Reversal of the Inflammatory Responses in Fabry Patient iPSC-Derived Cardiovascular Endothelial Cells by CRISPR/Cas9-Corrected Mutation
Abstract
1. Introduction
2. Results
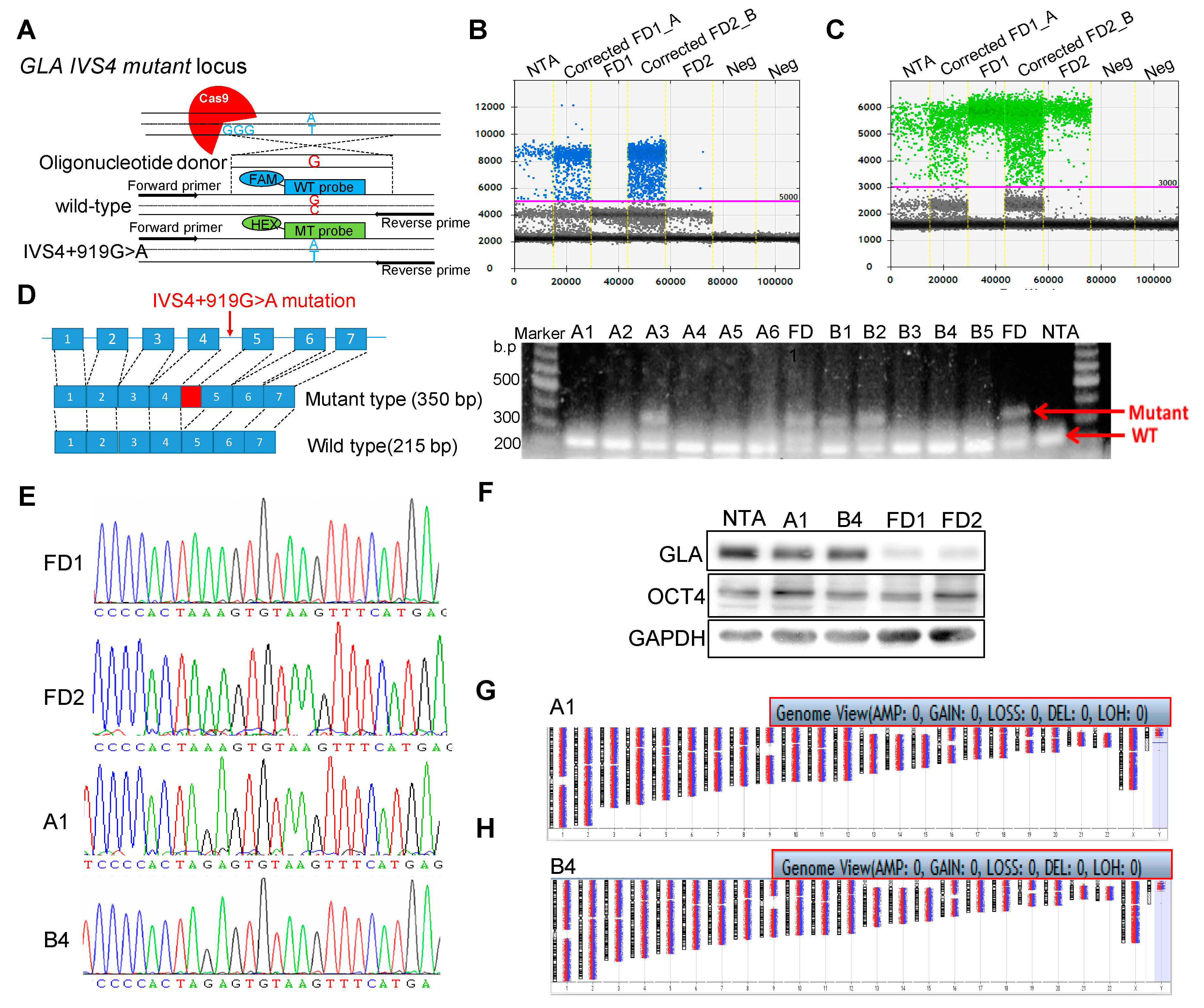
2.1. Correction of Fabry GLA IVS4 + 919 G > A Mutation in Isogenic Control iPSCs
2.2. Efficient Differentiation of Corrected FD-iPSCs into Functional Endothelial Cells
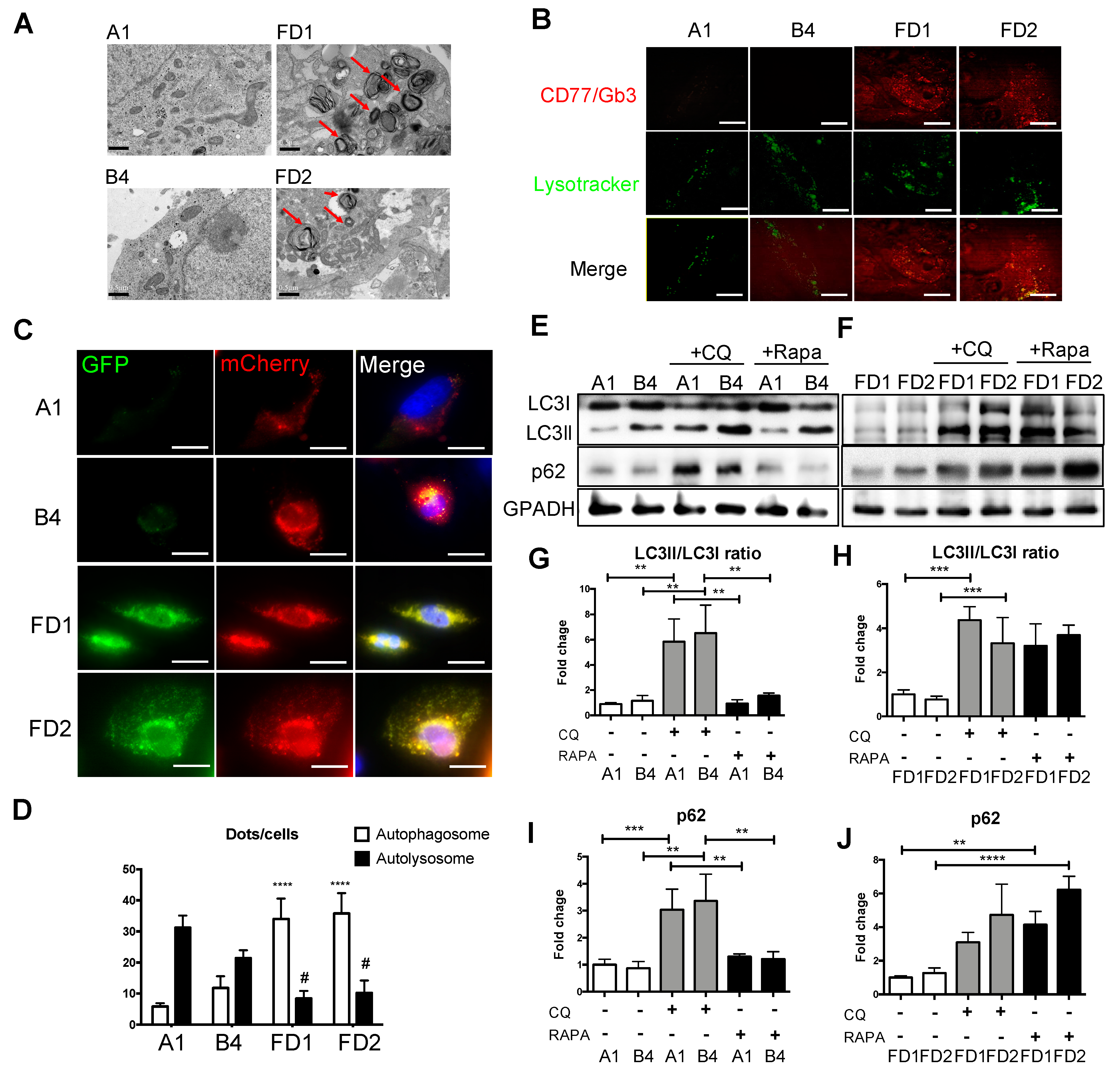
2.3. Attenuations of GLA-Deficient Phenotypic Abnormalities in the Corrected FD-ECs
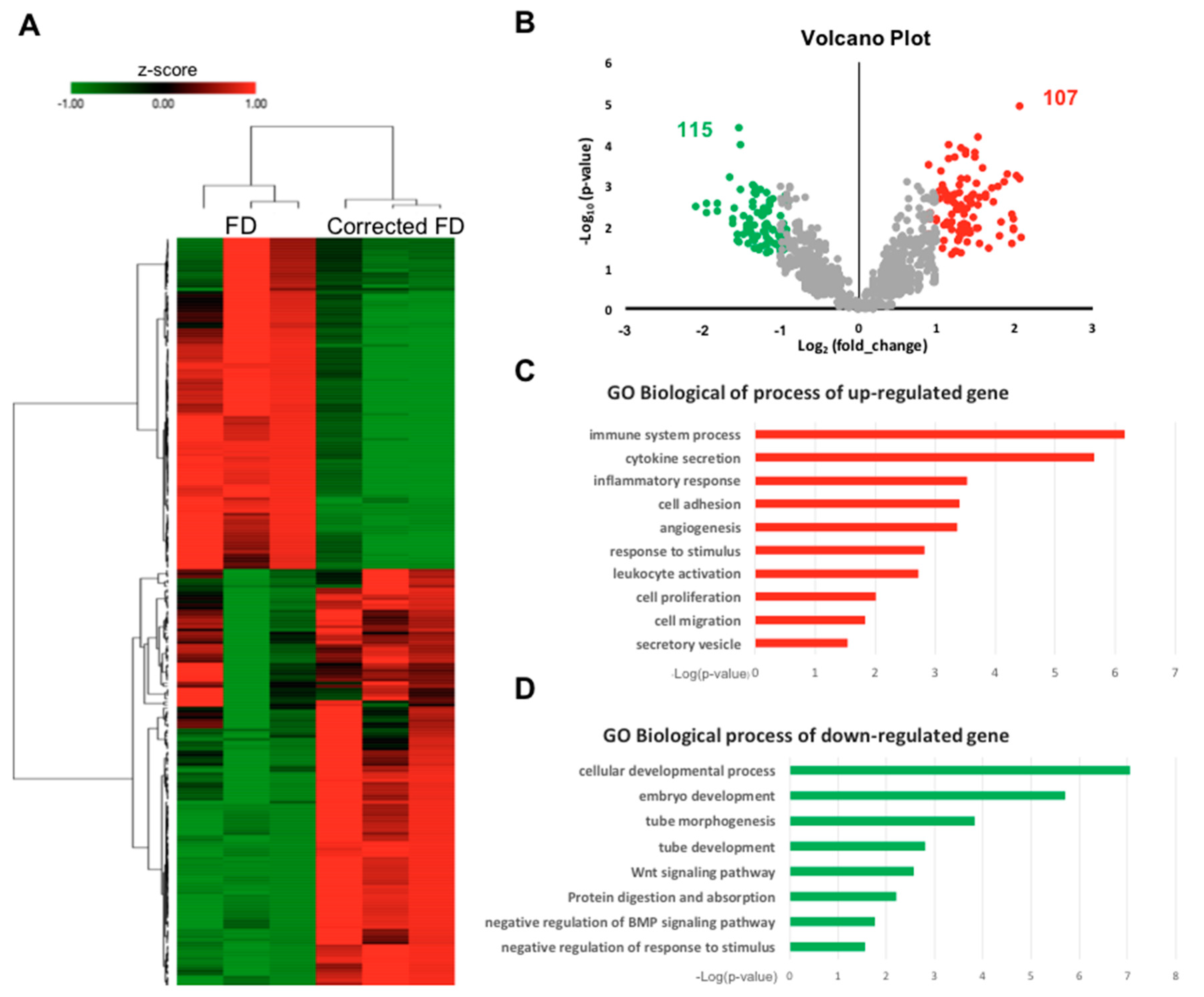
2.4. Enrichment of gene ontology (GO) Categories for Differentially Expressed Genes upon Correction of GLA IVS4 + 919G > A mutation
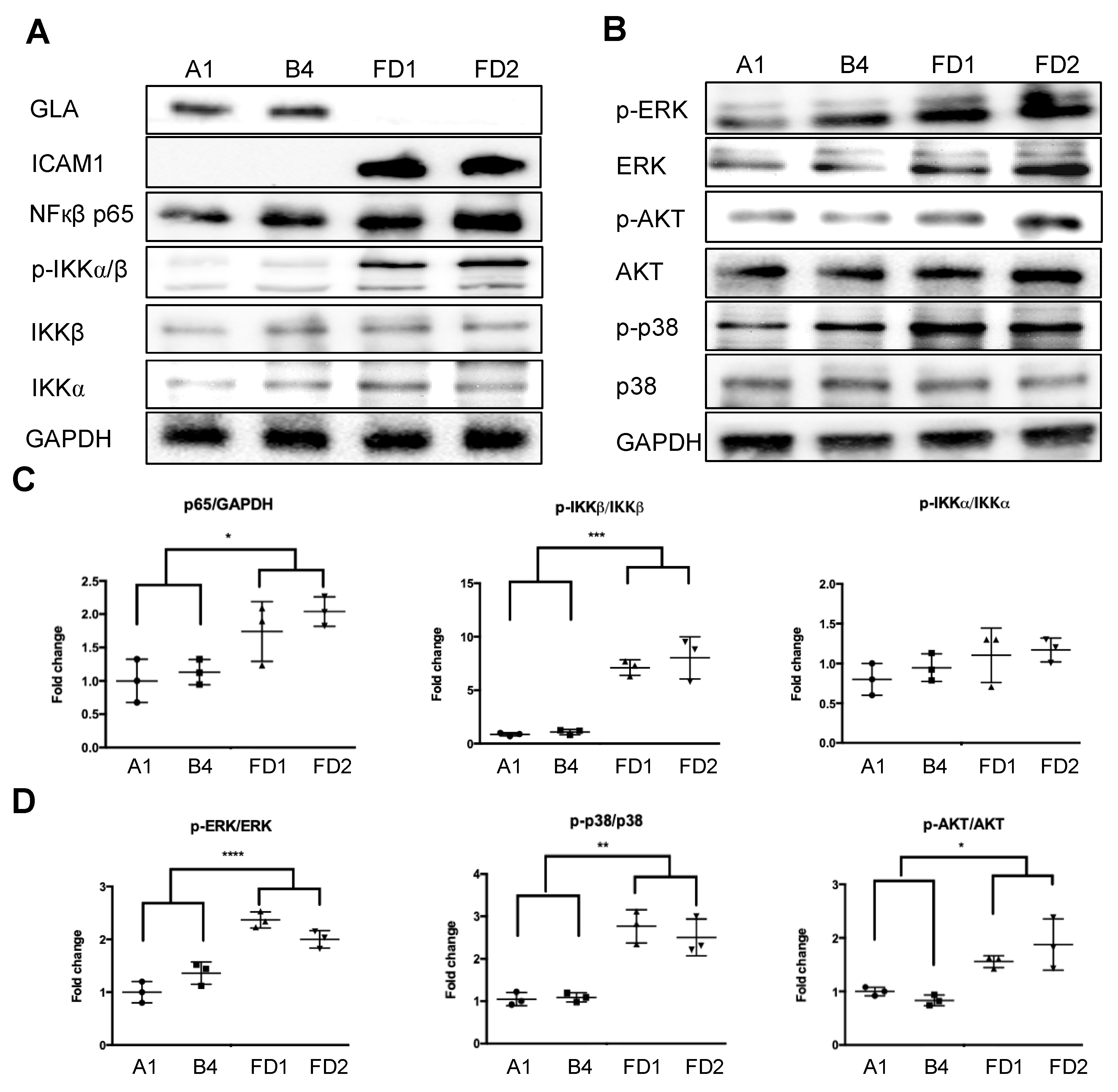
2.5. Validation of GLA Mutation Involved Inflammatory Response in FD and Corrected FD-ECs
3. Discussion
4. Materials and Methods
4.1. Genome Editing of FD GLA IVS4 + 919G > A iPSCs
4.2. Array-Based Comparative Genomic Hybridization (Array CGH)
4.3. Droplet Digital PCR (ddPCR)
4.4. Differentiation of iPSCs to Endothelial Cells (ECs)
4.5. Shear Stress Experiment
4.6. GLA Enzyme Activity
4.7. Immunofluorescence
4.8. Quantitative Real-Time PCR (qRT-PCR)
4.9. Immunoblotting
4.10. Transmission Electron Microscopy
4.11. Autophagy Flux Sensor
4.12. Microarray Analysis and Bioinformatics
4.13. Cytokine Array
4.14. Monocyte Adhesion Assay
4.15. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| LSD | Lysosomal storage disease |
| FD | Fabry disease |
| Gb3 | Globotriaosylceramide |
| Gal A | galactosidase A |
| ERT | Enzyme replacement therapy |
| CRISPR | Clustered regularly interspaced short palindromic repeats |
| sgRNA | Single-guide RNA |
| Cas9 | CRISPR-associated protein |
| ddPCR | Droplet digital PCR |
| ESCs | Embryonic stem cells |
| iPSCs | Induced pluripotent stem cells |
| CMs | Cardiomyocytes |
| ECs | Endothelial cells |
| HUVECs | Human umbilical vein endothelial cells |
| MVEs | Multivesicular endosomes |
| AVs | Autophagic vacuoles |
| ROS | Reactive oxygen species |
| EMT | Endothelial-to-mesenchymal transition |
References
- Peters, F.P.; Vermeulen, A.; Kho, T.L. Anderson-Fabry’s disease: Alpha-galactosidase deficiency. Lancet 2001, 357, 138–140. [Google Scholar] [CrossRef]
- Kotanko, P.; Kramar, R.; Devrnja, D.; Paschke, E.; Voigtlander, T.; Auinger, M.; Pagliardini, S.; Spada, M.; Demmelbauer, K.; Lorenz, M.; et al. Results of a nationwide screening for Anderson-Fabry disease among dialysis patients. J. Am. Soc. Nephrol. 2004, 15, 1323–1329. [Google Scholar] [CrossRef]
- Mehta, A.; Beck, M.; Eyskens, F.; Feliciani, C.; Kantola, I.; Ramaswami, U.; Rolfs, A.; Rivera, A.; Waldek, S.; Germain, D.P. Fabry disease: A review of current management strategies. QJM 2010, 103, 641–659. [Google Scholar] [CrossRef] [PubMed]
- Von Scheidt, W.; Eng, C.M.; Fitzmaurice, T.F.; Erdmann, E.; Hubner, G.; Olsen, E.G.; Christomanou, H.; Kandolf, R.; Bishop, D.F.; Desnick, R.J. An atypical variant of Fabry’s disease with manifestations confined to the myocardium. N. Engl. J. Med. 1991, 324, 395–399. [Google Scholar] [CrossRef]
- Sachdev, B.; Takenaka, T.; Teraguchi, H.; Tei, C.; Lee, P.; McKenna, W.J.; Elliott, P.M. Prevalence of Anderson-Fabry disease in male patients with late onset hypertrophic cardiomyopathy. Circulation 2002, 105, 1407–1411. [Google Scholar] [CrossRef]
- Ishii, S.; Nakao, S.; Minamikawa-Tachino, R.; Desnick, R.J.; Fan, J.Q. Alternative splicing in the alpha-galactosidase A gene: Increased exon inclusion results in the Fabry cardiac phenotype. Am. J. Hum. Genet. 2002, 70, 994–1002. [Google Scholar] [CrossRef] [PubMed]
- Hsu, T.R.; Sung, S.H.; Chang, F.P.; Yang, C.F.; Liu, H.C.; Lin, H.Y.; Huang, C.K.; Gao, H.J.; Huang, Y.H.; Liao, H.C.; et al. Endomyocardial biopsies in patients with left ventricular hypertrophy and a common Chinese later-onset Fabry mutation (IVS4 + 919G > A). Orphanet J. Rare Dis. 2014, 9, 96. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Li, C.F.; Lin, H.Y.; Lin, C.H.; Liu, H.C.; Tsai, S.F.; Niu, D.M. High-throughput detection of common sequence variations of Fabry disease in Taiwan using DNA mass spectrometry. Mol. Genet. Metab. 2014, 111, 507–512. [Google Scholar] [CrossRef]
- Liao, H.C.; Chiang, C.C.; Niu, D.M.; Wang, C.H.; Kao, S.M.; Tsai, F.J.; Huang, Y.H.; Liu, H.C.; Huang, C.K.; Gao, H.J.; et al. Detecting multiple lysosomal storage diseases by tandem mass spectrometry--a national newborn screening program in Taiwan. Clin. Chim. Acta 2014, 431, 80–86. [Google Scholar] [CrossRef]
- Liao, H.C.; Huang, Y.H.; Chen, Y.J.; Kao, S.M.; Lin, H.Y.; Huang, C.K.; Liu, H.C.; Hsu, T.R.; Lin, S.P.; Yang, C.F.; et al. Plasma globotriaosylsphingosine (lysoGb3) could be a biomarker for Fabry disease with a Chinese hotspot late-onset mutation (IVS4+919G>A). Clin. Chim. Acta 2013, 426, 114–120. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.Y.; Chong, K.W.; Hsu, J.H.; Yu, H.C.; Shih, C.C.; Huang, C.H.; Lin, S.J.; Chen, C.H.; Chiang, C.C.; Ho, H.J.; et al. High incidence of the cardiac variant of Fabry disease revealed by newborn screening in the Taiwan Chinese population. Circ. Cardiovasc. Genet. 2009, 2, 450–456. [Google Scholar] [CrossRef]
- Liu, H.C.; Lin, H.Y.; Yang, C.F.; Liao, H.C.; Hsu, T.R.; Lo, C.W.; Chang, F.P.; Huang, C.K.; Lu, Y.H.; Lin, S.P.; et al. Globotriaosylsphingosine (lyso-Gb3) might not be a reliable marker for monitoring the long-term therapeutic outcomes of enzyme replacement therapy for late-onset Fabry patients with the Chinese hotspot mutation (IVS4+919G>A). Orphanet J. Rare Dis. 2014, 9, 111. [Google Scholar] [CrossRef]
- Eng, C.M.; Guffon, N.; Wilcox, W.R.; Germain, D.P.; Lee, P.; Waldek, S.; Caplan, L.; Linthorst, G.E.; Desnick, R.J.; International Collaborative Fabry Disease Study Group. Safety and efficacy of recombinant human alpha-galactosidase A replacement therapy in Fabry’s disease. N. Engl. J. Med. 2001, 345, 9–16. [Google Scholar] [CrossRef]
- Rombach, S.M.; van den Bogaard, B.; de Groot, E.; Groener, J.E.; Poorthuis, B.J.; Linthorst, G.E.; van den Born, B.J.; Hollak, C.E.; Aerts, J.M. Vascular aspects of Fabry disease in relation to clinical manifestations and elevations in plasma globotriaosylsphingosine. Hypertension 2012, 60, 998–1005. [Google Scholar] [CrossRef]
- Satoh, K. Globotriaosylceramide induces endothelial dysfunction in fabry disease. Arter. Thromb. Vasc. Biol. 2014, 34, 2–4. [Google Scholar] [CrossRef]
- Chien, Y.; Chou, S.J.; Chang, Y.L.; Leu, H.B.; Yang, Y.P.; Tsai, P.H.; Lai, Y.H.; Chen, K.H.; Chang, W.C.; Sung, S.H.; et al. Inhibition of Arachidonate 12/15-Lipoxygenase Improves alpha-Galactosidase Efficacy in iPSC-Derived Cardiomyocytes from Fabry Patients. Int. J. Mol. Sci. 2018, 19, 1480. [Google Scholar] [CrossRef]
- Tseng, W.L.; Chou, S.J.; Chiang, H.C.; Wang, M.L.; Chien, C.S.; Chen, K.H.; Leu, H.B.; Wang, C.Y.; Chang, Y.L.; Liu, Y.Y.; et al. Imbalanced Production of Reactive Oxygen Species and Mitochondrial Antioxidant SOD2 in Fabry Disease-Specific Human Induced Pluripotent Stem Cell-Differentiated Vascular Endothelial Cells. Cell Transpl. 2017, 26, 513–527. [Google Scholar] [CrossRef]
- Ohshima, T.; Murray, G.J.; Swaim, W.D.; Longenecker, G.; Quirk, J.M.; Cardarelli, C.O.; Sugimoto, Y.; Pastan, I.; Gottesman, M.M.; Brady, R.O.; et al. alpha-Galactosidase A deficient mice: A model of Fabry disease. Proc. Natl. Acad. Sci. USA 1997, 94, 2540–2544. [Google Scholar] [CrossRef]
- Hockemeyer, D.; Jaenisch, R. Induced Pluripotent Stem Cells Meet Genome Editing. Cell Stem Cell 2016, 18, 573–586. [Google Scholar] [CrossRef] [PubMed]
- Chou, S.J.; Yu, W.C.; Chang, Y.L.; Chen, W.Y.; Chang, W.C.; Chien, Y.; Yen, J.C.; Liu, Y.Y.; Chen, S.J.; Wang, C.Y.; et al. Energy utilization of induced pluripotent stem cell-derived cardiomyocyte in Fabry disease. Int. J. Cardiol. 2017, 232, 255–263. [Google Scholar] [CrossRef]
- Patsch, C.; Challet-Meylan, L.; Thoma, E.C.; Urich, E.; Heckel, T.; O’Sullivan, J.F.; Grainger, S.J.; Kapp, F.G.; Sun, L.; Christensen, K.; et al. Generation of vascular endothelial and smooth muscle cells from human pluripotent stem cells. Nat. Cell Biol. 2015, 17, 994–1003. [Google Scholar] [CrossRef]
- Chiu, J.J.; Chien, S. Effects of disturbed flow on vascular endothelium: Pathophysiological basis and clinical perspectives. Physiol. Rev. 2011, 91, 327–387. [Google Scholar] [CrossRef]
- Dunn, J.; Thabet, S.; Jo, H. Flow-Dependent Epigenetic DNA Methylation in Endothelial Gene Expression and Atherosclerosis. Arter. Thromb. Vasc. Biol. 2015, 35, 1562–1569. [Google Scholar] [CrossRef]
- Peters, A.S.; Brunner, G.; Blumbach, K.; Abraham, D.J.; Krieg, T.; Eckes, B. Cyclic mechanical stress downregulates endothelin-1 and its responsive genes independently of TGFbeta1 in dermal fibroblasts. Exp. Derm. 2012, 21, 765–770. [Google Scholar] [CrossRef] [PubMed]
- Hervas, J.H.; Anton, Z.; Alonso, A. Biophysical Studies of LC3 Family Proteins. Methods Mol. Biol. 2019, 1880, 91–117. [Google Scholar] [CrossRef]
- Abdi, R.; Fiorina, P.; Adra, C.N.; Atkinson, M.; Sayegh, M.H. Immunomodulation by mesenchymal stem cells: A potential therapeutic strategy for type 1 diabetes. Diabetes 2008, 57, 1759–1767. [Google Scholar] [CrossRef]
- Garcia-Prat, L.; Martinez-Vicente, M.; Munoz-Canoves, P. Methods for Mitochondria and Mitophagy Flux Analyses in Stem Cells of Resting and Regenerating Skeletal Muscle. Methods Mol. Biol. 2016, 1460, 223–240. [Google Scholar] [CrossRef]
- Rozenfeld, P.; Feriozzi, S. Contribution of inflammatory pathways to Fabry disease pathogenesis. Mol. Genet. Metab. 2017, 122, 19–27. [Google Scholar] [CrossRef]
- Kawagoe, S.; Higuchi, T.; Otaka, M.; Shimada, Y.; Kobayashi, H.; Ida, H.; Ohashi, T.; Okano, H.J.; Nakanishi, M.; Eto, Y. Morphological features of iPS cells generated from Fabry disease skin fibroblasts using Sendai virus vector (SeVdp). Mol. Genet. Metab. 2013, 109, 386–389. [Google Scholar] [CrossRef]
- Itier, J.M.; Ret, G.; Viale, S.; Sweet, L.; Bangari, D.; Caron, A.; Le-Gall, F.; Benichou, B.; Leonard, J.; Deleuze, J.F.; et al. Effective clearance of GL-3 in a human iPSC-derived cardiomyocyte model of Fabry disease. J. Inherit. Metab. Dis. 2014, 37, 1013–1022. [Google Scholar] [CrossRef]
- Song, H.Y.; Chien, C.S.; Yarmishyn, A.A.; Chou, S.J.; Yang, Y.P.; Wang, M.L.; Wang, C.Y.; Leu, H.B.; Yu, W.C.; Chang, Y.L.; et al. Generation of GLA-Knockout Human Embryonic Stem Cell Lines to Model Autophagic Dysfunction and Exosome Secretion in Fabry Disease-Associated Hypertrophic Cardiomyopathy. Cells 2019, 8, 327. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.K.; Yu, J.H.; Min, S.H.; Park, S.W. Generation of a GLA knock-out human-induced pluripotent stem cell line, KSBCi002-A-1, using CRISPR/Cas9. Stem Cell Res. 2020, 42, 101676. [Google Scholar] [CrossRef] [PubMed]
- Park, C.Y.; Sung, J.J.; Choi, S.H.; Lee, D.R.; Park, I.H.; Kim, D.W. Modeling and correction of structural variations in patient-derived iPSCs using CRISPR/Cas9. Nat. Protoc. 2016, 11, 2154–2169. [Google Scholar] [CrossRef]
- Li, H.L.; Fujimoto, N.; Sasakawa, N.; Shirai, S.; Ohkame, T.; Sakuma, T.; Tanaka, M.; Amano, N.; Watanabe, A.; Sakurai, H.; et al. Precise correction of the dystrophin gene in duchenne muscular dystrophy patient induced pluripotent stem cells by TALEN and CRISPR-Cas9. Stem Cell Rep. 2015, 4, 143–154. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef]
- Miyaoka, Y.; Chan, A.H.; Judge, L.M.; Yoo, J.; Huang, M.; Nguyen, T.D.; Lizarraga, P.P.; So, P.L.; Conklin, B.R. Isolation of single-base genome-edited human iPS cells without antibiotic selection. Nat. Methods. 2014, 11, 291–293. [Google Scholar] [CrossRef]
- Park, S.; Kim, J.A.; Joo, K.Y.; Choi, S.; Choi, E.N.; Shin, J.A.; Han, K.H.; Jung, S.C.; Suh, S.H. Globotriaosylceramide leads to K(Ca)3.1 channel dysfunction: A new insight into endothelial dysfunction in Fabry disease. Cardiovasc. Res. 2011, 89, 290–299. [Google Scholar] [CrossRef]
- Nelson, M.P.; Tse, T.E.; O’Quinn, D.B.; Percival, S.M.; Jaimes, E.A.; Warnock, D.G.; Shacka, J.J. Autophagy-lysosome pathway associated neuropathology and axonal degeneration in the brains of alpha-galactosidase A-deficient mice. Acta Neuropathol. Commun. 2014, 2, 20. [Google Scholar] [CrossRef]
- Medzhitov, R. Inflammation 2010: New adventures of an old flame. Cell 2010, 140, 771–776. [Google Scholar] [CrossRef]
- Ben-Neriah, Y.; Karin, M. Inflammation meets cancer, with NF-kappaB as the matchmaker. Nat. Immunol. 2011, 12, 715–723. [Google Scholar] [CrossRef]
- Chevrier, M.; Brakch, N.; Celine, L.; Genty, D.; Ramdani, Y.; Moll, S.; Djavaheri-Mergny, M.; Brasse-Lagnel, C.; Annie Laquerriere, A.L.; Barbey, F.; et al. Autophagosome maturation is impaired in Fabry disease. Autophagy 2010, 6, 589–599. [Google Scholar] [CrossRef]
- Cai, Q.; Tammineni, P. Alterations in Mitochondrial Quality Control in Alzheimer’s Disease. Front. Cell. Neurosci. 2016, 10, 24. [Google Scholar] [CrossRef]
- Han, J.Y.; Kim, J.S.; Son, J.H. Mitochondrial homeostasis molecules: Regulation by a trio of recessive Parkinson’s disease genes. Exp. Neurobiol. 2014, 23, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Gregersen, N.; Bross, P. Protein misfolding and cellular stress: An overview. Methods Mol. Biol. 2010, 648, 3–23. [Google Scholar] [CrossRef]
- Shen, J.S.; Meng, X.L.; Moore, D.F.; Quirk, J.M.; Shayman, J.A.; Schiffmann, R.; Kaneski, C.R. Globotriaosylceramide induces oxidative stress and up-regulates cell adhesion molecule expression in Fabry disease endothelial cells. Mol. Genet. Metab. 2008, 95, 163–168. [Google Scholar] [CrossRef]
- Moore, D.F.; Scott, L.T.; Gladwin, M.T.; Altarescu, G.; Kaneski, C.; Suzuki, K.; Pease-Fye, M.; Ferri, R.; Brady, R.O.; Herscovitch, P.; et al. Regional cerebral hyperperfusion and nitric oxide pathway dysregulation in Fabry disease: Reversal by enzyme replacement therapy. Circulation 2001, 104, 1506–1512. [Google Scholar] [CrossRef] [PubMed]
- Biancini, G.B.; Vanzin, C.S.; Rodrigues, D.B.; Deon, M.; Ribas, G.S.; Barschak, A.G.; Manfredini, V.; Netto, C.B.; Jardim, L.B.; Giugliani, R.; et al. Globotriaosylceramide is correlated with oxidative stress and inflammation in Fabry patients treated with enzyme replacement therapy. Biochim. Biophys. Acta 2012, 1822, 226–232. [Google Scholar] [CrossRef]
- Chen, K.H.; Chien, Y.; Wang, K.L.; Leu, H.B.; Hsiao, C.Y.; Lai, Y.H.; Wang, C.Y.; Chang, Y.L.; Lin, S.J.; Niu, D.M.; et al. Evaluation of Proinflammatory Prognostic Biomarkers for Fabry Cardiomyopathy With Enzyme Replacement Therapy. Can. J. Cardiol. 2016, 32, 1221–1229. [Google Scholar] [CrossRef]
- Lambers, C.; Roth, M.; Zhong, J.; Campregher, C.; Binder, P.; Burian, B.; Petkov, V.; Block, L.H. The interaction of endothelin-1 and TGF-beta1 mediates vascular cell remodeling. PLoS ONE 2013, 8, e73399. [Google Scholar] [CrossRef]
- Lee, M.H.; Choi, E.N.; Jeon, Y.J.; Jung, S.C. Possible role of transforming growth factor-beta1 and vascular endothelial growth factor in Fabry disease nephropathy. Int. J. Mol. Med. 2012, 30, 1275–1280. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Nino, M.D.; Sanz, A.B.; Carrasco, S.; Saleem, M.A.; Mathieson, P.W.; Valdivielso, J.M.; Ruiz-Ortega, M.; Egido, J.; Ortiz, A. Globotriaosylsphingosine actions on human glomerular podocytes: Implications for Fabry nephropathy. Nephrol. Dial. Transpl. 2011, 26, 1797–1802. [Google Scholar] [CrossRef]
- Piera-Velazquez, S.; Li, Z.; Jimenez, S.A. Role of endothelial-mesenchymal transition (EndoMT) in the pathogenesis of fibrotic disorders. Am. J. Pathol. 2011, 179, 1074–1080. [Google Scholar] [CrossRef]
- Jeon, Y.J.; Jung, N.; Park, J.W.; Park, H.Y.; Jung, S.C. Epithelial-Mesenchymal Transition in Kidney Tubular Epithelial Cells Induced by Globotriaosylsphingosine and Globotriaosylceramide. PLoS ONE 2015, 10, e0136442. [Google Scholar] [CrossRef]
- Cho, S.W.; Kim, S.; Kim, J.M.; Kim, J.S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013, 31, 230–232. [Google Scholar] [CrossRef]
- Findlay, S.D.; Vincent, K.M.; Berman, J.R.; Postovit, L.M. A Digital PCR-Based Method for Efficient and Highly Specific Screening of Genome Edited Cells. PLoS ONE 2016, 11, e0153901. [Google Scholar] [CrossRef]
- Song, H.Y.; Chiang, H.C.; Tseng, W.L.; Wu, P.; Chien, C.S.; Leu, H.B.; Yang, Y.P.; Wang, M.L.; Jong, Y.J.; Chen, C.H.; et al. Using CRISPR/Cas9-Mediated GLA Gene Knockout as an In Vitro Drug Screening Model for Fabry Disease. Int. J. Mol. Sci. 2016, 17, 2089. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, H.-Y.; Yang, Y.-P.; Chien, Y.; Lai, W.-Y.; Lin, Y.-Y.; Chou, S.-J.; Wang, M.-L.; Wang, C.-Y.; Leu, H.-B.; Yu, W.-C.; et al. Reversal of the Inflammatory Responses in Fabry Patient iPSC-Derived Cardiovascular Endothelial Cells by CRISPR/Cas9-Corrected Mutation. Int. J. Mol. Sci. 2021, 22, 2381. https://doi.org/10.3390/ijms22052381
Song H-Y, Yang Y-P, Chien Y, Lai W-Y, Lin Y-Y, Chou S-J, Wang M-L, Wang C-Y, Leu H-B, Yu W-C, et al. Reversal of the Inflammatory Responses in Fabry Patient iPSC-Derived Cardiovascular Endothelial Cells by CRISPR/Cas9-Corrected Mutation. International Journal of Molecular Sciences. 2021; 22(5):2381. https://doi.org/10.3390/ijms22052381
Chicago/Turabian StyleSong, Hui-Yung, Yi-Ping Yang, Yueh Chien, Wei-Yi Lai, Yi-Ying Lin, Shih-Jie Chou, Mong-Lien Wang, Chien-Ying Wang, Hsin-Bang Leu, Wen-Chung Yu, and et al. 2021. "Reversal of the Inflammatory Responses in Fabry Patient iPSC-Derived Cardiovascular Endothelial Cells by CRISPR/Cas9-Corrected Mutation" International Journal of Molecular Sciences 22, no. 5: 2381. https://doi.org/10.3390/ijms22052381
APA StyleSong, H.-Y., Yang, Y.-P., Chien, Y., Lai, W.-Y., Lin, Y.-Y., Chou, S.-J., Wang, M.-L., Wang, C.-Y., Leu, H.-B., Yu, W.-C., & Chien, C.-S. (2021). Reversal of the Inflammatory Responses in Fabry Patient iPSC-Derived Cardiovascular Endothelial Cells by CRISPR/Cas9-Corrected Mutation. International Journal of Molecular Sciences, 22(5), 2381. https://doi.org/10.3390/ijms22052381





