Abstract
Saline-alkali soil has become an important environmental problem for crop productivity. One of the most effective approaches is to cultivate new stress-tolerant plants through genetic engineering. Through RNA-seq analysis and RT-PCR validation, a novel bZIP transcription factor ChbZIP1, which is significantly upregulated at alkali conditions, was obtained from alkaliphilic microalgae Chlorella sp. BLD. Overexpression of ChbZIP1 in Saccharomyces cerevisiae and Arabidopsis increased their alkali resistance, indicating ChbZIP1 may play important roles in alkali stress response. Through subcellular localization and transcriptional activation activity analyses, we found that ChbZIP1 is a nuclear-localized bZIP TF with transactivation activity to bind with the motif of G-box 2 (TGACGT). Functional analysis found that genes such as GPX1, DOX1, CAT2, and EMB, which contained G-box 2 and were associated with oxidative stress, were significantly upregulated in Arabidopsis with ChbZIP1 overexpression. The antioxidant ability was also enhanced in transgenic Arabidopsis. These results indicate that ChbZIP1 might mediate plant adaptation to alkali stress through the active oxygen detoxification pathway. Thus, ChbZIP1 may contribute to genetically improving plants’ tolerance to alkali stress.
1. Introduction
Microalgae have been considered as a promising feedstock for green renewable energy. Moreover, some microalgae also possess the ability to survive in extreme environments, such as alkaline conditions []. Recently, studies have reported that adversity tolerance of plants can be improved by genetic modification of heterologous genes [,]. Alkaliphilic microalgae is an ideal candidate resource for improving plant–alkaline tolerance []. Thus, understanding the microalgae alkali tolerance mechanisms and genetic engineering performance are the prerequisites for utilizing the alkaline environments for crop growth.
Transcription factors (TFs) play significant roles in response to plant defense and stress by activating or repressing the downstream target genes through specific binding with upstream cis-elements [,]. There are up to 320,370 TFs that have been identified in 165 plant species []. Based on the DNA-binding domains of TFs, they have been classified into 64 TF families in plant kingdoms []. The basic region-leucine zipper (bZIP) family is one of the most conserved and largest groups in the TF families. In plants, bZIPs play significant roles in metabolic processes, such as growth and development [], signaling transduction [,], and various biotic/abiotic stresses [,].
In microalgae research, there has been increased attention on TF function studies. To date, a total of 147 TFs have been found in Chlamydomonas reinhardtii, involving eleven MYBs, seven bZIPs, five C2H2s, four bHLHs, two MADSs, and one WRKY []. One study found that bZIP TFs in C. reinhardtii play an important role in oil accumulation under salt stress []. In the diatom Phaeodactylum tricornutum, some bZIP TFs, like bZIP14 participate in the regulation of TCA biosynthesis pathways []. However, functions remain unclear for the majority of bZIP TFs in other microalgae, especially the extreme algae.
In this study, a novel bZIP TF ChbZIP1 of alkaliphilic microalgae Chlorella sp. BLD has been identified with transcriptome analysis, which was upregulated under alkali conditions. A functional study showed that ChbZIP1 could improve the alkali-resistant ability of yeast and Arabidopsis, and the antioxidant ability of Arabidopsis was enhanced with ChbZIP1 overexpression. This study facilitates a better understanding of the molecular mechanism of bZIP TF in responding alkali conditions, which provides an important foundation to further guide genetic engineering with microalgae in improving crop tolerance with extreme environments.
2. Results
2.1. Changes of Transcription Factors in the Alkali Stress
In our previous study, a new alkaliphilic microalgae Chlorella sp. BLD, was isolated and transcriptome analysis was performed to explore the mechanism of alkali resistance under pH 10. Through transcriptome analysis, a total of 338 TFs have been identified in Chlorella sp. BLD and can be classified into 20 families, which contain 48 MYBs, 42 C2H2, 35 C3H, 34 AP2, 30 C2C2, 22 bHLH, 21 bZIP, 16 WRKY and other TFs (Figure S1). Under alkaline conditions, there were 35 TFs including 12 upregulated and 23 downregulated TFs differentially expressed (Figure 1A). These TFs covered 4 AP2s, 3 bZIPs, 4 GATAs, 6 MYBs, 3 SBPs, 7 ZFs and 8 other TFs. To validate the expression level, 12 upregulated TFs were selected with q-PCR analysis. The results showed that the variation tendency of the TFs validated by q-PCR was similar to the data from RNA-seq (Figure 1B). Among these, four TFs (AP2/3, bZIP1, SBP1, ZF7) were significantly upregulated at alkali condition, which means these TFs may participate in the regulation of alkali stress.
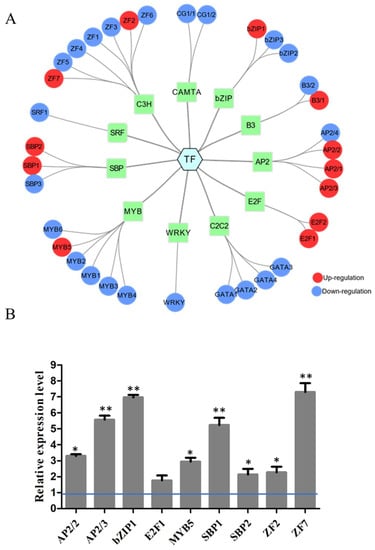
Figure 1.
Analysis the expression of transcription factors of Chlorella sp. BLD under alkali conditions. (A) RNA-seq analysis the regulation of transcription factors (TFs) expression of Chlorella sp. BLD at pH 10 compared to pH 7.5. The results of the network analysis including TFs were analyzed and visualized using CYTOSCAPE. The red nodes indicate upregulation, and the blue nodes indicate downregulation. (B) Validation of the RNA-seq results of transcription factors (TFs) in upregulated by q-PCR. * indicates p-value < 0.05, ** indicates p-value < 0.01.
2.2. Validation of the Alkali Resistance of TFs in S. cerevisiae
To understand the TFs’ function in alkali stress tolerance, the growth characteristics of wild type (WT) S. cerevisiae Y258 and strains overexpressing TFs (AP2/3, bZIP1, SBP1, and ZF7, respectively) at pH 8 YPD medium were surveyed after four days. As shown in Figure 2A, yeast with bZIP1 exhibited the best growth under alkaline conditions. More clones clearly appeared in bZIP1 transformed yeast than that of WT at dilution of 10−2 and 10−3 (Figure 2A). ZF7 and AP2/3 showed more colonies than WT at the dilution of 10−2 and 10−1, respectively. However, the transformants of SBP1 could not grow at pH 8 YPD medium with a dilution of 10−1, which similar to the growth tendency of WT. To further extensively explore the function of bZIP1 in alkaline condition, the growth of WT and bZIP1-overexpressed yeast cells were assayed under different concentrations of NaHCO3. The results showed that more colonies were found with bZIP1-overexpressed yeast than WT at 15 mM, 30 mM and 45 mM NaHCO3 with different dilutions (Figure. 2B). These results imply that the bZIP1, which is referred to as ChbZIP1 in the following, may play an important role in resisting alkali stress.
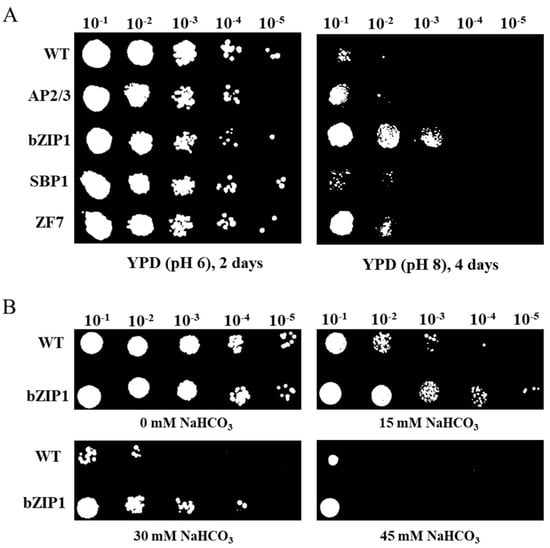
Figure 2.
Transcription factor screening from Chlorella sp. BLD with alkali tolerance analysis in S. cerevisiae. (A) TFs (AP2/3, bZIP1, SBP1 and ZF7) were constructed to the pEGH-A vector and transformed into Y258 yeast. Serial dilutions were spotted onto pH 8 yeast YPD solid plates as alkali treatment, and on normal YPD (pH 6) solid plates as control. (B) Yeast cells with bZIP1 overexpression and WT were diluted and spotted onto the YPD solid medium supplemented with 0 mM, 15 mM, 30 mM, and 45 mM NaHCO3.
2.3. Characterization of the ChbZIP1 Sequence
The ChbZIP1 TF belongs to the basic leucine zipper family, which is the third-largest TF family in Chlorella sp. []. Analysis of the ChbZIP1 sequence indicated that the ORF was 1188 nucleotides long and encoded a predicted protein of 396 amino acids. ChbZIP1 has a typical bZIP domain in the sequence (Figure 3A). Through neighbor-joining (NJ) tree analysis, ChbZIP1 showed a closer relationship with Chlorella variabilis (XP 005847) than other species. Meanwhile, the Chlorella sp. BLD first clustered with microalgae in the tree, then with plants (Figure 3B), which was in accordance with the expected taxonomy. To further explore the phylogenetic relationship of the ChbZIP1 in Chlorella sp. BLD, ten bZIP protein sequences from the Arabidopsis AtbZIP family were added into the tree and used as subgroup markers to classify the ChbZIP1. According to the circular NJ tree, ChbZIP1 was closer to AtbZIP41 of Arabidopsis (Figure 3C), which is classified into the G group.
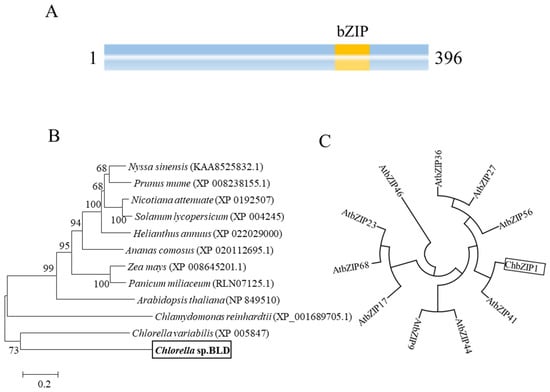
Figure 3.
Phylogenetic and sequence analyses of ChbZIP1 from Chlorella sp. BLD. (A) Schematic representation of ChbZIP1 with basic leucine zipper (bZIP) domain. (B) Neighbor-joining phylogenetic relationships among ChbZIP1 from Chlorella sp. BLD and various bZIPs from other species. Bootstrap values were calculated at 1000 ties. Chlorella sp. BLD is indicated in boldface in box. (C) Neighbor-joining phylogenetic relationships among ChbZIP1 and ten groups of bZIP families in Arabidopsis. Box indicates ChbZIP1.
2.4. Subcellular Localization and Transactivation Activity Analysis of ChbZIP1
Previous studies have reported that bZIP TFs were localized in nuclear [], cytoplasmic [], and endoplasmic reticulum []. Hence, to explore the subcellular localization of ChbZIP1 in plant cells, a ChbZIP1 sequence without the stop codon was constructed with the vector of pPZP211-GFP and transformed into Arabidopsis protoplasts. The result showed that the transformed protoplasts of ChbZIP1 exhibited GFP fluorescence in the nucleus (Figure 4A), suggesting that ChbZIP1 localized in the nucleus of Arabidopsis protoplasts.
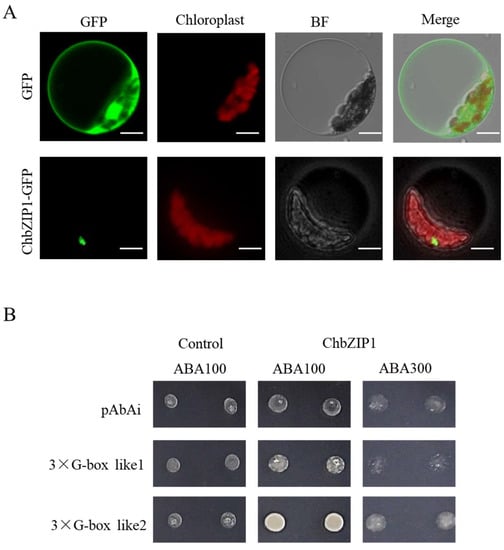
Figure 4.
Subcellular localization and transactivation activity analyses of ChbZIP1. (A) Subcellular localization of ChbZIP1:GFP in Arabidopsis protoplasts. GFP empty vector is shown at the top. Bars = 10 µm. (B) ChbZIP1 binds the artificial, triple-repeated G-box 2 motifs in yeast cells at 100 and 300 ng/L concentration of aureobasidin A (AbA), whereas it fails to bind the G-box 1 and AbAi control vector.
In order to further explore the transcriptional activation activity of ChbZIP1, the motif of G-box 1 (CCACGT) and G-box 2 (TGACGT), which was plant bZIPs’ protein-binding motif [], have been designed and verified by yeast one-hybrid (Y1H) experiments. To test the ChbZIP1 protein’s binding ability, triple repeated G-box 1 and G-box 2 motifs were designed and integrated into the genome of yeast cells. After introducing ChbZIP1 into the yeast strains, it was found that ChbZIP1 was preferred to bind the motif of G-box 2 (Figure 4B). Certainly, from the selection media SD/-L added with 100 ng/L and 300 ng/L Aureobasidin A (AbA), ChbZIP1 appears to have a higher affinity binding to the G-box 2 than to the G-box 1 motifs. The results of the yeast one-hybrid assay suggested that ChbZIP1 may participate in alkali responses by recognizing and interacting with the G-box2 motif.
2.5. Growth Analysis of ChbZIP1 Overexpression in Arabidopsis
To test the function of ChbZIP1 with alkali tolerance in plants, the cDNA of ChbZIP1 was overexpressed in Arabidopsis under the control of promoter CaMV 35S. The band of ChbZIP1 was detected with PCR screening in the six T1 (OX1-6) lines (Figure S2A). The above results confirmed that the ChbZIP1 gene was successfully integrated into the genome of Arabidopsis. Furthermore, the mRNA expression level of ChbZIP1 in T3 transgenic lines originated from the six identified T1 lines, indicating that ChbZIP1 was successfully overexpressed in the Arabidopsis (Figure S2B). Three T3 lines, which highly expressed in Arabidopsis, named OX1, OX2 and OX4 were used for further functional validation.
The NaHCO3 tolerance assay was used to detect the function of T3 lines (OX1, OX2 and OX4) of overexpressing ChbZIP1 in the Arabidopsis. The results showed that there were no significant changes between WT and transgenic lines (OX1, OX2 and OX4) in the growth on the 1/2 MS medium without NaHCO3 (Figure 5A). While under the treatment of NaHCO3, transgenic lines demonstrated the excellent alkali resistance ability compared with WT. Both the root growth (Figure 5A,B) and the entire seedlings fresh weight (Figure 5C) were significantly enhanced in transgenic lines compared with that in WT under 2 and 4 mM NaHCO3 treatment. Although the transgenic plants and WT all exhibited chlorosis at 6 mM NaHCO3 concentration, transgenic lines showed better adaptability with longer root length and higher fresh weight than WT.
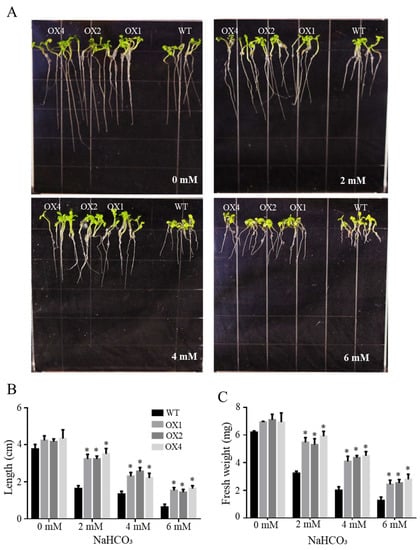
Figure 5.
Effects of NaHCO3 stress on the growth of wild-type (WT) and ChbZIP1-overexpressed transgenic Arabidopsis plants. (A) Phenotypes of ChbZIP1 transgenic and WT lines subjected to 0, 2, 4, and 6 mM NaHCO3 treatment, respectively. (B) Root length changes in ChbZIP1 transgenic and WT lines. (C) Fresh weight changes in ChbZIP1 transgenic and WT lines. * indicates p-value < 0.05.
2.6. The Antioxidant Ability in ChbZIP1-Overexpression Arabidopsis
Through yeast one-hybrid (Y1H) experiments, we found that ChbZIP1 showed high affinity binding to the G-box 2 (TGACGT). To further identify the function of ChbZIP1 in Arabidopsis, 1034 genes that contained G-box 2 motifs were obtained through Stress-Responsive Transcription Factor DataBase [] analysis in Arabidopsis. After GO analysis, 25 oxidant detoxification related genes, which contained G-box 2 and may interact with bZIP TF, were isolated (Figure 6A). The expression level of these oxidant detoxification-related genes in ChbZIP1-overexpression and WT Arabidopsis were validated by qPCR (Figure 6B). The results showed that 84% (21/25) genes were highly upregulated (Figure 6B). Among the upregulated genes, GPX1, DOX1, CAT2, EMB and APXS have the highest expression level, followed by FSD, PAP26, 24170 (AT3G24170.1), APX1, GPX2, and CAT1. These results indicated that ChbZIP1 may enhance the antioxidant ability of Arabidopsis by regulating the redox-related genes, thus improving the alkali tolerance in plants.
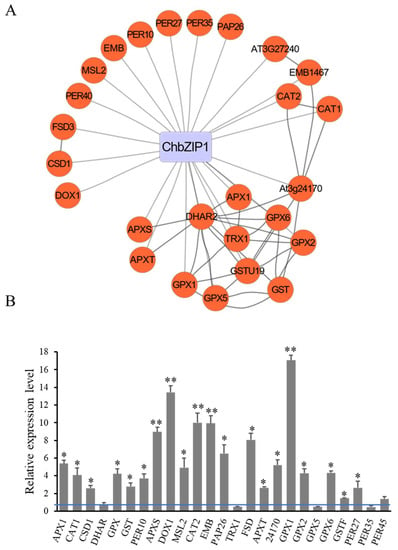
Figure 6.
Screening and validation of antioxidant related genes which contained G-box 2 and may be regulated by ChbZIP1. (A) Antioxidant-related genes were extracted from the G-box 2 motifs’ binding database with STRING analysis and drawn by CYTOSCAPE. (B) Antioxidant related genes were detected compared to ChbZIP1 transgenic lines with WT lines seedlings exposed to 200 mM NaHCO3 treatment by q-PCR. * indicates p-value < 0.05, ** indicates p-value < 0.01.
To verify the above inference, the activities of ROS scavenging enzymes like ascorbate peroxidase (APX), catalase (CAT), and peroxidase (POD) and ROS accumulation were measured. In this study, APX, CAT, and POD all exhibited higher enzyme activities in the ChbZIP1 transgenic line compared to WT (Figure 7A). The increase of antioxidant enzyme activity reduced the ROS content and thus improved the tolerance of plants to stressed environments []. The results of ROS accumulation are consistent with the activities of ROS scavenging enzymes. The content of malondialdehyde (MDA) and hydrogen peroxide (H2O2) showed no significant differences in ChbZIP1 transgenic plants compared with those in an untreated state in WT (Figure 7B). While under the NaHCO3 stress exposure, the content of MDA and H2O2 in the transgenic strains was dramatically decreased compared with that in WT (Figure 7B). However, there was no significant change in the Vitamin C (VC) content between WT and transgenic lines.
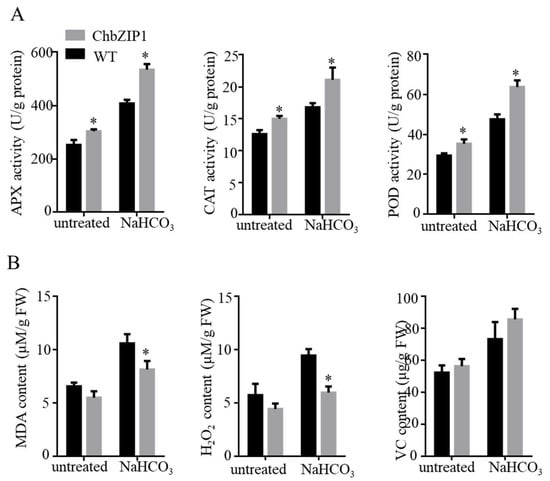
Figure 7.
Effects of NaHCO3 stress on the antioxidant ability in wild-type (WT) and ChbZIP1 overexpressed transgenic Arabidopsis plants. (A) Measure of the enzymatic activities of APX, CAT, and POD in ChbZIP1 transgenic and WT seedlings compared to NaHCO3 stress and control. (B) Measure of the content of MDA, H2O2, and Vc in ChbZIP1 transgenic and WT seedlings compared to NaHCO3 stress and control. * indicates p-value < 0.05.
3. Discussion
In recent years, the area of saline-alkali soil has gradually increased, which resulted in severe losses in agricultural production. Previously, we had isolated an alkaliphilic microalgae Chlorella sp. BLD, which can grow well in extreme alkali conditions at pH > 10. In transcriptome analysis, bZIP transcription factor ChbZIP1 was highly up regulated at pH 10 compared to normal conditions (Figure 1). The functional study also found that overexpression of ChbZIP1 in S. cerevisiae and Arabidopsis could improve their alkali resistance. Meanwhile, ChbZIP1 could also enhance the antioxidant ability of Arabidopsis with related genes and ROS-related enzymes. Together, these findings not only elucidate the unprecedented role of ChbZIP1 in regulating alkali resistance but also provide a new direction for plant genetic engineering.
3.1. ChbZIP1 May Play Important Roles in Resisting Alkali Stress
The ChbZIP1 gene was isolated from the cDNA of Chlorella sp. BLD, and structure analysis showed a typical bZIP domain in its sequence (Figure 3A). Evolutionary relationship analysis with Arabidopsis found that ChbZIP1 was classified into group G (Figure 3C). The homologous proteins of ChbZIP1 in Arabidopsis were AtbZIP16, AtbZIP41 (GBF1), AtbZIP54 (GBF2), and AtbZIP55 (GBF3) []. It was reported that the group G AtbZIP genes from Arabidopsis were mainly linked to light signal transduction, regulation of gibberellic acid (GA), and abscisic acid (ABA) response [,]. In our study, ChbZIP1 overexpressing in S. cerevisiae and Arabidopsis significantly improved their tolerance to alkali stresses (Figure 2 and Figure 4). Studies reported that bZIP TF could improve the survival of crop plants in alkaline conditions. A bZIP TF GsbZIP67, which was isolated from Glycine soja, overexpressed in alfalfa promoted plant growth under bicarbonate alkaline stress, as evidenced by longer roots and shoots []. DuanMu et al. also found that the bZIP TFs family was significantly induced in the alkaline stress responses of wild soybean roots through transcriptome analysis []. All these results indicated that bZIP TFs play an important role in response to the alkaline condition.
3.2. ChbZIP1 Is A Nuclear-Localized bZIP TF with Transactivation Activity
Members of the bZIP TF family participate in the regulation of various biological processes such as environmental responses, plant growth, and development. One study reported that bZIP TF can bind to elements that contains a functional ACGT core, like G-box, C-box, and A-box []. Our results showed that ChbZIP1 is a nuclear-localized bZIP TF with transactivation activity through subcellular localization and transcriptional activation activity analyses (Figure 4). ChbZIP1 preferentially binds with the G-box2 motif (TGACGT). Proteins (AtGBF1, AtGBF2, and AtGBF3) belonging to the same G-group as ChbZIP1 have been shown to bind with the G-box element, which mediates expression in response to different stimuli []. Thus, there is an extremely high probability that ChbZIP1 plays an important role in response to alkaline stress by binding with the G-box2 motif.
3.3. ChbZIP1 May Enhance the Antioxidant Ability through Regulating Oxidant Detoxification-Related Genes with G-Box 2 Motif
Genes related to oxidation reduction play significant roles in stress response, because they can scavenge the ROS and maintain the redox balance under stress []. In this study, we have found 25 oxidant detoxification related genes that contain G-box 2 and may interact with bZIP TF (Figure 6A). Through qPCR analysis, it is found that 84% (21/25) of genes related to oxidant detoxification are highly upregulated in ChbZIP1-overexpression Arabidopsis (Figure 6B). These results indicate that ChbZIP1 might enhance the antioxidant ability of Arabidopsis by regulating the redox-related genes, thus improving the alkali tolerance in plants. The activities of ROS scavenging enzymes (APX, CAT, and POD) and ROS accumulation were measured and verified this inference (Figure 7).
Superoxide dismutase (SOD), ascorbate peroxidase (APX), glutathione peroxidase (GPX), and catalase (CAT) are key antioxidative enzymes, which could counteract the damage of ROS. For example, the plant GPX family, which is upregulated by ChbZIP1 in our study, could detoxify H2O2 and organic hydroperoxides []. Besides that, the present study also finds that GPX is involved in regulating the cellular redox homeostasis by maintaining the thiol/disulfide or NADPH/NADP+ balance []. The APX as a component of the ascorbate-glutathione cycle could maintain in the appropriate level of H2O2 in organelles and cytosol []. Overexpression APX in potato and Arabidopsis exhibits high tolerance to abiotic stress, such as oxidative stress, chilling, salt and methyl viologen (MV) stress [,]. The CAT family, which contains three genes (CAT1-3) in most plant species, scavenges H2O2 in millimolar concentrations, especially in peroxisomes []. Some reports have implied that CAT1 plays an important role in signaling related to irradiance and abscisic acid (ABA) [,].
These results indicate that ChbZIP1 could regulate the oxidant detoxification-related genes with the G-box 2 motif, further enhance the activities of the enzymatic antioxidants, and reduce oxidative stress damage in the transgenic Arabidopsis lines (Figure 8). Thus, ChbZIP1 could be a potential gene for improving plant tolerance in alkali stress.
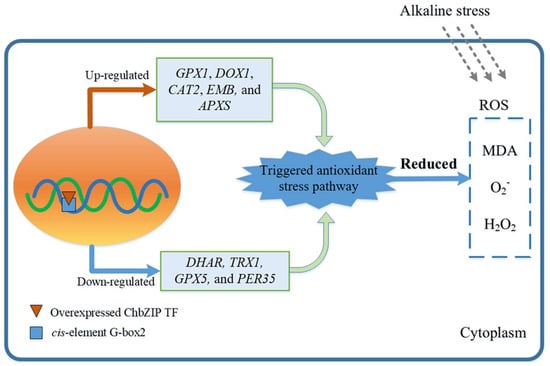
Figure 8.
The proposed alkaline tolerance mechanism in Arabidopsis with overexpressed ChbZIP1 TF.
4. Methods and Materials
4.1. Microalgae Cultivation
The algae Chlorella sp. BLD used in this study was presented by Ghopur Mijit (Xinjiang University) and cultivated with the BG-11 medium. The pH 7.5 (maintained with 25 mM Hepes) and pH 10 (maintained with 25 mM Caps) of the BG-11 medium was adjusted by NaOH. The temperature of cultivation was 25 ± 1 °C and light intensity was 160 μmoL photons m −2 s −1 in 1 L Erlenmeyer flask.
4.2. Validating the Expression of Genes with qPCR
Quantitative real-time PCR (qPCR) was used to verify the genes expression. RT-qPCR was measured by SYBR SuperReal PreMix Plus (Tiangen, Beijing, China) in Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). The housekeeping gene β-tubulin and UBQ10 (AT4g05320) were used as an internal standard for Chlorella sp. BLD and Arabidopsis, respectively. Primers used in this study for qPCR analysis are shown in Table S1. The relative expression levels were calculated using the 2−∆∆Ct method [].
4.3. Analysis of Resistance to Alkali Stress in S. cerevisiae with TFs Overexpression
To detect the related transcription genes’ response to alkali conditions, four highly-expressed genes were amplified and cloned into vector pEGH-A (yeast expression vector) with the Gateway cloning system. Using the LiAc method, the success clones were transformed into the S. cerevisiae strain Y258. To select the successful expressed plasmids, the medium of SD-Ura was used for yeast strains cultivation. The TFs’ overexpressed yeast cells and WT were adjusted from OD600 to 0.1 for alkali tolerance analysis. Then preparation for a serial of dilutions with ten-fold and was spotted on a solid YPD medium (pH 6 and pH 8) with 5 μL aliquots of each dilution for incubation at 30 °C for 2 days and 4 days, respectively.
4.4. Construction of Phylogenetic Trees
The sequence of ChbZIP1 was screened from the RNA-seq results. The homologous sequences of other species were searched using the program NCBI-pBLAST. For tree construction, the sequences that most closely matched the sequence of ChbZIP1 were obtained by BLAST from the GenBank database. Clustal X was used to align the similar sequences. The tree was constructed using the method of neighbor-joining with the MEGA5.0 software [].
4.5. Subcellular Localization Analysis of ChbZIP1 in Arabidopsis Protoplasts
The open reading frame (ORF) of the ChbZIP1 sequence without the stop codon was amplified and constructed into the pPZP211-GFP vector with the Kpn I and Xbal I sites. Then, the recombinant plasmid was transiently expressed in Arabidopsis protoplasts by PEG-mediated transformation according to the method of Yoo et al. []. The Arabidopsis protoplasts were then incubated at 23 °C for 16 h with protein expression and the fluorescence of GFP and autofluorescence of chlorophyll were observed with a confocal laser-scanning microscope at 488 nm and 561 nm, respectively.
4.6. Yeast One-Hybrid Assays
The TF–DNA interaction experiments were performed using the Gold Yeast One-Hybrid system []. G-box motif 1 and G-box motif 2 with triple tandem copies were inserted into the reporter plasmid pAbAi. The bait constructs were linearized and then integrated into the genome of the Y1H Gold yeast stain. Transformants were selected on a synthetic defined SD/-Ura agar medium plate and cultured at 30 °C for 2 days. The ChbZIP1 gene was constructed to the yeast activation vector of pGADT7 as prey. The prey construct was then transformed to the yeast cells, which contained the bait constructs. SD/-Leu medium with different concentrations of aureobasidin A (AbA) were used to culture the positively transformed clones.
4.7. Functional Analysis of ChbZIP1 Overexpression in Arabidopsis
The PCR product of ChbZIP1 was cloned into the expression vector pBI121. For Arabidopsis transformation, the pBI121::ChbZIP1 was first transformed into Agrobacterium tumefaciens GV3101 by chemical method. Then, the floral dip method was used to transform the Arabidopsis plants []. Half-strength Murashigeand Skoog (1/2 MS) medium was used for selecting the Arabidopsis transgenic seeds with 50 mg L−1 kanamycin. To further explore the function of ChbZIP1 with the resistance of alkali stress, the transgenic T3 generation and WT seeds were plated on the 1/2 MS medium containing 0, 2, 4, and 6 mmol L−1 NaHCO3 (concentration gradient was set up according to the preliminary experiment and previous studies []) for germination. After 10 days, the length of roots was measured. In addition, when the seedlings grew to the four-leaf stage, they were transplanted to the soil in the greenhouse at 23 °C under 12 h light/ 12 h dark cycles. After 30 days, the transgenic and wild type plants were treated with 200 mM NaHCO3 for one week. The content of MDA, H2O2 [] and Vc [], were measured. APX, CAT, and POD activities were measured as described in a previous study [].
4.8. Statistical Analysis
All data were presented as the means ± standard errors of at least three biological replicate samples (GraphPad Prism 7.03). One-way ANOVA followed by a T test were done for statistically significant results with p-value < 0.05 (IBM SPSS 24).
Supplementary Materials
The following are available online at https://www.mdpi.com/1422-0067/22/5/2387/s1, Figure S1: Transcription factors were founded in Chlorella sp. BLD through transcriptome analysis, Figure S2: Overexpressed ChbZIP1 in Arabidopsis. A. The band of ChbZIP1 was detected with PCR screening in the six T1 (OX1-6) lines. B. The mRNA expression level of ChbZIP1 in T3 transgenic lines, Table S1: Primers used in this study.
Author Contributions
D.Q. and X.M. designed the study. D.Q. executed the experiments, analyzed the data, and wrote the manuscript. P.-L.S. and X.M. made a critical revision of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was sponsored by National Key R&D Program of China (2020YFA0907700). It was also supported by Natural Science Foundation of Shanghai (No. 20ZR1426600). We are grateful to Ghopur Mijit (Xinjiang University) for providing microalgae strains.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Guadie, A.; Tizazu, S.; Melese, M.; Guo, W.; Ngo, H.H.; Xia, S. Biodecolorization of textile azo dye using Bacillus sp. strain CH12 isolated from alkaline lake. Biotechnol. Rep. 2017, 15, 92–100. [Google Scholar] [CrossRef]
- Guo, L.; Li, C.; Jiang, Y.; Luo, K.; Xu, C. Heterologous expression of poplar WRKY18/35 paralogs in Arabidopsis reveals their antagonistic regulation on pathogen resistance and abiotic stress tolerance via variable hormonal pathways. Int. J. Mol. Sci. 2020, 21, 5440. [Google Scholar] [CrossRef]
- Rajappa, S.; Krishnamurthy, P.; Kumar, P.P. Regulation of AtKUP2 Expression by bHLH and WRKY Transcription Factors Helps to Confer Increased Salt Tolerance to Arabidopsis thaliana Plants. Front. Plant Sci. 2020, 11, 1311. [Google Scholar] [CrossRef]
- Varshney, P.; Mikulic, P.; Vonshak, A.; Beardall, J.; Wangikar, P.P. Extremophilic micro-algae and their potential contribution in biotechnology. Bioresour. Technol. 2015, 184, 363–372. [Google Scholar] [CrossRef]
- Singh, K.B.; Foley, R.C.; Oñate-Sánchez, L. Transcription factors in plant defense and stress responses. Curr. Opin. Plant Biol. 2002, 5, 430–436. [Google Scholar] [CrossRef]
- Javed, T.; Shabbir, R.; Ali, A.; Afzal, I.; Zaheer, U.; Gao, S.-J. Transcription Factors in Plant Stress Responses: Challenges and Potential for Sugarcane Improvement. Plants 2020, 9, 491. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Tian, F.; Yang, D.-C.; Meng, Y.-Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef]
- Pérez-Rodríguez, P.; Riaño-Pachón, D.M.; Corrêa, L.G.G.; Rensing, S.A.; Kersten, B.; Mueller-Roeber, B. PlnTFDB: Updated content and new features of the plant transcription factor database. Nucleic Acids Res. 2009, 38, D822–D827. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Zhu, Q.; Dai, S.; Lamb, C.; Beachy, R.N. RF2a, a bZIP transcriptional activator of the phloem-specific rice tungro bacilliform virus promoter, functions in vascular development. EMBO J. 1997, 16, 5247–5259. [Google Scholar] [CrossRef]
- Uno, Y.; Furihata, T.; Abe, H.; Yoshida, R.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. USA 2000, 97, 11632–11637. [Google Scholar] [CrossRef] [PubMed]
- Fuhrmann-Aoyagi, M.B.; Ruas, C.D.F.; Barbosa, E.G.G.; Braga, P.; Moraes, L.A.C.; de Oliveira, A.C.B.; Kanamori, N.; Yamaguchi-Shinozaki, K.; Nakashima, K.; Nepomuceno, A.L.; et al. Constitutive expression of Arabidopsis bZIP transcription factor AREB1 activates cross-signaling responses in soybean under drought and flooding stresses. J. Plant Physiol. 2021, 257, 153338. [Google Scholar] [CrossRef]
- Sornaraj, P.; Luang, S.; Lopato, S.; Hrmova, M. Basic leucine zipper (bZIP) transcription factors involved in abiotic stresses: A molecular model of a wheat bZIP factor and implications of its structure in function. Biochim. Biophys. Acta BBA Gen. Subj. 2016, 1860, 46–56. [Google Scholar] [CrossRef] [PubMed]
- Herath, V.; Verchot, J. Insight into the bZIP Gene Family in Solanum tuberosum: Genome and Transcriptome Analysis to Understand the Roles of Gene Diversification in Spatiotemporal Gene Expression and Function. Int. J. Mol. Sci. 2020, 22, 253. [Google Scholar] [CrossRef]
- Riaño-Pachón, D.M.; Corrêa, L.G.G.; Trejos-Espinosa, R.; Mueller-Roeber, B. Green Transcription Factors: A Chlamydomonas Overview. Genetics 2008, 179, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Ji, C.; Mao, X.; Hao, J.; Wang, X.; Xue, J.; Cui, H.; Li, R. Analysis of bZIP Transcription Factor Family and Their Expressions under Salt Stress in Chlamydomonas reinhardtii. Int. J. Mol. Sci. 2018, 19, 2800. [Google Scholar] [CrossRef]
- Matthijs, M.; Fabris, M.; Obata, T.; Foubert, I.; Franco-Zorrilla, J.M.; Solano, R.; Fernie, A.R.; Vyverman, W.; Goossens, A. The transcription factor bZIP14 regulates the TCA cycle in the diatom Phaeodactylum tricornutum. EMBO J. 2017, 36, 1559–1576. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Zhu, P.; Jia, B.; Yang, J.; Shen, Y.; Cai, X.; Sun, X.; Zhu, Y.; Sun, M.; Zhu, P. A Glycine soja group S2 bZIP transcription factor GsbZIP67 conferred bicarbonate alkaline tolerance in Medicago sativa. BMC Plant Biol. 2018, 18, 234. [Google Scholar] [CrossRef]
- Kaminaka, H.; Nake, C.; Epple, P.; Dittgen, J.; Schütze, K.; Chaban, C.; Holt, B.F.; Merkle, T.; Schäfer, E.; Harter, K.; et al. bZIP10-LSD1 antagonism modulates basal defense and cell death in Arabidopsis following infection. EMBO J. 2006, 25, 4400–4411. [Google Scholar] [CrossRef]
- Liu, J.-X.; Srivastava, R.; Che, P.; Howell, S.H. Salt stress signaling in Arabidopsis thaliana involves a membrane-bound transcription factor AtbZIP17 as a signal transducer. Plant Signal. Behav. 2008, 3, 56–57. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jakoby, M.; Weisshaar, B.; Dröge-Laser, W.; Vicente-Carbajosa, J.; Tiedemann, J.; Kroj, T.; Parcy, F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002, 7, 106–111. [Google Scholar] [CrossRef]
- Naika, M.; Shameer, K.; Mathew, O.K.; Gowda, R.; Sowdhamini, R. STIFDB2: An Updated Version of Plant Stress-Responsive TranscrIption Factor DataBase with Additional Stress Signals, Stress-Responsive Transcription Factor Binding Sites and Stress-Responsive Genes in Arabidopsis and Rice. Plant Cell Physiol. 2013, 54, e8. [Google Scholar] [CrossRef]
- Djanaguiraman, M.; Prasad, P.V.; Seppanen, M. Selenium protects sorghum leaves from oxidative damage under high temper-ature stress by enhancing antioxidant defense system. Plant Physiol. Biochem. 2010, 48, 999–1007. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, W.-P.; Hsieh, H.-L.; Wu, S.-H. Arabidopsis bZIP16 Transcription Factor Integrates Light and Hormone Signaling Pathways to Regulate Early Seedling Development. Plant Cell 2012, 24, 3997–4011. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kircher, S.L.S.; Ledger, S.; Hayashi, H.; Weisshaar, B.; Schäfer, E.; Frohnmeyer, H. CPRF4a, a novel plant bZIP protein of the CPRF family: Comparative analyses of light-dependent expression, post-transcriptional regulation, nuclear import and heterodimerisation. Mol. Genet. Genom. 1998, 257, 595–605. [Google Scholar] [CrossRef]
- Duanmu, H.; Wang, Y.; Bai, X.; Cheng, S.; Deyholos, M.K.; Wong, G.K.-S.; Li, D.; Zhu, D.; Li, R.; Yu, Y.; et al. Wild soybean roots depend on specific transcription factors and oxidation reduction related genesin response to alkaline stress. Funct. Integr. Genom. 2015, 15, 651–660. [Google Scholar] [CrossRef] [PubMed]
- Foster, R.; Izawa, T.; Chua, N. Plant bZIP proteins gather at ACGT elements. FASEB J. 1994, 8, 192–200. [Google Scholar] [CrossRef]
- Schindler, U.; Terzaghi, W.; Beckmann, H.; Kadesch, T.; Cashmore, A. DNA binding site preferences and transcriptional activation properties of the Arabidopsis transcription factor GBF1. EMBO J. 1992, 11, 1275–1289. [Google Scholar] [CrossRef] [PubMed]
- Paz, R.C.; Rocco, R.A.; Jiménez-Bremont, J.F.; Rodríguez-Kessler, M.; Becerra-Flora, A.; Menéndez, A.B.; Ruiz, O.A. Identification of differentially expressed genes potentially involved in the tolerance of Lotus tenuis to long-term alkaline stress. Plant Physiol. Biochem. 2014, 82, 279–288. [Google Scholar] [CrossRef]
- Basantani, M.; Srivastava, A. Plant glutathione transferases—A decade falls short. Can. J. Bot. 2007, 85, 443–456. [Google Scholar] [CrossRef]
- Bela, K.; Horvath, E.; Gallé, Á.; Szabados, L.; Tari, I.; Csiszár, J. Plant glutathione peroxidases: Emerging role of the antioxidant enzymes in plant development and stress responses. J. Plant Physiol. 2015, 176, 192–201. [Google Scholar] [CrossRef]
- Shigeoka, S.; Ishikawa, T.; Tamoi, M.; Miyagawa, Y.; Takeda, T.; Yabuta, Y.; Yoshimura, K. Regulation and function of ascorbate peroxidase isoenzymes. J. Exp. Bot. 2002, 53, 1305–1319. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-H.; Lim, S.; Han, S.-H.; Lee, J.J.; Nam, K.J.; Jeong, J.C.; Lee, H.-S.; Kwak, S.-S. Expression of both CuZnSOD and APX in chloroplasts enhances tolerance to sulfur dioxide in transgenic sweet potato plants. Comptes Rendus Biol. 2015, 338, 307–313. [Google Scholar] [CrossRef]
- Shafi, A.; Chauhan, R.; Gill, T.; Swarnkar, M.K.; Sreenivasulu, Y.; Kumar, S.; Kumar, N.; Shankar, R.; Ahuja, P.S.; Singh, A.K. Expression of SOD and APX genes positively regulates secondary cell wall biosynthesis and promotes plant growth and yield in Arabidopsis under salt stress. Plant Mol. Biol. 2015, 87, 615–631. [Google Scholar] [CrossRef]
- Mittler, R. Abiotic stress, the field environment and stress combination. Trends Plant Sci. 2006, 11, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Song, L.; Yu, W.; Hu, Y.; Liu, Y.; Wu, J.; Ying, Y. Growth, physiological, and biochemical responses of Camptotheca acuminata seedlings to different light environments. Front. Plant Sci. 2015, 6, 321. [Google Scholar] [CrossRef]
- Tan, L.; Liu, Q.; Song, Y.; Zhou, G.; Luan, L.; Weng, Q.; He, C. Differential Function of Endogenous and Exogenous Abscisic Acid during Bacterial Pattern-Induced Production of Reactive Oxygen Species in Arabidopsis. Int. J. Mol. Sci. 2019, 20, 2544. [Google Scholar] [CrossRef] [PubMed]
- You, Z.; Zhang, Q.; Peng, Z.; Miao, X. Lipid Droplets Mediate Salt Stress Tolerance in Parachlorella kessleri. Plant Physiol. 2019, 181, 510–526. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Yoo, S.-D.; Cho, Y.-H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef]
- Shen, Q.; Lu, X.; Yan, T.; Fu, X.; Lv, Z.; Zhang, F.; Pan, Q.; Wang, G.; Sun, X.; Tang, K. The jasmonate-responsive AaMYC2 transcription factor positively regulates artemisinin biosynthesis in Artemisia annua. New Phytol. 2016, 210, 1269–1281. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Qiao, K.; Wang, M.; Takano, T.; Liu, S. Overexpression of Acyl-CoA-Binding Protein 1 (ChACBP1) from Saline-Alkali-Tolerant Chlorella sp. Enhances Stress Tolerance in Arabidopsis. Front. Plant Sci. 2018, 9, 1772. [Google Scholar] [CrossRef]
- Wang, L.; Su, H.; Han, L.; Wang, C.; Sun, Y.; Liu, F. Differential expression profiles of poplar MAP kinase kinases in response to abiotic stresses and plant hormones, and overexpression of PtMKK4 improves the drought tolerance of poplar. Gene 2014, 545, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Shalata, A.; Neumann, P.M. Exogenous ascorbic acid (vitamin C) increases resistance to salt stress and reduces lipid peroxidation. J. Exp. Bot. 2001, 52, 2207–2211. [Google Scholar] [CrossRef] [PubMed]
- Nakano, Y.; Asada, K. Hydrogen Peroxide is Scavenged by Ascorbate-specific Peroxidase in Spinach Chloroplasts. Plant Cell Physiol. 1981, 22, 867–880. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).