Regulation and Function of Defense-Related Callose Deposition in Plants
Abstract
1. Introduction
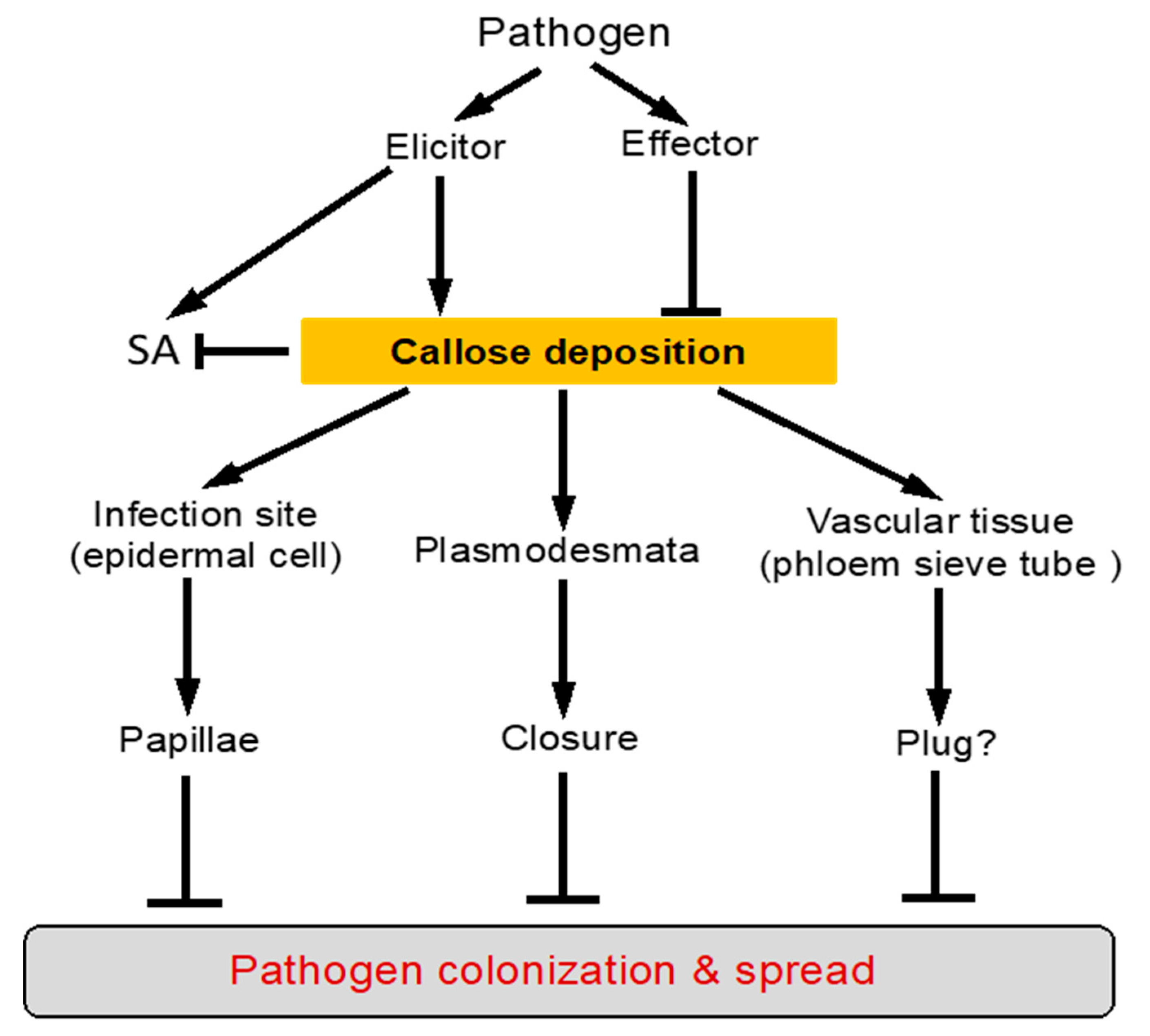
2. Defense-Related Callose Deposition in Plants
3. Regulation of Defense-Related Callose Deposition
3.1. Signaling Pathways Controlling Pathogen-Induced Callose Deposition
3.2. Biogenesis and Activation of Pathogen-Responsive Callose Synthases
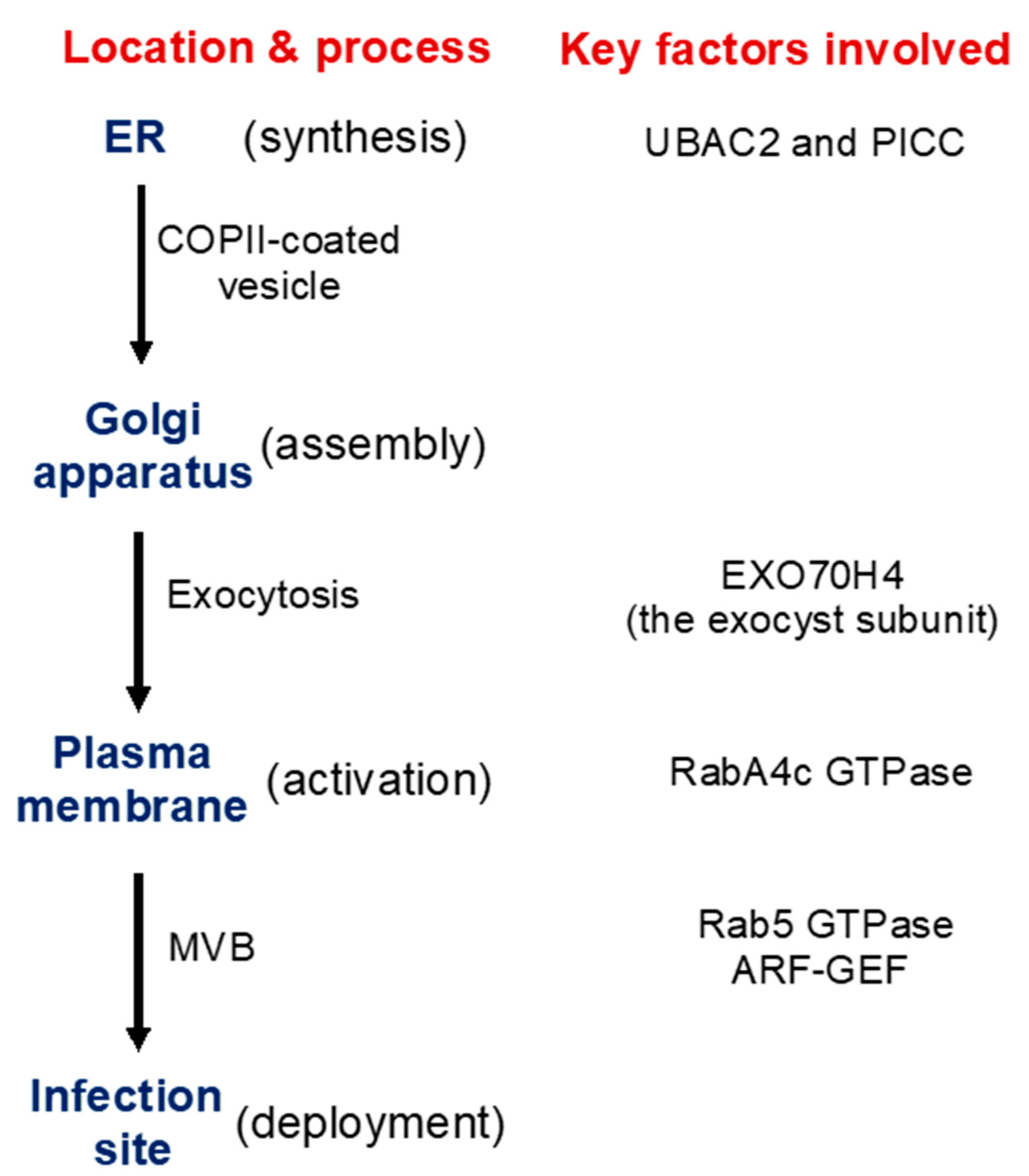
3.3. Transport Processes in Pathogen-Induced Callose Deposition
4. Function of Callose Deposition in Plant Defense
4.1. Effects of Callose Deposition on Plant Cell Wall
4.2. Association of Increased Callose Deposition with Plant Disease Resistance
4.3. Chemical Inhibition of Callose Synthesis Compromises Plant Disease Resistance
4.4. Genetic and Molecular Evidence for the Role of Callose Synthesis in Plant Disease Resistance
4.5. Undermining Callose-Mediated Defense by Pathogen Effectors
5. Conclusions and Prospects
Author Contributions
Funding
Conflicts of Interest
References
- Stone, B.A.; Clarke, A.C. Chemistry and Biology of (1->3)-b-Glucans; La Trobe University Press: Bundoora, Victoria, 1992. [Google Scholar]
- Hong, Z.L.; Delauney, A.J.; Verma, D.P.S. A cell plate specific callose synthase and its interaction with phragmoplastin. Plant Cell 2001, 13, 755–768. [Google Scholar] [CrossRef]
- Hong, Z.L.; Zhang, Z.M.; Olson, J.M.; Verma, D.P.S. A novel UDP-glucose transferase is part of the callose synthase complex and interacts with phragmoplastin at the forming cell plate. Plant Cell 2001, 13, 769–779. [Google Scholar] [CrossRef]
- Mccormick, S. Male Gametophyte Development. Plant Cell 1993, 5, 1265–1275. [Google Scholar] [CrossRef] [PubMed]
- Barratt, D.H.P.; Kolling, K.; Graf, A.; Pike, M.; Calder, G.; Findlay, K.; Zeeman, S.C.; Smith, A.M. Callose Synthase GSL7 Is Necessary for Normal Phloem Transport and Inflorescence Growth in Arabidopsis. Plant Physiol 2011, 155, 328–341. [Google Scholar] [CrossRef] [PubMed]
- Xie, B.; Wang, X.M.; Zhu, M.S.; Zhang, Z.M.; Hong, Z.L. CalS7 encodes a callose synthase responsible for callose deposition in the phloem. Plant J. 2011, 65, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.W.; Kumar, R.; Iswanto, A.B.B.; Kim, J.Y. Callose balancing at plasmodesmata. J. Exp. Bot. 2018, 69, 5325–5339. [Google Scholar] [CrossRef]
- Voigt, C.A. Callose-mediated resistance to pathogenic intruders in plant defense-related papillae. Front. Plant Sci. 2014, 5, 168. [Google Scholar] [CrossRef]
- Osbourn, A.; Carter, J.; Daniels, M.; Dufresne, M.; Hugouvieux, V.; Leggett, M.; Martin-Hernandez, A.; Melton, R.; Morrissey, J.; Papadopoulou, K. Preformed antifungal compounds and plant defence. Biol. Plant-Microbe Interact. 2000, 2, 170–174. [Google Scholar]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Zipfel, C. Plant pattern-recognition receptors. Trends Immunol. 2014, 35, 345–351. [Google Scholar] [CrossRef]
- Dou, D.; Zhou, J.M. Phytopathogen effectors subverting host immunity: Different foes, similar battleground. Cell Host Microbe 2012, 12, 484–495. [Google Scholar] [CrossRef]
- Bonardi, V.; Cherkis, K.; Nishimura, M.T.; Dangl, J.L. A new eye on NLR proteins: Focused on clarity or diffused by complexity? Curr. Opin. Immunol. 2012, 24, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, K.; Katagiri, F. Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr. Opin. Plant Biol. 2010, 13, 459–465. [Google Scholar] [CrossRef]
- Tsuda, K.; Somssich, I.E. Transcriptional networks in plant immunity. New Phytol. 2015, 206, 932–947. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, A.K.; Lipka, V.; Burton, R.A.; Panstruga, R.; Strizhov, N.; Schulze-Lefert, P.; Fincher, G.B. An Arabidopsis Callose Synthase, GSL5, Is Required for Wound and Papillary Callose Formation. Plant Cell 2003, 15, 2503–2513. [Google Scholar] [CrossRef]
- Nurnberger, T.; Kemmerling, B. PAMP-Triggered Basal Immunity in Plants. Adv. Bot. Res. 2009, 51, 1–38. [Google Scholar] [CrossRef]
- Iriti, M.; Faoro, F. Chemical Diversity and Defence Metabolism: How Plants Cope with Pathogens and Ozone Pollution. Int. J. Mol. Sci. 2009, 10, 3371–3399. [Google Scholar] [CrossRef]
- Hou, S.G.; Liu, Z.Y.; Shen, H.X.; Wu, D.J. Damage-Associated Molecular Pattern-Triggered Immunity in Plants. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Conrath, U. Systemic Acquired Resistance. Plant Signal. Behav. 2006, 1, 179–184. [Google Scholar] [CrossRef]
- Kohler, A.; Schwindling, S.; Conrath, U. Benzothiadiazole-induced priming for potentiated responses to pathogen infection, wounding, and infiltration of water into leaves requires the NPR1/NIM1 gene in Arabidopsis. Plant Physiol. 2002, 128, 1046–1056. [Google Scholar] [CrossRef]
- Ton, J.; Mauch-Mani, B. Beta-amino-butyric acid-induced resistance against necrotrophic pathogens is based on ABA-dependent priming for callose. Plant J. 2004, 38, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Perez-de-Luque, A.; Tille, S.; Johnson, I.; Pascual-Pardo, D.; Ton, J.; Cameron, D.D. The interactive effects of arbuscular mycorrhiza and plant growth-promoting rhizobacteria synergistically enhance host plant defences against pathogens. Sci. Rep. 2017, 7, 16409. [Google Scholar] [CrossRef]
- Sanmartin, N.; Pastor, V.; Pastor-Fernandez, J.; Flors, V.; Pozo, M.J.; Sanchez-Bel, P. Role and mechanisms of callose priming in mycorrhiza-induced resistance. J. Exp. Bot. 2020, 71, 2769–2781. [Google Scholar] [CrossRef]
- Underwood, W. The plant cell wall: A dynamic barrier against pathogen invasion. Front. Plant Sci. 2012, 3, 85. [Google Scholar] [CrossRef]
- Micali, C.O.; Neumann, U.; Grunewald, D.; Panstruga, R.; O’Connell, R. Biogenesis of a specialized plant-fungal interface during host cell internalization of Golovinomyces orontii haustoria. Cell Microbiol. 2011, 13, 210–226. [Google Scholar] [CrossRef]
- Amsbury, S.; Kirk, P.; Benitez-Alfonso, Y. Emerging models on the regulation of intercellular transport by plasmodesmata-associated callose. J. Exp. Bot. 2018, 69, 105–115. [Google Scholar] [CrossRef]
- Lee, J.Y.; Lu, H. Plasmodesmata: The battleground against intruders. Trends Plant Sci. 2011, 16, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Sager, R.E.; Lee, J.Y. Plasmodesmata at a glance. J. Cell Sci. 2018, 131. [Google Scholar] [CrossRef]
- Iglesias, V.A.; Meins, F. Movement of plant viruses is delayed in a beta-1,3-glucanase-deficient mutant showing a reduced plasmodesmatal size exclusion limit and enhanced callose deposition. Plant J. 2000, 21, 157–166. [Google Scholar] [CrossRef]
- Galletti, R.; Denoux, C.; Gambetta, S.; Dewdney, J.; Ausubel, F.M.; De Lorenzo, G.; Ferrari, S. The AtrbohD-Mediated Oxidative Burst Elicited by Oligogalacturonides in Arabidopsis Is Dispensable for the Activation of Defense Responses Effective against Botrytis cinerea. Plant Physiol. 2008, 148, 1695–1706. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Shao, F.; Cui, H.; Chen, L.J.; Li, H.T.; Zou, Y.; Long, C.Z.; Lan, L.F.; Chai, J.J.; Chen, S.; et al. A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-Induced immunity in plants. Cell Host Microbe 2007, 1, 175–185. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Q.Q.; Zhang, J.G.; Wu, L.; Qi, Y.J.; Zhou, J.M. Identification of MicroRNAs Involved in Pathogen-Associated Molecular Pattern-Triggered Plant Innate Immunity. Plant Physiol. 2010, 152, 2222–2231. [Google Scholar] [CrossRef]
- Clay, N.K.; Adio, A.M.; Denoux, C.; Jander, G.; Ausubel, F.M. Glucosinolate Metabolites Required for an Arabidopsis Innate Immune Response. Science 2009, 323, 95–101. [Google Scholar] [CrossRef]
- Luna, E.; Pastor, V.; Robert, J.; Flors, V.; Mauch-Mani, B.; Ton, J. Callose Deposition: A Multifaceted Plant Defense Response. Mol. Plant Microbe Interact. 2011, 24, 183–193. [Google Scholar] [CrossRef]
- Wang, X.; Sager, R.; Cui, W.E.; Zhang, C.; Lu, H.; Lee, J.Y. Salicylic Acid Regulates Plasmodesmata Closure during Innate Immune Responses in Arabidopsis. Plant Cell 2013, 25, 2315–2329. [Google Scholar] [CrossRef]
- Gamir, J.; Pastor, V.; Sanchez-Bel, P.; Agut, B.; Mateu, D.; Garcia-Andrade, J.; Flors, V. Starch degradation, abscisic acid and vesicular trafficking are important elements in callose priming by indole-3-carboxylic acid in response to Plectosphaerella cucumerina infection. Plant J. 2018, 96, 518–531. [Google Scholar] [CrossRef]
- Bashline, L.; Li, S.; Gu, Y. The trafficking of the cellulose synthase complex in higher plants. Ann. Bot. 2014, 114, 1059–1067. [Google Scholar] [CrossRef]
- Wang, Z.; Li, X.; Wang, X.; Liu, N.; Xu, B.; Peng, Q.; Guo, Z.; Fan, B.; Zhu, C.; Chen, Z. Arabidopsis Endoplasmic Reticulum-Localized UBAC2 Proteins Interact with PAMP-INDUCED COILED-COIL to Regulate Pathogen-Induced Callose Deposition and Plant Immunity. Plant Cell 2019, 31, 153–171. [Google Scholar] [CrossRef]
- Ellinger, D.; Naumann, M.; Falter, C.; Zwikowics, C.; Jamrow, T.; Manisseri, C.; Somerville, S.C.; Voigt, C.A. Elevated early callose deposition results in complete penetration resistance to powdery mildew in Arabidopsis. Plant Physiol. 2013, 161, 1433–1444. [Google Scholar] [CrossRef] [PubMed]
- Calonge, T.M.; Arellano, M.; Coll, P.M.; Perez, P. Rga5p is a specific Rho1p GTPase-activating protein that regulates cell integrity in Schizosaccharomyces pombe. Mol. Microbiol. 2003, 47, 507–518. [Google Scholar] [CrossRef]
- Qadota, H.; Python, C.P.; Inoue, S.B.; Arisawa, M.; Anraku, Y.; Zheng, Y.; Watanabe, T.; Levin, D.E.; Ohya, Y. Identification of yeast Rho1p GTPase as a regulatory subunit of 1,3-beta-glucan synthase. Science 1996, 272, 279–281. [Google Scholar] [CrossRef]
- Ellinger, D.; Glockner, A.; Koch, J.; Naumann, M.; Sturtz, V.; Schutt, K.; Manisseri, C.; Somerville, S.C.; Voigt, C.A. Interaction of the Arabidopsis GTPase RabA4c with its effector PMR4 results in complete penetration resistance to powdery mildew. Plant Cell 2014, 26, 3185–3200. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.L.; Guo, W. The exocyst complex in exocytosis and cell migration. Protoplasma 2012, 249, 587–597. [Google Scholar] [CrossRef]
- Kulich, I.; Vojtikova, Z.; Glanc, M.; Ortmannova, J.; Rasmann, S.; Zarsky, V. Cell wall maturation of Arabidopsis trichomes is dependent on exocyst subunit EXO70H4 and involves callose deposition. Plant Physiol. 2015, 168, 120–131. [Google Scholar] [CrossRef]
- Kulich, I.; Vojtikova, Z.; Sabol, P.; Ortmannova, J.; Nedela, V.; Tihlarikova, E.; Zarsky, V. Exocyst Subunit EXO70H4 Has a Specific Role in Callose Synthase Secretion and Silica Accumulation. Plant Physiol. 2018, 176, 2040–2051. [Google Scholar] [CrossRef]
- An, Q.; Ehlers, K.; Kogel, K.H.; van Bel, A.J.; Huckelhoven, R. Multivesicular compartments proliferate in susceptible and resistant MLA12-barley leaves in response to infection by the biotrophic powdery mildew fungus. New Phytol. 2006, 172, 563–576. [Google Scholar] [CrossRef] [PubMed]
- An, Q.; Huckelhoven, R.; Kogel, K.H.; van Bel, A.J. Multivesicular bodies participate in a cell wall-associated defence response in barley leaves attacked by the pathogenic powdery mildew fungus. Cell. Microbiol. 2006, 8, 1009–1019. [Google Scholar] [CrossRef] [PubMed]
- An, Q.; van Bel, A.J.; Huckelhoven, R. Do plant cells secrete exosomes derived from multivesicular bodies? Plant Signal. Behav. 2007, 2, 4–7. [Google Scholar] [CrossRef]
- Marchant, R.; Robards, A.W. Membrane systems associated with the plasmalemma of plant cells. Ann. Bot. 1968, 32, 457–471. [Google Scholar] [CrossRef]
- Bohlenius, H.; Morch, S.M.; Godfrey, D.; Nielsen, M.E.; Thordal-Christensen, H. The multivesicular body-localized GTPase ARFA1b/1c is important for callose deposition and ROR2 syntaxin-dependent preinvasive basal defense in barley. Plant Cell 2010, 22, 3831–3844. [Google Scholar] [CrossRef]
- Meyer, D.; Pajonk, S.; Micali, C.; O’Connell, R.; Schulze-Lefert, P. Extracellular transport and integration of plant secretory proteins into pathogen-induced cell wall compartments. Plant J. Cell Mol. Biol. 2009, 57, 986–999. [Google Scholar] [CrossRef]
- Nielsen, M.E.; Feechan, A.; Bohlenius, H.; Ueda, T.; Thordal-Christensen, H. Arabidopsis ARF-GTP exchange factor, GNOM, mediates transport required for innate immunity and focal accumulation of syntaxin PEN1. Proc. Natl. Acad. Sci. USA 2012, 109, 11443–11448. [Google Scholar] [CrossRef]
- Berkey, R.; Zhang, Y.; Ma, X.; King, H.; Zhang, Q.; Wang, W.; Xiao, S. Homologues of the RPW8 Resistance Protein Are Localized to the Extrahaustorial Membrane that Is Likely Synthesized De Novo. Plant Physiol. 2017, 173, 600–613. [Google Scholar] [CrossRef] [PubMed]
- Bozkurt, T.O.; Belhaj, K.; Dagdas, Y.F.; Chaparro-Garcia, A.; Wu, C.H.; Cano, L.M.; Kamoun, S. Rerouting of plant late endocytic trafficking toward a pathogen interface. Traffic 2015, 16, 204–226. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, M.E.; Jurgens, G.; Thordal-Christensen, H. VPS9a Activates the Rab5 GTPase ARA7 to Confer Distinct Pre- and Postinvasive Plant Innate Immunity. Plant Cell 2017, 29, 1927–1937. [Google Scholar] [CrossRef]
- Nomura, K.; DebRoy, S.; Lee, Y.H.; Pumplin, N.; Jones, J.; He, S.Y. A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science 2006, 313, 220–223. [Google Scholar] [CrossRef] [PubMed]
- Abou-Saleh, R.H.; Hernandez-Gomez, M.C.; Amsbury, S.; Paniagua, C.; Bourdon, M.; Miyashima, S.; Helariutta, Y.; Fuller, M.; Budtova, T.; Connell, S.D.; et al. Interactions between callose and cellulose revealed through the analysis of biopolymer mixtures. Nat. Commun. 2018, 9. [Google Scholar] [CrossRef]
- Eggert, D.; Naumann, M.; Reimer, R.; Voigt, C.A. Nanoscale glucan polymer network causes pathogen resistance. Sci. Rep. 2014, 4. [Google Scholar] [CrossRef]
- Chowdhury, J.; Henderson, M.; Schweizer, P.; Burton, R.A.; Fincher, G.B.; Little, A. Differential accumulation of callose, arabinoxylan and cellulose in nonpenetrated versus penetrated papillae on leaves of barley infected with Blumeria graminis f. sp. hordei. New Phytol. 2014, 204, 650–660. [Google Scholar] [CrossRef]
- Li, W.L.; Zhao, Y.S.; Liu, C.J.; Yao, G.B.; Wu, S.S.; Hou, C.Y.; Zhang, M.C.; Wang, D.M. Callose deposition at plasmodesmata is a critical factor in restricting the cell-to-cell movement of Soybean mosaic virus. Plant Cell Rep. 2012, 31, 905–916. [Google Scholar] [CrossRef]
- Bayles, C.J.; Ghemawat, M.S.; Aist, J.R. Inhibition by 2-Deoxy-D-Glucose of Callose Formation, Papilla Deposition, and Resistance to Powdery Mildew in an Ml-O Barley Mutant. Physiol. Mol. Plant Pathol. 1990, 36, 63–72. [Google Scholar] [CrossRef]
- Nishimura, M.T.; Stein, M.; Hou, B.H.; Vogel, J.P.; Edwards, H.; Somerville, S.C. Loss of a callose synthase results in salicylic acid-dependent disease resistance. Science 2003, 301, 969–972. [Google Scholar] [CrossRef]
- Huibers, R.P.; Loonen, A.E.H.M.; Gao, D.L.; Van den Ackerveken, G.; Visser, R.G.F.; Bai, Y.L. Powdery Mildew Resistance in Tomato by Impairment of SlPMR4 and SlDMR1. PLoS ONE 2013, 8, e67467. [Google Scholar] [CrossRef]
- Martinez, M.S.I.; Bracuto, V.; Koseoglou, E.; Appiano, M.; Jacobsen, E.; Visser, R.G.F.; Wolters, A.M.A.; Bai, Y.L. CRISPR/Cas9-targeted mutagenesis of the tomato susceptibility gene PMR4 for resistance against powdery mildew. BMC Plant Biol. 2020, 20, 284. [Google Scholar] [CrossRef]
- Kim, M.G.; da Cunha, L.; McFall, A.J.; Belkhadir, Y.; DebRoy, S.; Dangl, J.L.; Mackey, D. Two Pseudomonas syringae type III effectors inhibit RIN4-regulated basal defense in Arabidopsis. Cell 2005, 121, 749–759. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, J.; Schober, M.S.; Shirley, N.J.; Singh, R.R.; Jacobs, A.K.; Douchkov, D.; Schweizer, P.; Fincher, G.B.; Burton, R.A.; Little, A. Down-regulation of the glucan synthase-like 6 gene (HvGsl6) in barley leads to decreased callose accumulation and increased cell wall penetration by Blumeria graminis f. sp hordei. New Phytol. 2016, 212, 434–443. [Google Scholar] [CrossRef] [PubMed]
- Zavaliev, R.; Levy, A.; Gera, A.; Epel, B.L. Subcellular Dynamics and Role of Arabidopsis beta-1,3-Glucanases in Cell-to-Cell Movement of Tobamoviruses. Mol. Plant Microbe Interact. 2013, 26, 1016–1030. [Google Scholar] [CrossRef]
- Alfano, J.R.; Collmer, A. Type III secretion system effector proteins: Double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 2004, 42, 385–414. [Google Scholar] [CrossRef]
- Bestwick, C.S.; Bennett, M.H.; Mansfield, J.W. Hrp Mutant of Pseudomonas-Syringae Pv Phaseolicola Induces Cell-Wall Alterations but Not Membrane Damage Leading to the Hypersensitive Reaction in Lettuce. Plant Physiol. 1995, 108, 503–516. [Google Scholar] [CrossRef]
- Brown, I.; Mansfield, J.; Bonas, U. Hrp genes in Xanthomonas campestris pv vesicatoria determine ability to suppress papilla deposition in pepper mesophyll cells. Mol. Plant Microbe Interact. 1995, 8, 825–836. [Google Scholar] [CrossRef]
- Brown, I.; Trethowan, J.; Kerry, M.; Mansfield, J.; Bolwell, G.P. Localization of components of the oxidative cross-linking of glycoproteins and of callose synthesis in papillae formed during the interaction between non-pathogenic strains of Xanthomonas campestris and French bean mesophyll cells. Plant J. 1998, 15, 333–343. [Google Scholar] [CrossRef]
- Hauck, P.; Thilmony, R.; He, S.Y. A Pseudomonas syringae type III effector suppresses cell wall-based extracellular defense in susceptible Arabidopsis plants. Proc. Natl. Acad. Sci. USA 2003, 100, 8577–8582. [Google Scholar] [CrossRef] [PubMed]
- Koh, S.; Andre, A.; Edwards, H.; Ehrhardt, D.; Somerville, S. Arabidopsis thaliana subcellular responses to compatible Erysiphe cichoracearum infections. Plant J. 2005, 44, 516–529. [Google Scholar] [CrossRef]
- Chowdhury, R.N.; Lasky, D.; Karki, H.; Zhang, Z.Y.; Goyer, A.; Halterman, D.; Rakotondrafara, A.M. HCPro Suppression of Callose Deposition Contributes to Strain-Specific Resistance Against Potato Virus Y. Phytopathology 2020, 110, 164–173. [Google Scholar] [CrossRef]
- Tomczynska, I.; Stumpe, M.; Doan, T.G.; Mauch, F. A Phytophthora effector protein promotes symplastic cell-to-cell trafficking by physical interaction with plasmodesmata-localised callose synthases. New Phytol. 2020, 227, 1467–1478. [Google Scholar] [CrossRef]
- Granato, L.M.; Galdeano, D.M.; D’Alessandre, N.D.; Breton, M.C.; Machado, M.A. Callose synthase family genes plays an important role in the Citrus defense response to Candidatus Liberibacter asiaticus. Eur. J. Plant Pathol. 2019, 155, 25–38. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Li, X.; Fan, B.; Zhu, C.; Chen, Z. Regulation and Function of Defense-Related Callose Deposition in Plants. Int. J. Mol. Sci. 2021, 22, 2393. https://doi.org/10.3390/ijms22052393
Wang Y, Li X, Fan B, Zhu C, Chen Z. Regulation and Function of Defense-Related Callose Deposition in Plants. International Journal of Molecular Sciences. 2021; 22(5):2393. https://doi.org/10.3390/ijms22052393
Chicago/Turabian StyleWang, Ying, Xifeng Li, Baofang Fan, Cheng Zhu, and Zhixiang Chen. 2021. "Regulation and Function of Defense-Related Callose Deposition in Plants" International Journal of Molecular Sciences 22, no. 5: 2393. https://doi.org/10.3390/ijms22052393
APA StyleWang, Y., Li, X., Fan, B., Zhu, C., & Chen, Z. (2021). Regulation and Function of Defense-Related Callose Deposition in Plants. International Journal of Molecular Sciences, 22(5), 2393. https://doi.org/10.3390/ijms22052393





