) | |
| 1 | 59 (44.70) |
| 2 | 71 (53.79) |
| 3 | 2 (1.52) |
| Stage, No. (%) | |
| I | 59 (44.70) |
| II | 23 (17.42) |
| III | 15 (11.36) |
| IV | 35 (26.52) |
| Parameters | n | POLB Expression, n (%) | p-Value | |
|---|---|---|---|---|
| Low | High | |||
| Total | 133 | 63 (47.37) | 70 (52.63) | |
| Age | 0.1893 | |||
| ≤40 yrs | 5 | 4 (6.35) | 1 (1.43) | |
| >40 yrs | 128 | 59 (93.65) | 69 (98.57) | |
| Gender | 0.0068 * | |||
| female | 8 | 0 (0.00) | 8 (11.43) | |
| male | 125 | 63 (100.00) | 62 (88.57) | |
| Tumor size | 0.0961 | |||
| ≤2.0 cm | 65 | 26 (41.27) | 39 (55.71) | |
| >2.0 cm | 68 | 37 (58.73) | 31 (44.29) | |
| Grade | 0.6009 | |||
| I | 59 | 26 (41.94) | 33 (47.14) | |
| II/III | 73 | 36 (58.06) | 37 (52.86) | |
| Tumor stage | 0.0176 * | |||
| T1/T2 | 101 | 42 (66.67) | 59 (84.29) | |
| T3/T4 | 32 | 21 (33.33) | 11 (15.71) | |
| Nodal stage | 0.5822 | |||
| N0 | 100 | 46 (73.02) | 54 (77.14) | |
| N1/N2/N3 | 33 | 17 (26.98) | 16 (22.86) | |
| Metastatic stage | 0.3410 | |||
| M0 | 132 | 63 (100.00) | 69 (98.57) | |
| M1 | 1 | 0 (0.00) | 1 (1.43) | |
| Tumor metastasis | 0.1907 | |||
| Absent | 123 | 56 (88.89) | 67 (95.1) | |
| Present | 10 | 7 (11.11) | 3 (4.29) | |
| Tumor recurrent | 0.1045 | |||
| Absent | 82 | 34 (54.84) | 48 (68.57) | |
| Present | 50 | 28 (45.16) | 22 (31.43) | |
| Survival status | 0.0325 * | |||
| Survival | 98 | 41 (65.08) | 57 (81.43) | |
| Death | 35 | 22 (34.92) | 13 (18.57) | |
| Alcohol | 0.9242 | |||
| N | 48 | 23 (36.51) | 25 (35.71) | |
| Y | 85 | 40 (63.49) | 45 (64.29) | |
| Areca nuts | 0.8968 | |||
| N | 31 | 15 (23.81) | 16 (22.86) | |
| Y | 102 | 48 (76.19) | 54 (77.14) | |
| Smoking | 0.146 | |||
| N | 21 | 13 (20.63) | 8 (11.43) | |
| Y | 112 | 50 (79.37) | 62 (88.57) | |
| Factors | Univariate | Multivariate | ||
|---|---|---|---|---|
| HR (95% CI) | p-Value | HR (95% CI) | p-Value | |
| POLB expression | 0.0688 | 0.0360 * | ||
| Low | 1.0 | 1.0 | ||
| High | 0.526 (0.263–1.051) | 0.464 (0.226–0.951) | ||
| Tumor Size | 0.0055 * | 0.1436 | ||
| ≤2.0 cm | 1.0 | 1.0 | ||
| >2.0 cm | 2.945 (1.373–6.315) | 1.846 (0.812–4.198) | ||
| Grade | 0.0081 * | 0.1823 | ||
| I | 1.0 | 1.0 | ||
| II/III | 0.356 (0.166–0.765) | 0.578 (0.259–1.293) | ||
| Nodal stage | 0.0002 * | 0.0065 * | ||
| N0 | 1.0 | 1.0 | ||
| N1/N2/N3 | 3.534 (1.799–6.942) | 2.777 (1.330–5.796) | ||
| Alcohol | 0.0688 | 0.3797 | ||
| N | 1.0 | 1.0 | ||
| Y | 2.164 (0.942–4.969) | 1.620 (0.552–4.750) | ||
| Areca nut | 0.3419 | 0.0365 * | ||
| N | 1.0 | 1.0 | ||
| Y | 0.699 (0.334–1.462) | 0.413 (0.181–0.946) | ||
| Smoking | 0.2833 | 0.4950 | ||
| N | 1.0 | 1.0 | ||
| Y | 1.913 (0.585–6.260) | 1.678 (0.380–7.413) | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.-C.; Chan, L.-P.; Wu, C.-C.; Chang, S.-J.; Moi, S.-H.; Luo, C.-W.; Pan, M.-R. Silencing DNA Polymerase β Induces Aneuploidy as a Biomarker of Poor Prognosis in Oral Squamous Cell Cancer. Int. J. Mol. Sci. 2021, 22, 2402. https://doi.org/10.3390/ijms22052402
Wang H-C, Chan L-P, Wu C-C, Chang S-J, Moi S-H, Luo C-W, Pan M-R. Silencing DNA Polymerase β Induces Aneuploidy as a Biomarker of Poor Prognosis in Oral Squamous Cell Cancer. International Journal of Molecular Sciences. 2021; 22(5):2402. https://doi.org/10.3390/ijms22052402
Chicago/Turabian StyleWang, Hui-Ching, Leong-Perng Chan, Chun-Chieh Wu, Shu-Jyuan Chang, Sin-Hua Moi, Chi-Wen Luo, and Mei-Ren Pan. 2021. "Silencing DNA Polymerase β Induces Aneuploidy as a Biomarker of Poor Prognosis in Oral Squamous Cell Cancer" International Journal of Molecular Sciences 22, no. 5: 2402. https://doi.org/10.3390/ijms22052402
APA StyleWang, H.-C., Chan, L.-P., Wu, C.-C., Chang, S.-J., Moi, S.-H., Luo, C.-W., & Pan, M.-R. (2021). Silencing DNA Polymerase β Induces Aneuploidy as a Biomarker of Poor Prognosis in Oral Squamous Cell Cancer. International Journal of Molecular Sciences, 22(5), 2402. https://doi.org/10.3390/ijms22052402






